Abstract
Duchenne/Becker muscular dystrophies (D/BMD) are X-linked recessive disorders resulting from dystrophin gene mutations. Intragenic recombination in the dystrophin gene occurs with a high frequency. Therefore, determination of the location and frequency of recombination improves D/BMD carrier detection and prenatal diagnosis in families in which the disease-causing mutation cannot be detected by most conventional methods. We describe herein a linkage analysis performed using a fast method based on capillary gel electrophoresis of fluorescent-labeled amplified alleles of 15 intragenic short tandem repeats spanning the entire dystrophin gene. On characterization of recombination events in 93 unrelated D/BMD families from southern Italy, we mapped 25 intragenic recombinations out of 273 informative meioses analyzed. The terminal regions of a gene are notoriously challenging for linkage analysis because some recombination events could be missed in case of lack of informativeness of the outermost markers. Many recombination events (10/25) identified in this study were located at the terminal regions of the dystrophin gene, and some were found by typing of several informative short tandem repeats located in these regions. Moreover, about 24% of the recombination events found in this study mapped to the 3′ region of the gene, in contrast with the low frequency (4 to 15%) reported by others.
Duchenne/Becker muscular dystrophies (D/BMD) are X-linked recessive genetic disorders resulting from mutations in the dystrophin gene located at the Xp21 region. The locus is very unstable: one-third of all D/BMD cases are due to new mutations, which occur in patients without a family history of the disease.1 Intragenic deletions and duplications together account for over two-thirds of the mutations that cause D/BMD; the remaining cases are due to point mutations or small insertions/deletions scattered along the entire gene (Leiden Muscular Dystrophy, http://www.dmd.nl, accessed January 2006). Several methods have been devised to identify deletions, duplications, and point mutations in affected males and in female carriers.2,3,4,5,6,7 Linkage analysis, based on short tandem repeats (STRs) at polymorphic loci in the dystrophin gene, is widely used for carrier detection and in the prenatal diagnosis of D/BMD families in which the causative mutations cannot be or were not determined in the proband.8,9,10,11 In recent years, many novel STRs have been identified and mapped in the dystrophin gene (Leiden Muscular Dystrophy, http://www.dmd.nl, accessed January 2006). However, intragenic recombination in the dystrophin gene has been shown to occur with a frequency of 10 to 12%,12,13,14 which is about four times that expected on the basis of the length of the gene. This high recombination rate is not restricted to families segregating for D/BMD but occurs also in healthy families.12,13 Therefore, determination of the location and frequency of recombination improves D/BMD carrier detection and prenatal diagnosis in families in which the disease-causing mutation cannot be detected by most conventional methods.
Here, we report a linkage analysis and the characterization of recombination events in 93 unrelated D/BMD families from southern Italy. The analysis is based on a fast method based on the separation by capillary gel electrophoresis of the fluorescent-labeled amplified alleles of 15 intragenic STRs that span the entire dystrophin gene.
Materials and Methods
The D/BMD families studied in this article were two- or three-generation families that were previously characterized for the presence of exon deletions or duplications.5,15 Deletions were detected in 62 families (67% of the families). Informed consent was obtained for each patient/family according to the procedure established by the local Institutional Bioethics Committee.
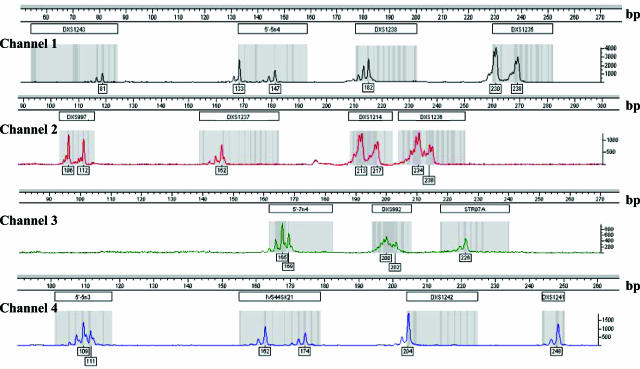
Genomic DNA was extracted from peripheral blood samples with the Nucleon procedure (Amersham, Buckinghamshire, UK). Five fluorescent-labeled multiplex PCR reactions were designed to amplify 15 dystrophin intragenic STRs (ie, DXS1242, 5′-5n3, IVS44SK21, in multiplex A; DXS997, DXS1214, DXS992 in multiplex B; 5′-5n4, DXS1238, DXS1235 in multiplex C; DXS1237 and DXS1236 in multiplex D; DXS1243, STR07A, 5′-7n4, and DXS1241 in multiplex E) using previously reported primer oligonucleotides (Leiden Muscular Dystrophy, http://www.dmd.nl, accessed January 2006). The forward primers were labeled with 5-carboxyfluorescein (FAM), PET, NED, or VIC fluorochromes (Figure 1). Multiplex polymerase chain reaction (PCR) mixtures (25 μl) contained 200 ng of genomic DNA, 0.25 mmol/L deoxynucleoside triphosphate mixture, 10 mmol/L Tris-HCl, pH 8.3, 50 mmol/L KCl, 1 nl/μl gelatin, 2.5 mmol/L MgCl2 (2 mmol/L MgCl2 in multiplex reaction B), 30 nl/μl dimethyl sulfoxide (in multiplex reactions B, C, and E), and 0.5 U of Taq polymerase. Amplification reactions were performed using a touch-down protocol (denaturation at 95°C for 3 minutes; 39 cycles with denaturation at 95°C for 20 seconds, annealing at 62°C for 40 seconds, −0.5°C per cycle for 14 cycles and at 55°C for 40 seconds for 25 cycles, and polymerization at 72°C for 45 seconds; final extension: 72°C for 7 minutes) and a thermal cycler PCR System 9600 (Applied Biosystems, Foster City, CA). These PCR conditions were designed to provide a robust amplification of the 15 STRs under the same thermal profile. PCR products (0.5 μl from each multiplex reaction) were mixed with 0.5 μl of Gene-Scan-500 LIZ size standard (Applied Biosystems) and were separated by capillary gel electrophoresis (15 kV at 60°C for 30 minutes) on the ABI Prism 310 Genetic Analyzer (Applied Biosystems) using the POP-4 polymer. We used the Genotyper 3.7 (Applied Biosystems) software for data analysis and created a macro that allowed us to label the peaks and identify the alleles of each marker automatically.
Figure 1.
Electropherogram of the 15 intragenic STRs labeled with NED (channel 1), PET (channel 2), VIC (channel 3), or FAM (channel 4) fluorochromes.
Results
A typical electropherogram of the 15 STRs is shown in Figure 1. The automated analysis is highly reproducible: the interindividual variability, calculated as the experimental CV for each allele of each STR, was between 0.6 and 5.5% (n ≥ 15), and the between-run variability ranged between 3 × 10−3% and 4 × 10−3% (n = 5). The method we used for linkage analysis in D/BMD families is fast (all of the multiplex PCRs are amplified in ∼2.5 hours under the same thermal conditions, and the capillary gel electrophoretic run takes ∼30 minutes), accurate, and easy to perform. For several STRs, we identified a higher number of alleles (Table 1) than reported earlier (Leiden Muscular Dystrophy, http://www.dmd.nl, accessed January 2006). However, the observed heterozygosity values did not substantially differ from those previously reported (Leiden Muscular Dystrophy, http://www.dmd.nl, accessed January 2006).16,17,18
Table 1.
Heterozygosity (HT) Values and Allele Numbers of the 15 Intragenic STRs in the Dystrophin Gene
| Markers (STRs) | HT found in this study | HT reported by others | Allele number identified in this study | Allele number reported by others* |
|---|---|---|---|---|
| DXS1242 | 0.79 | 0.63* | 15 | 8 |
| DXS1243 | 0.43 | 0.57* | 7 | 10 |
| 5′-5n3 | 0.80 | 0.7616,17 | 13 | 9 |
| 5′-5n4 | 0.81 | Not reported (PIC 0.64)17 | 13 | 8 |
| STR07A | 0.62 | 0.68* | 8 | 12 |
| 5′-7n4 | 0.54 | Not reported (PIC 0.52)17 | 5 | 4 |
| DXS1238 | 0.84 | 0.85* | 15 | 12 |
| IVS44SK21 | 0.80 | 0.8418 | 12 | 10 |
| DXS1237 | 0.84 | 0.87* | 15 | 13 |
| DXS997 | 0.75 | 0.65* | 7 | 4 |
| DXS1236 | 0.83 | 0.91* | 19 | 19 |
| DXS1235 | 0.70 | 0.70* | 14 | 6 |
| DXS1241 | 0.40 | 0.51* | 6 | 5 |
| DXS1214 | 0.83 | 0.79* | 10 | 7 |
| DXS992 | 0.71 | 0.87* | 9 | 11 |
Leiden Muscular Dystrophy page (http://www.dmd.nl).
PIC, polymorphism information content.
Twelve STRs had high heterozygosity values (0.6 to 0.83) (Table 1). We included in the panel the DXS1243, 5′-7n4, and DXS1241 markers, which have heterozygosity values of about 0.4 to 0.5 (Table 1), so that the STR loci investigated were evenly spread along the dystrophin gene (Figure 2 A). All of the subjects were typed with the 15 STRs. We were thus able to locate some recombination events more accurately: the DXS1243 STR marks the start of recombination in family FD# 73 and the 5′-7n4 STR marks the end or the start of recombination in family FD# 228 and in families FD# 134 and FD# 40, respectively (Figure 2).
Figure 2.
A: Simplified map of the dystrophin gene showing the location of the 15 STRs (below) and of exons (above). B: Intragenic recombination events detected in this study and exon deletions present in some families15; the black bars indicate the intervals where recombination events occurred; the red bars indicate the deletion extent and position. The family identification numbers are indicated.
The unequivocal haplotype reconstruction in 93 D/BMD families allowed us to map 25 intragenic recombinations (Figure 2B) out of 273 informative meioses analyzed. Thus, the recombination frequency was about 9%. Table 2 lists the recombination events found in our population, the informative meioses analyzed for each STR pair and the distances between the marker pairs. All recombinants were checked by retyping individuals. No double recombinants were identified, whereas in one family (FD# 73), we found two different recombinants in one generation, and in another family (FD# 241), we found two different recombinants in two generations. A deletion was detected in 13 of the 23 recombinant families. In three deleted families, one relative has a recombination event mapped in a region corresponding to one of the deletion breakpoints segregating in the pedigree (FD# 89: deletion of exons 3 to 7, recombination STR07A/DXS1238; FD# 145: deletion of exons 45 to 55, recombination IVS44SK21/DXS1237; FD# 245: deletion of exons 45 to 47, recombination IVS44SK21/DXS1237) (Figure 2A). In the remaining 10 deleted families, there was no correlation between the deletion site and the recombination site (Figure 2A). Nineteen of the 25 recombinations (76%) mapped in the 1.492-mb region between the STRs DXS1242 (located 1.2 kb from the 5′ of the exon 1 B) and DXS1236 (at intron 49), six (24%) in the region of at least 1 mb between the STRs DXS1235 (at intron 50) and DXS992 (>20 kb from the 3′ region of exon 79) (Figure 2). Figure 3 shows the haplotypes of two pedigrees in which no deletions or duplications segregated; the use of several markers located at the 5′ and 3′ regions of the gene allowed us to identify recombination events also in case of lack of informativeness of some STRs at the terminal regions of the gene. In pedigree FD# 73, the DXS1242 marker is not informative, but two independent recombinations were mapped (Figure 3A) subsequent to segregation of the DXS1243, 5′-5n3 and 5′-5n4 marker alleles, which are not included in the STR panels reported by other groups.11,19,20 A similar picture is depicted for the recombination event mapped at the 3′ region of the gene between the DXS1214 and DXS992 markers in pedigree FD# 8 (Figure 3B). Although subject II:2 carried the maternal not-at-risk haplotype from the DXS1242 to the DXS1214 markers, the identification of the recombination between DXS1214 and DXS992 precluded a conclusive diagnosis about the carrier/noncarrier status of subject II:2 because this region could contain a mutation.
Table 2.
Informative Meioses and Recombination Events between the Intragenic STRs in the Dystrophin Gene
| Marker (STRs) pairs | Informative meioses for STR pairs (n) | Recombination events (n) | Distance (kb) |
|---|---|---|---|
| DXS1242/DXS1243 | 81 | 0 | 144 |
| DXS1242/5′-5n3 | 191 | 2 | 223 |
| DXS1242/5′-5n4 | 191 | 1 | 508 |
| DXS1243/5′-5n3 | 105 | 1 | 78 |
| 5′-5n3/5′-5n4 | 197 | 2 | 285 |
| 5′-5n3/DXS1238 | 209 | 1 | 912 |
| 5′-5n4/STR07A | 167 | 1 | 44 |
| 5′-5n4/IVS44SK21 | 182 | 1 | 675 |
| STR07A/5′-7n4 | 142 | 1 | 331 |
| STR07A/DXS1238 | 175 | 2 | 582 |
| 5′-7n4/DXS1238 | 165 | 2 | 251 |
| DXS1238/IVS44SK21 | 198 | 1 | 49 |
| IVS44SK21/DXS1237 | 200 | 3 | 188 |
| DXS1237/DXS997 | 171 | 0 | 105 |
| DXS997/DXS1236 | 200 | 1 | 39 |
| DXS1236/DXS1235 | 193 | 0 | 43 |
| DXS1235/DXS1241 | 87 | 0 | 302 |
| DXS1235/DXS1214 | 178 | 1 | 537 |
| DXS1241/DXS1214 | 95 | 1 | 235 |
| DXS1214/DXS992 | 174 | 4 | 321 |
See also Figure 2.
Figure 3.
Pedigree and haplotype analysis of two D/BMD families showing recombination events located at the 5′ (A) or at the 3′ (B) region of the dystrophin gene. The thin black bar in FD# 73 (II:1 subject) indicates the region that cannot unambiguously be attributed to one haplotype. The arrows indicate the proband of each family.
Discussion
The estimate of the recombination frequency in a population and precise mapping of the recombination events are critical for genetic counseling and prenatal diagnosis. Given the high intragenic recombination frequency of the dystrophin gene, the analysis of a large STR panel is crucial for a faithful characterization of the gene segregation. This analysis can be easily performed thanks to an automated, fast, and accurate method based on the separation by capillary gel electrophoresis of the fluorescent-labeled amplified alleles of STRs that span the entire dystrophin gene. The terminal regions of a gene are notoriously challenging for linkage analysis because some recombination events could be missed in case of lack of informativeness of the outermost markers. We identified many recombination events by typing several informative markers located at these regions of the dystrophin gene. In fact, the use of several STRs in the 5′ region of the gene (from DXS1242 to DMDSTR07A) allowed us to identify six recombination events that could be missed in case of lack of informativeness of the DXS1242 marker using smaller STR panels, which include only few markers, as reported by others.11,19,20 Furthermore, the use of several STRs with high heterozygosity in the 3′ region (from intron 50) of the dystrophin gene enabled us to identify about 24% of all the recombinations found in our population. In contrast, a low number of recombinants (4 to 15% of total recombinants) was identified in this region in other populations,13,14 probably because of the typing of a smaller STR panel.
In conclusion, by detecting and mapping recombination events along the entire dystrophin gene, STR haplotyping with 15 intragenic markers could minimize diagnostic errors in linkage analysis of D/BMD families. Moreover, an accurate linkage analysis using several heterozygous STRs spanning the whole dystrophin gene could also help to detect hidden germline mosaicisms,21 thereby improving genetic counseling.
Acknowledgments
We are grateful to Jean A. Gilder for revising and editing the text.
Footnotes
Supported by a grant from Regione Campania (Convenzione CEINGE-Regione Campania, G.R. 20/12/2004 N. 2495) and from Ministero dell’Istruzione, dell’Università e della Ricerca-Rome PS35-126/IND.
References
- Worton RG, Molnar MJ, Brais B, Karpati G. The muscular dystrophies. Scriver CR, Beaudet AL, Sly WS, Valle D, editors. New York: McGraw Hill,; The Metabolic and Molecular Bases of Inherited Disease. 2001:5493–5523. [Google Scholar]
- Yau SC, Bobrow M, Mathew CG, Abbs SJ. Accurate diagnosis of deletions and duplications in Duchenne/Becker muscular dystrophy by fluorescent dosage analysis. J Med Genet. 1996;33:550–558. doi: 10.1136/jmg.33.7.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S, Kalf M, Liu Q, Villerius M, Engelsma D, Kriek M, Vollebregt E, Bakker B, van Ommen GJ, Breuning MH, den Dunnen JT. Comprehensive detection of genomic duplications and deletions in the DMD gene, by use of multiplex amplifiable probe hybridization. Am J Hum Genet. 2002;71:365–374. doi: 10.1086/341942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joncourt F, Neuhaus B, Jostarndt-Foegen K, Kleinle S, Steiner B, Gallati S. Rapid identification of female carriers of DMD/BMD by quantitative real-time PCR. Hum Mutat. 2004;23:385–391. doi: 10.1002/humu.20007. [DOI] [PubMed] [Google Scholar]
- Frisso G, Carsana A, Tinto N, Calcagno G, Salvatore F, Sacchetti L. Direct detection of exon deletions/duplications in female carriers of and male patients with Duchenne/Becker muscular dystrophy. Clin Chem. 2004;50:1435–1438. doi: 10.1373/clinchem.2004.033795. [DOI] [PubMed] [Google Scholar]
- Flanigan KM, von Niederhausern A, Dunn DM, Alder J, Mendell JR, Weiss RB. Rapid direct sequence analysis of the dystrophin gene. Am J Hum Genet. 2003;72:931–939. doi: 10.1086/374176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent KM, Dunn DM, von Niederhausern C, Aoyagi AT, Kerr L, Bromberg MB, Hart KJ, Tuohy T, White S, den Dunnen JT, Weiss RB, Flanigan KM. Improved molecular diagnosis of dystrophinopathies in an unselected clinical cohort. Am J Hum Genet. 2005;134A:295–298. doi: 10.1002/ajmg.a.30617. [DOI] [PubMed] [Google Scholar]
- Clemens PR, Fenwick RG, Chamberlain JS, Gibbs RA, de Andrade M, Chakraborty R, Caskey CT. Carrier detection and prenatal diagnosis in Duchenne and Becker muscular dystrophy families, using dinucleotide repeat polymorphisms. Am J Hum Genet. 1991;49:951–960. [PMC free article] [PubMed] [Google Scholar]
- Feener CA, Boyce FM, Kunkel LM. Rapid detection of CA polymorphisms in cloned DNA: application to the 5′ region of the dystrophin gene. Am J Hum Genet. 1991;48:621–627. [PMC free article] [PubMed] [Google Scholar]
- Shiroshita Y, Katayama S. Prenatal diagnosis of Duchenne muscular dystrophy in the Japanese population by fluorescent CA repeat polymorphism analysis. J Obstet Gynaecol Res. 1997;23:453–461. doi: 10.1111/j.1447-0756.1997.tb00873.x. [DOI] [PubMed] [Google Scholar]
- Ferreiro V, Giliberto F, Francipane L, Szijan I. The role of polymorphic short tandem (CA)(n) repeat loci segregation analysis in the detection of Duchenne muscular dystrophy carriers and prenatal diagnosis. Mol Diagn. 2005;9:67–80. doi: 10.1007/BF03260074. [DOI] [PubMed] [Google Scholar]
- Abbs S, Roberts RG, Mathew CG, Bentley DR, Bobrow M. Accurate assessment of intragenic recombination frequency within the Duchenne muscular dystrophy gene. Genomics. 1990;7:602–606. doi: 10.1016/0888-7543(90)90205-9. [DOI] [PubMed] [Google Scholar]
- Oudet C, Hanauer A, Clemens P, Caskey T, Mandel JL. Two hot spots of recombination in the DMD gene correlate with the deletion prone regions. Hum Mol Genet. 1992;1:599–603. doi: 10.1093/hmg/1.8.599. [DOI] [PubMed] [Google Scholar]
- Florentin L, Bili C, Kekou K, Tripodis N, Mavrou A, Metaxotou C. Mapping dystrophin gene recombinants in Greek DMD/BMD families: low recombination frequencies in the STR region. Hum Genet. 1995;96:423–426. doi: 10.1007/BF00191800. [DOI] [PubMed] [Google Scholar]
- Carsana A, Frisso G, Tremolaterra MR, Lanzillo R, Vitale DF, Santoro L, Salvatore F. Analysis of dystrophin gene deletions indicates that the hinge III region of the protein correlates with disease severity. Ann Hum Genet. 2005;69:253–259. doi: 10.1046/j.1529-8817.2005.00160.x. [DOI] [PubMed] [Google Scholar]
- King SC, Stapleton PM, Walker AP, Love DR. Two dinucleotide repeat polymorphisms at the DMD locus. Hum Mol Genet. 1994;3:523. doi: 10.1093/hmg/3.3.523. [DOI] [PubMed] [Google Scholar]
- King SC, Roche AL, Passos-Bueno MR, Takata R, Zatz M, Cockburn DJ, Seller A, Stapleton PM, Love DR. Molecular characterization of further dystrophin gene microsatellites. Mol Cell Probes. 1995;9:361–370. doi: 10.1016/s0890-8508(95)91700-4. [DOI] [PubMed] [Google Scholar]
- Köchling S, den Dunnen JT, Dworniczak B, Horst J. Two polymorphic dinucleotide repeats in intron 44 of the dystrophin gene. Hum Genet. 1995;95:475–477. doi: 10.1007/BF00208985. [DOI] [PubMed] [Google Scholar]
- Percesepe A, Ferrari M, Coviello D, Zanussi M, Castagni M, Neri I, Travi M, Forabosco A, Tedeschi S. Detection of a novel dystrophin gene mutation through carrier analysis performed during prenatal diagnosis in a case with intragenic recombination. Prenat Diagn. 2005;25:1011–1014. doi: 10.1002/pd.1238. [DOI] [PubMed] [Google Scholar]
- Chaturvedi LS, Mittal RD, Srivastasa S, Mukherjee M, Mittal B. Analysis of dinucleotide repeat loci of dystrophin gene for carrier detection, germline mosaicism and de novo mutations in Duchenne muscular dystrophy. Clin Genet. 2000;58:234–236. doi: 10.1034/j.1399-0004.2000.580312.x. [DOI] [PubMed] [Google Scholar]
- Ferreiro V, Szijan I, Giliberto F. Detection of germline mosaicism in two Duchenne muscular dystrophy families using polymorphic dinucleotide (CA)n repeat loci within the dystrophin gene. Mol Diagn. 2004;8:115–121. doi: 10.1007/BF03260054. [DOI] [PubMed] [Google Scholar]