Abstract
We herein describe the development of a sensitive microarray hybridization method called competitive DNA hybridization (CDH) and its use for analysis of BRAF somatic mutations. These mutations have been identified in many human cancers, and fast, reliable BRAF mutation detection may one day facilitate directed therapy of BRAF-mutated tumors. Our fast, reliable mutation detection by CDH is based on the principle that competition among multiple fluorescent-labeled samples for binding to shared wild-type sequences should reduce nonspecific results and increase the positive signals of unshared mutated sequences. The positive signals can then be discriminated based on the labeling of each sample (ie, with Cy3, Cy5, or Alexa-594). For testing of this method, we developed a BRAF oligonucleotide microarray containing 65 mutation types (more than 95% of the known BRAF mutations) and validated this microarray with 20 colorectal cancer tissues/cancer cell lines with BRAF mutations and 60 BRAF-negative samples. In sum, we were able to screen up to nine cancer samples on a single BRAF microarray (three per CDH on three regions per slide), indicating that this method may dramatically decrease the experimental time, cost, and effort of mutation detection in BRAF and other genes amenable to microarray analysis.
The past decade has seen great technical advancements in our strategies for analyzing mutations, single nucleotide polymorphisms, and genotypes. For mutational analysis, automatic dideoxy sequencing is still regarded as the gold standard, but its application is limited by high cost, numerous experimental steps, and low throughput. Denaturing high-performance liquid chromatography1 overcomes some of these limitations in that it is simple, sensitive,2 and allows real-time analysis of polymerase chain reaction (PCR) products without additional purification and treatment steps.1 However, abnormal (possibly mutated) samples identified by denaturing high-performance liquid chromatography must still be confirmed by expensive, time-consuming dideoxy sequencing.
A promising option for robust, high-throughput mutation or single nucleotide polymorphism screening is the use of microarray techniques,3,4 such as those based on the commercially available Affymetrix GeneChips.5 GeneChips have been used for mutation detection in the human mitochondrial genome and in genes such as TP53, BRCA1, and ATM.5 The various oligonucleotide microarray technologies differ from each other in terms of fabrication, sample preparation, and/or hybridization methods. Although sample-pooling methods have been investigated, these techniques require additional hybridization and/or sequencing of the positive samples. Thus, researchers are currently seeking to develop and streamline methods for multiple sample analysis in microarrays, which will facilitate high-throughput microarray analysis and dramatically reduce experimental time and cost. It was reported that two-color hybridization microarrays detected two different types of samples for mutational analysis.6,7,8 Previously, we also investigated multiple sample analysis on a small-scale K-ras oligonucleotide microarray.9 Here, we report the development of a competitive DNA hybridization (CDH)-based microarray technique with three-color hybridization involving simultaneous hybridization of multiple samples, and we assess its use for BRAF mutational analysis
BRAF is one of three serine/threonine kinases (ARAF, BRAF, and CRAF/RAF1) that act within the RAS/RAF/MEK/ERK/MAPK signaling pathway.10 Somatic mutations in BRAF have been reported in various human cancers,11 including 80% of primary melanomas, 68% of metastatic melanomas,12 36% of papillary thyroid carcinoma,10 14 to 33% of ovarian carcinomas,11,13 and 5.1 to 18% of colorectal cancers.11,14 In terms of mutational analysis, a hot spot mutation (V600E) in exon 15 of BRAF accounts for more than 80% of the BRAF mutations identified to date, with all other mutations being found in exons 11 and 15.11 Because BRAF mutations have been found in most human cancers and the mutation frequencies are relatively high (comparable with the rates of K-ras mutations), the development of BRAF inhibitors may have therapeutic potential in cancers harboring BRAF mutations.13,15,16 Several BRAF inhibitors have already entered into clinical trials.17 Because BRAF mutation detection may prove valuable for characterizing cancer types and directing BRAF inhibitor therapy, it is of clinical and therapeutic importance that we develop a quick, efficient, and effective tool for high-throughput detection of BRAF mutations. Thus, we herein used BRAF mutation detection as a model for evaluating the effectiveness of our CDH-based microarray technique, which allows for high-throughput analysis of multiple samples and may form the basis for new mutation detection strategies in the clinic and the lab.
Materials and Methods
Cancer Samples
We collected 16 colorectal cancer tissues harboring BRAF mutations18 and 60 BRAF-negative tissues from the Seoul National University Hospital and the National Cancer Center (Korea). Four cancer cell lines previously reported to harbor BRAF mutations were used as controls for our BRAF mutation screening. The WM-266-4 melanoma cell line containing the V600D BRAF mutation,11 the NCI-H1666 NSCLC cell line harboring the G466V mutation, and the NCI-H2087 NSCLS cell line harboring the L597V mutation were obtained from the American Type Culture Collection (Rockville, MD). The SNU-18 thyroid cell line harboring the G469A mutation was obtained from the Korean Cell Line Bank (Seoul, Korea). Genomic DNA was extracted from frozen specimens using the Trizol reagent (Invitrogen, Carlsbad, CA).
BRAF Oligonucleotide Microarray Design and Fabrication
Mutated and wild-type versions of 16 codons (439, 440, 459, 462, 463, 464, 466, 468, 469, 594, 595, 596, 597, 599, 600, and 601) from BRAF exons 11 and 15 were identified from database searches and the literature (Table 1). The oligonucleotide sequences were designed to be 23-mers containing the designated mutation in the middle (nucleotide 11) as previously described.4 These oligonucleotides were synthesized along with a 12-carbon spacer (Metabion, Martinsried, Bremen, Germany), 5′-modified with amino residues, purified by HPLC, and confirmed by denaturing high-performance liquid chromatography (WAVE; Transgenomic, Omaha, NE) and matrix-assisted laser desorption ionization/time of flight mass spectrometry (Ultraflex; Bruker Daltonics, Bremen, Germany). They were 5′-modified with amino residues for Schiff’s base reaction with an aldehyde group on a glass slide. Carbon spacers were used to increase the efficiency of the hybridization and to make the target sample (labeled with fluorescent dye) approach the spotted oligonucleotides more easily. Oligonucleotide probes were designed as 5′-NH2-(CH2)12-oligonucleotides. During the spotting procedure, the humidity was adjusted (ie, 60%) to prevent oligonucleotide evaporation, and a cooling nest was used to cool the well plate containing the oligonucleotides. Sample evaporation causes concentration differences in the spotted oligonucleotides, which may be a reason for later nonspecific results. Unreacted aldehyde groups were reduced to nonreactive primary alcohol by treatment with sodium borohydride (NaBH4). If this blocking step was omitted, specific signals were difficult to obtain, and much fluorescent debris will be shown. The oligonucleotides were diluted to 40 pmol/μl in microspotting solution (TeleChem International Inc., Sunnyvale, CA) and printed on an aldehyde-coated glass slide (26 × 76 × 1 mm Superadehyde and CEL slide; ArrayIt, Sunnyvale, CA) using a pin microarrayer (Cartesian Microsys 5100; Cartesian Technologies Inc., Irvine, CA). The 81 oligonucleotides were each printed in quadruplicate in horizontally adjacent spots, beginning at the bottom of the slide. This microarray was repeated three times on the slide, allowing at least three different samples to be hybridized per slide.
Table 1.
Oligonucleotide Sequences of the BRAF Oligonucleotide Microarray
| No. | Probe name | Exon | Codon | Sequence |
|---|---|---|---|---|
| 1 | 439W | 11 | 439 | 5′-TTTTATCAAGAAAACACTTGGTA-3′ |
| 2 | 439M1 | 11 | 439 | 5′-TTTTTATCAAGTAAACACTTGGT-3′ |
| 3 | 439M2 | 11 | 439 | 5′-TTTTTATCAAGGAAACACTTGGT-3′ |
| 4 | K439Q | 11 | 439 | 5′-TTTTTATCAAGCAAACACTTGGT-3′ |
| 5 | 439M4 | 11 | 439 | 5′-TTTTATCAAGATAACACTTGGTA-3′ |
| 6 | 439M5 | 11 | 439 | 5′-TTTTATCAAGAGAACACTTGGTA-3′ |
| 7 | K439T | 11 | 439 | 5′-TTTTATCAAGACAACACTTGGTA-3′ |
| 8 | 439M7 | 11 | 439 | 5′-TTTATCAAGAATACACTTGGTAG-3′ |
| 9 | 439M8 | 11 | 439 | 5′-TTTATCAAGAACACACTTGGTAG-3′ |
| 10 | 440W | 11 | 440 | 5′-TTATCAAGAAAACACTTGGTAGA-3′ |
| 11 | T440P | 11 | 440 | 5′-TTATCAAGAAACCACTTGGTAGA-3′ |
| 12 | 459W | 11 | 459 | 5′-GGCAGATTACAGTGGGACAAAGA-3′ |
| 13 | V459L | 11 | 459 | 5′-GGCAGATTACACTGGGACAAAGA-3′ |
| 14 | 462W | 11 | 462 | 5′-AGTGGGACAAAGAATTGGATCTG-3′ |
| 15 | R462I | 11 | 462 | 5′-AGTGGGACAAATAATTGGATCTG-3′ |
| 16 | 463W | 11 | 463 | 5′-GGGACAAAGAATTGGATCTGGAT-3′ |
| 17 | I463S | 11 | 463 | 5′-GGGACAAAGAAGTGGATCTGGAT-3′ |
| 18 | 464W | 11 | 464 | 5′-ACAAAGAATTGGATCTGGATCAT-3′ |
| 19 | 464M1 | 11 | 464 | 5′-GACAAAGAATTAGATCTGGATCA-3′ |
| 20 | 464M2 | 11 | 464 | 5′-GACAAAGAATTTGATCTGGATCA-3′ |
| 21 | 464M3 | 11 | 464 | 5′-GACAAAGAATTCGATCTGGATCA-3′ |
| 22 | G464E | 11 | 464 | 5′-ACAAAGAATTGAATCTGGATCAT-3′ |
| 23 | G464V | 11 | 464 | 5′-ACAAAGAATTGTATCTGGATCAT-3′ |
| 24 | 464M6 | 11 | 464 | 5′-ACAAAGAATTGCATCTGGATCAT-3′ |
| 25 | 466W | 11 | 466 | 5′-AATTGGATCTGGATCATTTGGAA-3′ |
| 26 | 466M1 | 11 | 466 | 5′-GAATTGGATCTAGATCATTTGGA-3′ |
| 27 | 466M2 | 11 | 466 | 5′-GAATTGGATCTTGATCATTTGGA-3′ |
| 28 | 466M3 | 11 | 466 | 5′-GAATTGGATCTCGATCATTTGGA-3′ |
| 29 | G466E | 11 | 466 | 5′-AATTGGATCTGAATCATTTGGAA-3′ |
| 30 | G466V | 11 | 466 | 5′-AATTGGATCTGTATCATTTGGAA-3′ |
| 31 | G466A | 11 | 466 | 5′-AATTGGATCTGCATCATTTGGAA-3′ |
| 32 | 468W | 11 | 468 | 5′-ATCTGGATCATTTGGAACAGTCT-3′ |
| 33 | F468C | 11 | 468 | 5′-ATCTGGATCATGTGGAACAGTCT-3′ |
| 34 | 469W | 11 | 469 | 5′-TGGATCATTTGGAACAGTCTACA-3′ |
| 35 | 469M1 | 11 | 469 | 5′-CTGGATCATTTAGAACAGTCTAC-3′ |
| 36 | 469M2 | 11 | 469 | 5′-CTGGATCATTTTGAACAGTCTAC-3′ |
| 37 | 469M3 | 11 | 469 | 5′-CTGGATCATTTCGAACAGTCTAC-3′ |
| 38 | G469E | 11 | 469 | 5′-TGGATCATTTGAAACAGTCTACA-3′ |
| 39 | 469M5 | 11 | 469 | 5′-TGGATCATTTGTAACAGTCTACA-3′ |
| 40 | G469A | 11 | 469 | 5′-TGGATCATTTGCAACAGTCTACA-3′ |
| 41 | 469MD | 11 | 469 | 5′-CTGGATCATTTTCAACAGTCTAC-3′ |
| 42 | 594W | 15 | 594 | 5′-AAAAATAGGTGATTTTGGTCTAG-3′ |
| 43 | 594M1 | 15 | 594 | 5′-TAAAAATAGGTAATTTTGGTCTA-3′ |
| 44 | 594M2 | 15 | 594 | 5′-TAAAAATAGGTTATTTTGGTCTA-3′ |
| 45 | 594M3 | 15 | 594 | 5′-TAAAAATAGGTCATTTTGGTCTA-3′ |
| 46 | D594V | 15 | 594 | 5′-AAAAATAGGTGTTTTTGGTCTAG-3′ |
| 47 | D594G | 15 | 594 | 5′-AAAAATAGGTGGTTTTGGTCTAG-3′ |
| 48 | 594M6 | 15 | 594 | 5′-AAAAATAGGTGCTTTTGGTCTAG-3′ |
| 49 | 594M7 | 15 | 594 | 5′-AAAATAGGTGAATTTGGTCTAGC-3′ |
| 50 | 594M8 | 15 | 594 | 5′-AAAATAGGTGAGTTTGGTCTAGC-3′ |
| 51 | 595W: | 15 | 595 | 5′-ATAGGTGATTTTGGTCTAGCTAC-3′ |
| 52 | F595L | 15 | 595 | 5′-ATAGGTGATTTGGGTCTAGCTAC-3′ |
| 53 | 596W | 15 | 596 | 5′-TAGGTGATTTTGGTCTAGCTACA-3′ |
| 54 | G596R | 15 | 596 | 5′-TAGGTGATTTTCGTCTAGCTACA-3′ |
| 55 | 597W | 15 | 597 | 5′-TGATTTTGGTCTAGCTACAGTGA-3′ |
| 56 | 597M1 | 15 | 597 | 5′-GTGATTTTGGTATAGCTACAGTG-3′ |
| 57 | 597M2 | 15 | 597 | 5′-GTGATTTTGGTTTAGCTACAGTG-3′ |
| 58 | L597V | 15 | 597 | 5′-GTGATTTTGGTGTAGCTACAGTG-3′ |
| 59 | 597M4 | 15 | 597 | 5′-TGATTTTGGTCAAGCTACAGTGA-3′ |
| 60 | L597R-2 | 15 | 597 | 5′-TGATTTTGGTCGAGCTACAGTGA-3′ |
| 61 | 597M6 | 15 | 597 | 5′-TGATTTTGGTCCAGCTACAGTGA-3′ |
| 62 | 599W | 15 | 599 | 5′-TGGTCTAGCTACAGTGAAATCTC-3′ |
| 63 | T599I | 15 | 599 | 5′-TGGTCTAGCTATAGTGAAATCTC-3′ |
| 64 | 600W | 15 | 600 | 5′-TCTAGCTACAGTGAAATCTCGAT-3′ |
| 65 | V600M | 15 | 600 | 5′-GTCTAGCTACAATGAAATCTCGA-3′ |
| 66 | 600M2 | 15 | 600 | 5′-GTCTAGCTACATTGAAATCTCGA-3′ |
| 67 | 600M3 | 15 | 600 | 5′-GTCTAGCTACACTGAAATCTCGA-3′ |
| 68 | V600E | 15 | 600 | 5′-TCTAGCTACAGAGAAATCTCGAT-3′ |
| 69 | 600M5 | 15 | 600 | 5′-TCTAGCTACAGGGAAATCTCGAT-3′ |
| 70 | 600M6 | 15 | 600 | 5′-TCTAGCTACAGCGAAATCTCGAT-3′ |
Table 1.
Continued
| No. | Probe name | Exon | Codon | Sequence |
|---|---|---|---|---|
| 71 | V600D | 15 | 600 | 5′-CTAGCTACAGATAAATCTCGATG-3′ |
| 72 | 600MD2 | 15 | 600 | 5′-GTCTAGCTACAAGGAAATCTCGA-3′ |
| 73 | 601W | 15 | 601 | 5′-AGCTACAGTGAAATCTCGATGGA-3′ |
| 74 | 601M1 | 15 | 601 | 5′-TAGCTACAGTGTAATCTCGATGG-3′ |
| 75 | K601E | 15 | 601 | 5′-TAGCTACAGTGGAATCTCGATGG-3′ |
| 76 | 601M3 | 15 | 601 | 5′-TAGCTACAGTGCAATCTCGATGG-3′ |
| 77 | 601M4 | 15 | 601 | 5′-AGCTACAGTGATATCTCGATGGA-3′ |
| 78 | 601M5 | 15 | 601 | 5′-AGCTACAGTGAGATCTCGATGGA-3′ |
| 79 | 601M6 | 15 | 601 | 5′-AGCTACAGTGACATCTCGATGGA-3′ |
| 80 | K601N | 15 | 601 | 5′-GCTACAGTGAATTCTCGATGGAG-3′ |
| 81 | 601M8 | 15 | 601 | 5′-GCTACAGTGAACTCTCGATGGAG-3′ |
Microarray Treatments: Washing before and after Hybridization
After being spotted, the BRAF microarrays were rinsed three times with 0.2% sodium dodecyl sulfate for 1 minute each and once in distilled water for 1 minute; they were then denatured at 95°C for 2 minutes. Each slide was then rinsed once with sodium borohydride (Sigma Chemical Co., St. Louis, MO) for 5 minutes and three times with 0.2% sodium dodecyl sulfate for 1 minute each. Finally, the slide was washed in distilled water for 1 minute and stored at −20°C until use. After hybridization, the slide was rinsed three times at room temperature in a buffer of 2× standard saline citrate and 0.2% sodium dodecyl sulfate. The hybridization and washing steps were all performed in the dark.
Sample Preparation for Conventional (Non-CDH) Microarray Hybridization
Genomic DNA (100 ng) from the indicated cancer tissues and cell lines was amplified by PCR using the previously described primers for amplification of BRAF exons 11 and 15.11 Each PCR reaction was performed with an optimized Cy5-containing mixture (8 μmol/L Cy5-dCTP and dCTP; and 40 μmol/L dATP, dGTP, and dTTP) and 0.5 unit of Taq polymerase (Qiagen Inc., Valencia, CA); a GeneAmp PCR system 9700 (Applied Biosystems Inc., Foster City, CA) was used to amplify the samples. PCR conditions consisted of 40 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute, with a final elongation of 7 minutes at 72°C. The Cy5-dCTP-labeled PCR products were resolved in a 2% acrylamide gel, purified with a Qiagen purification kit (Qiagen), and eluted in 45 μl of distilled water. Each sample was then digested with 0.25 unit of DNase I (Takara, Shiga, Japan) at 25°C for 10 minutes, and the enzyme was inactivated at 85°C for 10 minutes. The digested PCR products were dried in a concentrator (Eppendorf, Hamburg, Germany) at 60°C for 20 minutes. Each DNA pellet was resuspended in 3.5 μl of prewarmed 5× Hybit Solution (TeleChem International) and hybridized onto the microarray in a saturated vapor tube at 60°C for 3 hours. The hybridized samples were washed and scanned with a red 632.8-nm excitation laser (appropriate for visualization of Cy5).
CDH
The PCR primers and conditions were identical to those used for conventional hybridization, except that two additional cancer tissue or cell line samples were amplified with Cy3-dCTP and Alexa-594 5UTP, respectively. The Cy5-, Cy3- and Alexa-594-labeled amplicons were mixed before the purification step, and hybridization was performed as above. We then used a ScanArray 5000 (Perkin Elmer, Boston, MA) equipped with red, green, and yellow lasers to sequentially scan the microarray for detection of Cy5, Cy3, and Alexa-594 signals, respectively.
Automatic Dideoxy Sequencing
Bi-directional sequencing of genomic DNA was performed using the original amplification primers, a Taq dideoxy terminator cycle sequencing kit, and an ABI 3730 DNA sequencer (Applied Biosystems).
CDH with 10 Different Samples
To test the possibility of using more than three samples for CDH, we hybridized the BRAF microarray with a mixture of 10 different samples and examined mutation detection with a single fluorescently labeled sample. DNA from one cancer cell line (WM-266-4 or NCI-H2087) was amplified with Cy5 and mixed with amplified samples from nine different normal sources before purification. The CDH procedure was performed as described above, except that the DNase digestion was performed at 25°C for 15 minutes rather than 10 minutes.
Results
Principle of CDH
CDH is based on competition among multiple samples for binding to each oligonucleotide spot. Traditional cDNA microarrays use competitive hybridization between two samples, wherein one sample (ie, a normal sample) is labeled with Cy3, and the other sample (ie, a cancer sample) is labeled with Cy5. These two samples compete for binding to the cDNA spots. In contrast, traditional oligonucleotide microarray techniques (such as those involving the Affymetrix GeneChip) tend not to involve competition because a single sample is generally hybridized with a single oligonucleotide microarray. Nonspecific signal is the problem of “signal-to-noise ratio.” We previously observed that nonspecific signals (false positives) in oligonucleotide microarrays showed similar patterns in the same batch of oligonucleotide microarrays or on the same day. Moreover, because these nonspecific signals in certain probes have been observed in most of samples, we hypothesized that the nonspecific signals should be shared and reduced by simultaneous hybridization of multiple samples. The wild-type sequences should be shared among the samples, whereas the mutated sequences should not be shared. As shown in Figure 1, we tested this hypothesis using DNA extracted from three different types of samples (a cell line, a cancer tissue, and blood) and amplified with three different fluorescent dyes (Cy3-dCTP, Cy5-dCTP, and Alexa-594-dUTP). Because the three different fluorescent dyes have different emission and excitation wavelengths, the hybridization results could be easily differentiated. All CDH and non-CDH results were confirmed by automatic direct sequencing.
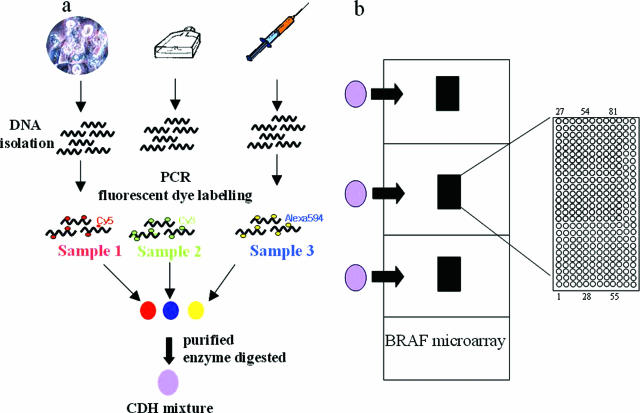
Figure 1.
Schematic depiction of CDH analysis of our BRAF oligonucleotide microarray with three samples. a: DNA was extracted from a cancer tissue sample, a cell line, and blood. The isolated DNA was amplified with three different fluorescent dyes (Cy5, Cy3, and Alexa-594), and the fluorescently labeled PCR products were mixed, purified, enzyme digested, and concentrated. The pellet was then resuspended in hybridization solution and hybridized to the spotted areas. b: Our BRAF oligonucleotide microarray contained three copies of the full array on one slide, allowing three hybridizations per slide. Because CDH allowed hybridization of three different samples per area, this technique could allow hybridization of nine total samples per microarray. The enlarged spotted area shows that a total of 324 (81 oligonucleotides in quadruplicate) were spotted per area. Each oligonucleotide was printed four times horizontally, in an upward direction starting from position 1, and the printing was divided into three groups. Positions 1 to 27 were printed first, then positions 28 to 54, and finally, positions 55 to 81.
Development of a BRAF Oligonucleotide Microarray and Mutation Detection of Colorectal Cancer Samples Using CDH
A total of 81 different BRAF oligonucleotides were designed, synthesized, and spotted in quadruplicate onto three different regions of the same slide. These included 16 wild-type sequences and 65 mutated exon 11 and 15 sequences identified from mutation databases (http://www.sanger.ac.uk/cosmic/) and the literature.11,12,13,14,15,16,17 We then used a conventional hybridization method to screen 76 colorectal cancer tissues using a BRAF oligonucleotide microarray, and we could detect 16 BRAF somatic mutations (Table 2). We then sought to determine whether our new CDH technique could be used for effective multisample hybridization of our BRAF microarray. DNA from NCI-H2087 cells and cancer tissue samples 640 and 194 was labeled with Cy5, Cy3, and Alexa-594, respectively. The BRAF microarray was hybridized with a mixture of the three probes and then scanned with a ScanArray 5000 (Perkin Elmer) using the following wavelengths: Cy5, excitation 649 nm and scanning 632.8 nm (red); Cy3, excitation 550 nm and scanning 543.5 nm (green); and Alexa-594, excitation 590 nm and scanning 594 nm (yellow). The red (632.8 nm) scan revealed a positive signal for the L597V mutation, which is consistent with the known mutation in the Cy5-labeled NCI-H2087 DNA (positive control). The green (534.5 nm) scan revealed that sample 640 contained a G464V (codon 464, Glu→Val, exon 11) mutation, and the yellow (594 nm) scan showed that sample 194 harbored a V600E mutation (codon 600, Val→Glu, exon 15), which is the most frequent BRAF hot spot mutation. As shown in Figure 2, each wavelength detected the appropriate dye without overlapping. These results indicate that CDH is suitable for multiple sample analysis and does not require further experiments to confirm mutated samples.
Table 2.
BRAF Mutations Identified from Colorectal Cancer Tissues
| Sample
|
Location | BRAF mutation | |||
|---|---|---|---|---|---|
| Name | Type | Codon | Exon | Mutation | |
| 640 | Tumor | Distal | 464 | 11 | G464V, GGA→GTA |
| 670 | Tumor | Distal | 469 | 11 | G469A, GGA→GCA |
| 172 | Tumor | Proximal | 600 | 15 | V600E, GTG→GAG |
| 194 | Tumor | Proximal | 600 | 15 | V600E, GTG→GAG |
| 233 | Tumor | Proximal | 600 | 15 | V600E, GTG→GAG |
| 296 | Tumor | Distal | 600 | 15 | V600E, GTG→GAG |
| 305 | Tumor | Distal | 601 | 15 | K601N, AAA→AAT |
| 474 | Tumor | Proximal | 600 | 15 | V600E, GTG→GAG |
| 481 | Tumor | Distal | 594 | 15 | D594G, GAT→GGT |
| 497 | Tumor | Distal | 600 | 15 | V600E, GTG→GAG |
| 523 | Tumor | Distal | 600 | 15 | V600D, GAG→GAT |
| 590 | Tumor | Distal | 600 | 15 | V600E, GTG→GAG |
| 626 | Tumor | Proximal | 600 | 15 | V600E, GTG→GAG |
| 653 | Tumor | Proximal | 600 | 15 | V600E, GTG→GAG |
| 656 | Tumor | Proximal | 600 | 15 | V600E, GTG→GAG |
| 734 | Tumor | Proximal | 600 | 15 | V600E, GTG→GAG |
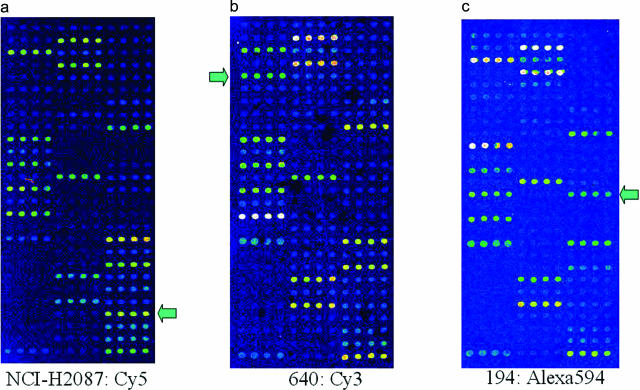
Figure 2.
Results of CDH analysis of the BRAF oligonucleotide microarray using cell line and colorectal cancer DNA samples. Wild-type (positive control) signals were shown in positions of 1, 10, 12, 14, 16, 18, 25, 32, 34, 42, 51, 53, 55, 62, 64, and 73. a: NCI-H2087 cell line DNA labeled with Cy5 showed a mutated signal in the 58th microarray position, confirming the previously reported L597V mutation. The wild-type signal at position 55 was also positive, indicating a heterozygous mutation. b: Cancer tissue 604 labeled with Cy3 showed a mutated signal at the 23rd position and a wild-type signal at position 18, indicating a heterozygous G464V mutation. c: Cancer tissue 194 labeled with Alexa-594 showed a mutated signal at position 68 and a wild-type signal at 64, indicating a heterozygous V600E mutation (the most common BRAF mutation). Each mutation signal was unique to the relevant scan, indicating that there was no overlapping of the fluorescent dye signals and providing evidence that CDH may be used for multiple sample analysis. The arrows indicate the mutation signals in each case.
Comparison of Conventional Microarray and CDH Analysis
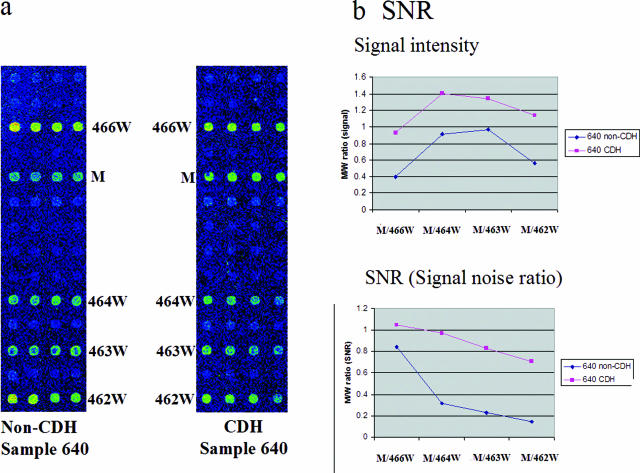
We examined whether CDH increased the sensitivity and specificity of mutation detection in comparison with conventional microarray hybridization. In the conventional (non-CDH) microarray experiment, DNA from colorectal cancer sample 640 was labeled with Cy3-dCTP. In the CDH experiments, we labeled sample 640 with Cy3-dCTP, labeled two normal samples with Cy5-dCTP and Alexa-594-dUTP, respectively, and then mixed and co-hybridized the three samples. We then selected the mutation based on the mutation/wild-type (M/W) ratios of G464V/464W (mutation/codon 464 wild type),4,9 G464V/G466W, G464V/G463W, and G464V/G462W using either the signal value or the signal-to-noise ratio value calculated with the QuantArray software (Packard Instrument, Wellesley, MA). When we calculated the signal intensity in the non-CDH hybridization (Figure 3a), we obtained M/W ratios ranging from 0.4 to 0.97 (Figure 3b). All of the M/W ratios were below 1. In contrast, the M/W ratio of G464V obtained from the CDH hybridization was over 1 and could thus be easily discriminated as a mutation (Figure 3b). Specifically, in the CDH experiment, the four wild-type signals (466W, 464W, 463W, and 462W) were reduced (Figure 3a), whereas the mutation signal was unchanged, leading to an M/W almost twice that seen in the non-CDH experiment (although the patterns were similar). When we examined the signal-to-noise ratios, the M/W ratio differences between non-CDH and CDH values were even greater. The M/464W ratio, the most important ratio for determining a mutation, was three times higher in the CDH-based experiment (0.97) than in the non-CDH experiment (0.32), whereas the difference was smaller when the CDH and non-CDH M/W463 ratios were compared using signal values (1.41 versus 0.91, respectively). Importantly, when the M/W ratio was calculated by signal/noise ratio in the non-CDH experiment, the M/W ratio of the G464V mutation (0.32) was less than 0.4, meaning that it would not be considered a positive mutation signal under our criteria.4 For the calculation of M/W ratios, the signal-to-noise ratio value (or signal value) of each spot was provided with QuantArray software, and each signal/noise ratio value (or signal value) from mutant spots was divided with that of wild-type spot in each codon. M/W ratio is the concept that compares mutant signals within those of its own wild-type codon. Collectively, these results indicate that CDH increases not only sample throughput but also the sensitivity and specificity of microarray hybridization.
Figure 3.
Comparison of the mutation-discriminating efficiencies of non-CDH (conventional) and CDH microarray analyses. a: Colorectal cancer tissue 640 was Cy3-labeled and hybridized according to the protocol of conventional microarray analysis. The mutated signal (G464V; M) is similar to that of wild-type codon 464. In contrast, when sample 640 was subjected to CDH (right) with normal DNA samples labeled with Cy5 and Alexa-594, the mutated signal (M) was increased and other wild-type signals were reduced. b: The ratio of mutation to wild-type signal (M/W ratio) was calculated for the CDH (pink) and non-CDH (blue) experiments. When calculated using the signal value, the M/W ratios of the CDH experiments were almost twofold higher than those of the non-CDH experiments. When calculated using the signal-to-noise ratio (calculated by the Quantarray software), the M/W ratios of the CDH experiments were even higher than those of the non-CDH experiments, indicating that CDH is more efficient than non-CDH for mutation detection under these conditions.
Evaluation of CDH for Analyzing Larger Sample Numbers (10 Samples)
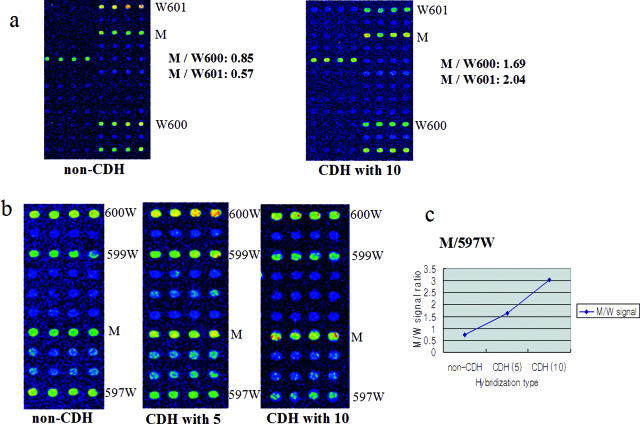
Here, we investigated whether 10 different amplified samples could be analyzed in a singly labeled CDH experiment, to evaluate whether CDH could be used for larger sample numbers once additional scanner lasers and fluorescent dyes are developed. We labeled DNA from a WM-266-4 melanoma cell line (containing a BRAF V600D mutation) with Cy5, amplified nine normal samples without fluorescent dye, mixed the 10 samples, and performed CDH (Figure 4). A control non-CDH hybridization of WM-266-4 yielded M/W600 and M/W601 ratios of 0.85 and 0.57, respectively (Figure 4a). The CDH experiments using 10 samples showed increased M/W600 and M/W601 ratios of 1.69 and 2.04, respectively, corresponding to ∼two- and fourfold increases in the discriminatory power.
Figure 4.
Evaluation of CDH with 10 different cancer samples: nine unlabeled DNAs lacking BRAF mutations and one labeled mutant DNA. a: DNA from WM-266-4 melanoma cells (harboring the V600D mutation) was labeled with Cy5, and nine other cancer tissue amplicons without BRAF mutations were mixed with the labeled DNA and then used for CDH. We compared the M/W ratios for this experiment with those obtained with non-CDH conventional microarray analysis; the M/W ratios were two- to fourfold higher in the CDH-based experiment. b: We also compared the results of microarray analyses using non-CDH, CDH with five samples (one labeled), and CDH with 10 samples (one labeled). DNA from NCI-H2087 cells (harboring the L597V mutation) was labeled with Cy5 and hybridized alone (left), co-hybridized with four different unlabeled samples (middle), or co-hybridized with nine different unlabeled samples (right). c: The M/W ratios (based on signal values) indicate that as the CDH sample numbers increased, better mutation detecting results were obtained.
To further examine the dose responsiveness of this effect, we examined DNA from NCI-H 2087 cells (harboring an L597V BRAF mutation) using non-CDH microarray analysis, CDH with five samples (one labeled) and CDH with 10 samples (one labeled) (Figure 4b). Increased CDH sample numbers were associated with decreases in the wild-type signals (597W, 599W, and 600W), leading to a more easily identified mutation signal (M). The M/W ratio derived from the non-CDH experiment was 0.73 (Figure 4c), whereas that from the CDH with five samples was 1.64 and that from CDH with 10 different samples was 3.03. These results show that as larger numbers of different samples were used for CDH, better results were obtained. Further work with additional dyes and scanning lasers will be necessary to determine the maximum number of samples that can be mixed and discriminated from each other in CDH, but this preliminary work suggests that even higher throughputs may be possible in the future.
Discussion
Here, we report the refinement of our microarray mutation detection method, CDH, which increases the sensitivity and specificity of microarray analysis, as well as the number of samples analyzed per experiment (and thus the cost effectiveness). Experiment time taken is dependent on the number of samples. If we use three samples, we can reduce the experiment time by one-third. For the experiment cost, the use of spotted microarrays can be saved. Because we can hybridize nine different samples in one microarray slide, we can reduce the spotted microarray cost by one-ninth. The other expenses for reagents are the same with those of non-CDH. CDH was not affected by labeling methods or DNA preparation because each sample was separately labeled by PCR amplification, and PCR purification was also performed with the same purification kit (Qiagen) used in non-CDH method. When we compared conventional and CDH microarray analyses using cancer tissues (Figure 3), we found that the signal-based M/W ratio patterns were very similar between the two. However, the CDH M/W ratio was almost twofold that obtained from the conventional microarray, indicating that CDH increased our ability to identify the mutation signal. We spotted three regions per microarray, allowing three different samples to be analyzed per conventional hybridization. CDH allowed simultaneous co-hybridization of three samples per region, meaning that nine different samples could be analyzed per microarray. In addition, CDH does not require further analysis to confirm the mutation, providing a clear improvement on previous methods. CDH also has the potential to greatly increase sample throughput. Here, we tested samples labeled with three fluorescent dyes because our microarray scanner was equipped with three different lasers. However, we also found that as we included a greater number of different samples in the hybridization (up to a total of 10 different samples), the M/W discrimination of a single fluorescently labeled sample was dramatically improved (up to fourfold versus the M/W of a conventional hybridization). Although there is no commercially available microarray scanner with 10 different lasers, future development of such equipment and additional fluorescent dyes may allow larger-scale CDH experiments. For example, at least 30 different samples per experiment could theoretically be analyzed using our BRAF oligonucleotide microarrays. It will be interesting to determine whether the observed high specificity of mutation detection is maintained with additional fluorophores and lasers.
For the determination of sensitivity and specificity between non-CDH and CDH, we checked 16 mutant and 60 wild-type cancer tissues. We could see 100% specificity for both non-CDH and CDH. For the sensitivity, two samples (samples 590 and 640) did not pass the cut-off level (0.4) in non-CDH and were regarded as nonspecific signals. On the other hand, these two samples showed higher ratios (more than 0.4) with CDH analysis. Taken together, specificity was the same in both non-CDH and CDH, and sensitivity was lower in non-CDH than CDH. However, if we raise the cut-off value to get more stringent results, sensitivity of non-CDH may be lower than with the CDH method. Because we used cancer tissues with normal cell contamination, the cut-off level (0.4) may increase when using pure cancer cells or cancer cell lines. The general basis of CDH relies on the premise that nonspecific and wild-type signals are shared among multiple samples, but the specific mutation signal is not. Although it is very rare in real genetic screening to have different cancer tissues with the same mutation types in one microarray, we tested whether CDH could detect a mutation shared among three different samples. We labeled DNA from three different colorectal cancer tissues harboring the V600E BRAF mutation with Cy5, Cy3, or Alexa-594 and co-hybridized these samples onto our BRAF microarray. Under these conditions, we were able to detect the V600E mutation in all three cases to levels comparable with traditional non-CDH microarray analysis (data not shown). Thus, our results indicate that CDH can be widely used regardless of sample or mutation combinations. In summary, we demonstrated that multiple sample analysis using CDH increased not only sample analysis numbers but also the sensitivity and specificity of the microarray results. CDH may prove to be an efficient, effective, robust, and high-throughput improvement over standard microarray mutation detection techniques for clinical and experimental applications.
Footnotes
Supported by a research grant from the National Cancer Center, Korea and the BK21 project for Medicine, Dentistry, and Pharmacy.
I.-J.K. and H.C.K. contributed equally to this work.
References
- Kim IJ, Shin Y, Kang HC, Park JH, Ku JL, Park HW, Park HR, Lim SB, Jeong SY, Kim WH, Park JG. Robust microsatellite instability (MSI) analysis by denaturing high-performance liquid chromatography (DHPLC). J Hum Genet. 2003;48:525–530. doi: 10.1007/s10038-003-0070-y. [DOI] [PubMed] [Google Scholar]
- Kang HC, Kim IJ, Park JH, Kwon HJ, Won YJ, Heo SC, Lee SY, Kim KH, Shin Y, Noh DY, Yang DH, Choe KJ, Lee BH, King SB, Park JG. Germline mutations of BRCA1 and BRCA2 in Korean breast and/or ovarian cancer families. Hum Mutat. 2002;20:235. doi: 10.1002/humu.9059. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Kim IJ, Kang HC, Park JH, Shin Y, Ku JL, Yoo BC, Park JG. Development and application of an oligonucleotide microarray for mutational analysis. Hardiman G, editor. Eagleville: PA, DNA Press,; Microarrays Methods and ApplicationsNuts & Bolts. 2003: 249–272. [Google Scholar]
- Hacia JG, Sun B, Hunt N, Edgemon K, Mosbrook D, Robbins C, Fodor SP, Tagle DA, Collins FS. Strategies for mutational analysis of the large multiexon ATM gene using high-density oligonucleotide arrays. Genome Res. 1998;8:1245–1258. doi: 10.1101/gr.8.12.1245. [DOI] [PubMed] [Google Scholar]
- Hacia JG, Edgemon K, Sun B, Stern D, Fodor SP, Collins FS. Two color hybridization analysis using high density oligonucleotide arrays and energy transfer dyes. Nucleic Acids Res. 1998;26:3865–3866. doi: 10.1093/nar/26.16.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortina P, Delgrosso K, Sakazume T, Santacroce R, Moutereau S, Su HJ, Graves D, McKenzie S, Surrey S. Simple two-color array-based approach for mutation detection. Eur J Hum Genet. 2000;8:884–894. doi: 10.1038/sj.ejhg.5200558. [DOI] [PubMed] [Google Scholar]
- Hacia JG, Edgemon K, Fang N, Mayer RA, Sudano D, Hunt N, Collins FS. Oligonucleotide microarray based detection of repetitive sequence changes. Hum Mutat. 2000;16:354–363. doi: 10.1002/1098-1004(200010)16:4<354::AID-HUMU8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Park JH, Kim IJ, Kang HC, Shin Y, Park HW, Jang SG, Ku JL, Lim SB, Jeong SY, Park JG. Oligonucleotide microarray-based mutation detection of the K-ras gene in colorectal cancers with use of competitive DNA hybridization. Clin Chem. 2004;50:1688–1691. doi: 10.1373/clinchem.2004.034017. [DOI] [PubMed] [Google Scholar]
- Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res. 2003;63:1454–1457. [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, Moses TY, Hostetter G, Wagner U, Kakareka J, Salem G, Pohida T, Heenan P, Duray P, Kallioniemi O, Hayward NK, Trent JM, Meltzer PS. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- Singer G, Oldt R, III, Cohen Y, Wang BG, Sidransky D, Kurman RJ, Shih IEM. Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma. J Natl Cancer Inst. 2003;95:484–486. doi: 10.1093/jnci/95.6.484. [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: rAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- Tannapfel A, Sommerer F, Benicke M, Katalinic A, Uhlmann D, Witzigmann H, Hauss J, Wittekind C. Mutations of the BRAF gene in cholangiocarcinoma but not in hepatocellular carcinoma. Gut. 2003;52:706–712. doi: 10.1136/gut.52.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, Barker MA, Arnold S, McGivern A, Matsubara N, Tanaka N, Higuchi T, Young J, Jass JR, Leggett BA. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoki K, Chen TH, Richards WG, Sugarbaker DJ, Meyerson M. Missense mutations of the BRAF gene in human lung adenocarcinoma. Cancer Res. 2002;62:7001–7003. [PubMed] [Google Scholar]
- Kim IJ, Kang HC, Jang SG, Kim K, Ahn SA, Yoon HJ, Yoon SN, Park JG. Oligonucleotide microarray analysis of distinct gene expression patterns in colorectal cancer tissues harboring BRAF and K-ras mutations. Carcinogenesis. 2006;27:392–404. doi: 10.1093/carcin/bgi237. [DOI] [PubMed] [Google Scholar]