Abstract
Gastrointestinal stromal tumors (GISTs) are the most common gastrointestinal mesenchymal tumors driven by KIT or PDGFRA mutations. A majority of these mutations affect KIT exon 11 and represent deletions or point mutations, but insertions and duplications have also been reported. The latter have been found exclusively in the 3′ part of KIT exon 11. Reported frequency of duplications varies, and a higher frequency has been reported in studies based on frozen tissue. Recently, we have hypothesized that in some cases, the duplications might remain undetected in formalin-fixed, paraffin-embedded GISTs because of the preferential polymerase chain reaction (PCR) amplification of wild-type KIT over the mutant allele. In this study, 16 GISTs initially diagnosed as a wild-type KIT were evaluated using PCR assay amplifying only the 3′ part of KIT exon 11, the region commonly affected by duplications. Denaturing high-pressure liquid chromatography and direct sequencing analyses revealed duplications in 4 (25%) of 16 analyzed cases. Use of the PCR assay amplifying the specific region affected by duplications and yielding 129 bp in wild-type KIT can substantially improve the detection of these mutations in formalin-fixed, paraffin-embedded GISTs.
Gastrointestinal stromal tumors (GISTs) are the most common mesenchymal tumors of gastrointestinal tract driven by KIT or platelet-derived growth factor α (PDGFRA) gain-of-function mutations.1,2,3 A great majority of these mutations affects KIT juxtamembrane domain encoded by exon 11 and represent deletions and point mutations; however, insertions and duplications have also been reported.4,5 Although most of the deletions and point mutations cluster in the 5′ part, duplications have been found almost exclusively in the 3′ part of KIT exon 11.6,7 In addition, duplications are predominantly associated with gastric tumor location.6,7,8,9 Reported frequency of duplications varies from 3.6 to 13.6%.7,10 This variation could be attributed to lower or higher number of gastric GISTs in analyzed tumor cohorts and to methodological differences in mutation detection. Recently, we have hypothesized that lower frequency of duplication in formalin-fixed, paraffin-embedded (FFPE) GISTs could also be related to preferential polymerase chain reaction (PCR) amplification of the wild-type (WT) KIT over the mutant allele because of severe DNA degradation in FFPE tissue.3 In this study, we found support for this hypothesis and show that duplication in KIT can be more accurately detected with PCR assay tailored to amplify the 3′ part of KIT exon 11 involved in the duplications.
Materials and Methods
Sixteen GISTs analyzed in this study were obtained from the Department of Pathology, University Hospital of Tromsoe. DNA was extracted from FFPE tissues as previously reported.4 All tumors were considered KIT- and PDGFRA-WT after screening of KIT exon 9, 11, 13, and 17 and PDGFRA exon 12, 14, and 18 mutation “hot spots” using previously reported PCR assays.11
A region commonly affected by duplications in the 3′ part of KIT exon 11 was identified based on analysis of 50 duplications, including 18 that were previously published,7 available at our GIST mutation database and 46 cases reported by others in the literature.5,6,8,12,13,14,15,16,17,18,19,20,21,22,23 All analyzed duplications, except two (2.1%), have been reported in the KIT region c.1729_1794 (g.75722_75787). Primers CK11.1F and CK11.3R closely flanking this region were used for PCR amplification of smaller products, yielding 129 bp in KIT-WT. For comparative purposes, the entire KIT exon 11 was subsequently amplified using two PCR assays based on intronic primers and generating 271- and 234-bp PCR products in selected cases. Primer sequences, PCR conditions, and length of the PCR products are listed in Table 1.
Table 1.
PCR Assays Used to Evaluate KIT Exon 11
| Primer | Nucleotide position* | Primer sequence | PCR | Temperature (°C) | Size (bp) |
|---|---|---|---|---|---|
| CK10.7F | g.75575_75595 | 5′-CCATTTATTTGTTCTCTCTCC-3′ | CK10.7F/CK11.3R | 50 | 234 |
| CK11.1F | g.75670_75690 | 5′-CAGTGGAAGGTTGTTGAGGAG-3′ | CK11.1F/CK11.3R | 55 | 129 |
| CK11.3R | g.75789_75808 | 5′-AGCCCCTGTTTCATACTGAC-3′ | |||
| CK11.3.5R | g.75825_75845 | 5′-AGTCACTGTTATGTGTACCCA-3′ | CK10.7F/CK11.3.5R | 50 | 271 |
F, forward; R, reverse.
Nucleotide position based on human KIT (locus HSU63834) sequence available in GenBank at http:www.ncbi.nlm.nih.gov.
All PCR products were screened for mutations using denaturing high-pressure liquid chromatography (DHPLC) and WAVE System 3500HT (Transgenomic Ltd., Elancourt, France). Partially denaturing conditions for mutation detection were predicted using Navigator Software v.1.6.0. PCR products were analyzed under at least two different temperatures. Simultaneously and independently from DHPLC, PCR products were screened for mutations by direct sequencing as previously reported.24
Nomenclature of the mutations identified at the DNA level and deduced mutations at the protein level are based on the recommendations of the Human Genome Mutation Society (http:www.hgvs.org). The following KIT reference sequences, HSU63834 and XO6182, were obtained from GenBank (http:www.ncbi.nlm.nih.gov).
Results
Sixteen KIT and PDGFRA-WT GISTs from a series of 89 tumors were screened for KIT duplication using PCR assay designed to amplify a region in the 3′ part of KIT exon 11 commonly affected by duplications. PCR amplification was successful in all cases. DHPLC analyses suggested mutations in four (25%) tumors. Direct sequencing of PCR products identified duplications in all four GISTs indicated by DHPLC. Mutant KIT sequences are shown in Figure 1. Identified duplications ranged from 18 to 42 nucleotides and involved region c.1735_1782; one duplication, c.1741_1779dup, has been previously reported in a few cases.9,25
Figure 1.
Genomic sequences of mutated KIT alleles identified in this study. Part of the genomic and amino acid sequence of WT KIT exon 11 is shown above. Duplicated sequences are boxed. Nomenclature of the mutations identified at the DNA level and deduced mutations at the protein level are shown in the second column.
There were three male patients and one female patient with ages ranging from 41 to 59 years. Three tumors were gastric GISTs, and one was diagnosed in omentum. Histologically, all four tumors were spindle cell GISTs with very low (1 to 2 mitosis/50 high-power fields) or no mitotic activity. Tumor size varied from 4 to 26 cm. The demographic and clinicopathologic data of the four cases are summarized in Table 2.
Table 2.
The Demographic and Clinicopathologic Data of Four GISTs with Duplications
| Case | Age | Sex | Surgery (year) | Location | Size (cm) | Mitosis/50HPF | Histology |
|---|---|---|---|---|---|---|---|
| 1 | 58 | F | 2001 | Stomach | 26 | 0 | Spindle |
| 2 | 41 | M | 1988 | Stomach | 4 | 2 | Spindle |
| 3 | 59 | M | 1986 | Stomach | 5 | 0 | Spindle |
| 4 | 49 | M | 1994 | Omentum | 11 | 1 | Spindle |
F, female; M, male.
To understand the nature of previous false-negative results, we subsequently studied these tumors using two different PCR assays based on intronic primers with target amplicons of 271 and 234 bp of genomic DNA. PCR amplification of 271 bp was achieved in three of four analyzed GISTs. DHPLC analysis of these PCR products indicated mutation only in two cases. Review of previous KIT mutation studies based on amplification and direct sequencing of 271-bp PCR products revealed in the cases indicated by DHPLC weak “tail sequence,” suggesting possible duplication. These sequences were previously interpreted as a sequencing background.
PCR amplification of 234 bp was possible in all four analyzed cases. Mutations were indicated in three cases and confirmed by direct sequencing. However, accurate identification of mutant allele sequence was possible only in two cases. Although an extended “tail sequence” of longer mutant KIT allele was seen in the third case, elevated peaks of mutant KIT allele could not be easily distinguished from the elevated background. Figures 2 to 4 show representative data obtained by DHPLC and direct sequencing of PCR products.
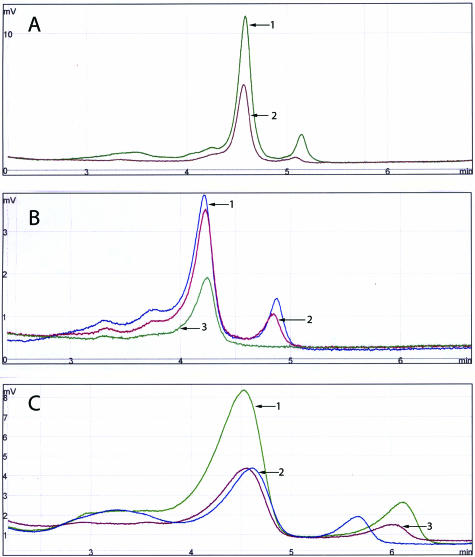
Figure 2.
DHPLC analyses of 271-bp (A), 234-bp (B), and 129-bp (C) PCR products amplified from GISTs 1, 2, and 3. Left and right peaks represent products amplified from WT and mutant KIT alleles, respectively. A: Note the presence of WT and mutant KIT peaks in case 1, no indication of mutation (one WT peak) in case 2, and the lack of PCR amplification products in case 3. B: Note the presence of WT and mutant KIT peaks in case 1 and 2 and no indication of mutation (one WT peak) in case 3. C: Note the presence of WT and mutant KIT peaks in all three cases.
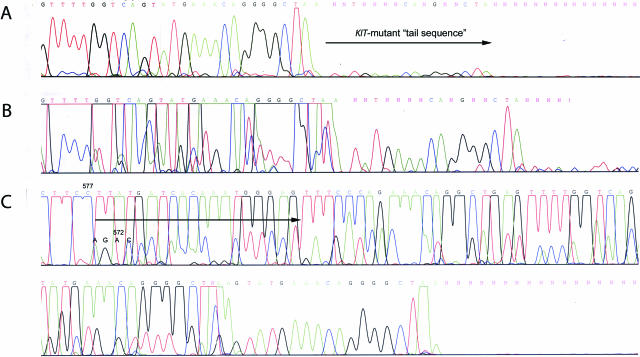
Figure 3.
Direct sequencing of 234-bp (A and B) and 129-bp (C) PCR products from GIST 2. Note the hardly visible (A) and artificially elevated (B) “tail sequence” of mutant KIT allele. Elevated peaks of mutant KIT allele could not be easily distinguished from the elevated background. C: A shift of sequence detected in 129-bp PCR assay indicating duplication is emphasized by an arrow. WT and mutant KIT allele codons demarcating beginning of duplication are indicated by codon numbers.
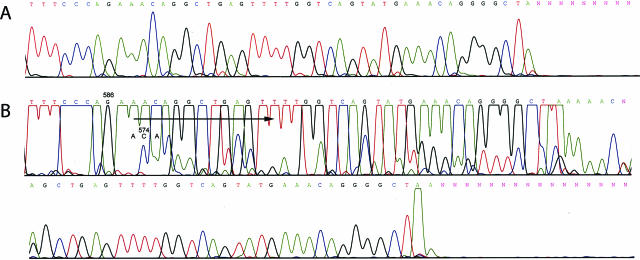
Figure 4.
Direct sequencing of 234-bp (A) and 129-bp (B) PCR products from GIST 3. A: Note the lack of mutant KIT allele sequence in the products from 234-bp PCR assay. B: A shift of sequence detected in 129-bp PCR assay indicating duplication is emphasized by an arrow. WT and mutant KIT allele codons demarcating beginning of duplication are indicated by codon numbers.
Discussion
GISTs are common mesenchymal tumors of gastrointestinal tract, driven by KIT or PDGFRA gain-of-function mutations.1,2 The frequency of these mutations varies between the studies and may depend to some extent on the constellation of tumor type and location in different series. KIT exon 9 mutations have been found almost exclusively in intestinal GISTs,6,24,25 whereas PDGFRA exon 18 mutations are linked to gastric tumors with epithelioid cell morphology.19,26,27,28 Although a majority of KIT exon 11 mutations have been found in GISTs from different locations, duplications affecting 3′KIT exon 11 occur predominantly in gastric tumors; 59 of 67 (88%) GISTs with KIT exon 11 duplications reported with clinical data were of gastric origin.6,7,8,9,10,17,18,19,22,25 Also in this study, three of four analyzed GISTs were gastric, and one was diagnosed in the omentum. Omental GISTs show histological and prognostic features similar to gastric tumors.29
Recent studies based on archival FFPE material reported independently by two different groups revealed decreasing mutation detection rate with increasing age of paraffin blocks.9,30 Although specific factors causing this phenomenon have not been identified yet, there is a possibility that long-term storage of FFPE tissue might increase degradation of nucleic acids. In the current study, two GISTs (cases 2 and 3), which generated most serious diagnostic problems, were prepared by FFPE 18 and 20 years ago, respectively. However, one of the analyzed tumors (case 4) was prepared by FFPE only 5 years ago. Several factors including time between surgery and tissue fixation, prolonged tissue exposure to room temperature, and use of suboptimal formalin and paraffin may increase degradation of nucleic acids and lead to preferential amplification of the KIT allele not containing a duplication, thereby leading to false-negative results.
KIT exon 11 duplications reported in GISTs ranged from 3 to 57 nucleotides.7,8 In this study, three of four analyzed GISTs had relatively large duplications of 39 to 42 nucleotides, whereas one tumor had only 18 nucleotides duplicated. However, this tumor was prepared by FFPE 20 years ago. Size of duplication and degree of degradation (more severe in older paraffin blocks) are two important factors leading to preferential amplification of WT versus mutant KIT allele with duplication.
In this study, 4 of 16 GISTs initially diagnosed KIT-WT were found to have duplications when evaluated with PCR assay exclusively yielding the amplification of a significantly shorter PCR product from the region commonly affected by duplication. Subsequently, we reevaluated all of these tumors using KIT exon 11 PCR assays generating larger products. Although DHPLC analysis indicated mutations in some cases, identification of mutant KIT sequences was often impossible because of the low signal, which could not be separated from the elevated background. Potentially, this could be overcome by DHPLC in-line fraction collector to isolate low-signal heteroduplex peaks followed by their direct sequencing to reveal the mutation.31
Recent studies have shown that DHPLC-based KIT mutation screening is more sensitive than direct sequencing of the PCR products.32 In this study, one of the duplications was undetected by DHPLC when larger PCR products were analyzed. This observation indicates that both direct sequencing and DHPLC could create false-negative results due to suboptimal amplification of the longer mutant KIT allele with duplication when analyzing severely degraded DNA.
Imatinib mesylate, a KIT tyrosine kinase inhibitor also known as Gleevec/Glivec, has been used in the treatment of metastatic GISTs.33,34 Although the majority of GISTs respond well to imatinib treatment, some cases are variably resistant.35 Tumors with KIT exon 11 mutations respond to Gleevec treatment better than GISTs with Ala502_Tyr503dup (KIT exon 9 mutation) or those with WT KIT; also, GISTs with PDGFRA Asp842Val are primarily resistant to Gleevec treatment.14,36,37 Because of these findings, accurate detection of KIT and PDGFRA mutations in GISTs are clinically significant.
In summary, our study confirmed that screening with DHPLC helps to eliminate misinterpretation of the direct sequencing of PCR products but, in some cases, could also create false-negative results due to suboptimal amplification of the longer mutant KIT allele with duplication. Both DHPLC- and direct sequencing-based mutation detection of KIT exon 11 duplications in GISTs could be improved by using PCR assays targeting smaller amplicons including the region commonly affected by duplication.
Footnotes
Supported by the Polish Committee for the Scientific Research (grant 3PO5C05925) and by the Foundation for Polish Science (grant to B.W.).
The opinions and assertions contained herein are the expressed views of the authors and are not to be construed as official or reflecting the views of the Departments of the Army or Defense.
References
- Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, Muhammad Tunio G, Matsuzawa Y, Kanakura Y, Shinomura Y, Kitamura Y. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A, Demetri GD, Fletcher CD, Fletcher JA. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Lasota J. Gastrointestinal stromal tumors (GISTs): definition, occurrence, pathology, differential diagnosis and molecular genetics. Pol J Pathol. 2003;54:3–24. [PubMed] [Google Scholar]
- Lasota J, Jasinski M, Sarlomo-Rikala M, Miettinen M. Mutations in exon 11 of c-kit occur preferentially in malignant versus benign gastrointestinal stromal tumors and do not occur in leiomyomas or leiomyosarcomas. Am J Pathol. 1999;154:53–60. doi: 10.1016/S0002-9440(10)65250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskaluk CA, Tian Q, Marshall CR, Rumpel CA, Franquemont DW, Frierson HF., Jr Mutations of c-kit JM domain are found in a minority of human gastrointestinal stromal tumors. Oncogene. 1999;18:1897–1902. doi: 10.1038/sj.onc.1202496. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Sommer G, Sarran L, Tschernyavsky SJ, Riedel E, Woodruff JM, Robson M, Maki R, Brennan MF, Ladanyi M, DeMatteo RP, Besmer P. Association of KIT exon 9 mutations with non-gastric primary site and aggressive behavior: KIT mutation analysis and clinical correlates of 120 gastrointestinal stromal tumors. Clin Cancer Res. 2003;15:3329–3337. [PubMed] [Google Scholar]
- Lasota J, Dansonka-Mieszkowska A, Stachura T, Schneider-Stock R, Kallajoki M, Steigen SE, Sarlomo-Rikala M, Boltze C, Kordek R, Roessner A, Stachura J, Miettinen M. Gastrointestinal stromal tumors (GIST) with internal tandem duplications in 3′ end of KIT juxtamembrane domain occur predominantly in stomach and generally seem to have a favorable course. Mod Pathol. 2003;16:1257–1264. doi: 10.1097/01.MP.0000097365.72526.3E. [DOI] [PubMed] [Google Scholar]
- Martín J, Poveda A, Llombart-Bosch A, Ramos R, Lopez-Guerrero JA, Garcia del Muro J, Maurel J, Calabuig S, Gutierrez A, Gonzalez de Sande JL, Martinez J, De Juan A, Lainez N, Losa F, Alija V, Escudero P, Casado A, Garcia P, Blanco R, Buesa JM, Spanish Group for Sarcoma Research Deletions affecting codons 557–558 of the c-KIT gene indicate a poor prognosis in patients with completely resected gastrointestinal stromal tumors: a study by the Spanish group of sarcoma research (GEIS). J Clin Oncol. 2005;23:6190–6198. doi: 10.1200/JCO.2005.19.554. [DOI] [PubMed] [Google Scholar]
- Andersson J, Bumming P, Meis-Kindblom JM, Sihto H, Nupponen N, Joensuu H, Oden A, Gustavsson B, Kindblom L-G, Nilsson B. Gastrointestinal stromal tumors with KIT exon 11 deletions are associated with poor prognosis. Gastroenterology. 2006;130:1573–1581. doi: 10.1053/j.gastro.2006.01.043. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Nam SW, Kim H, Rhee H, Kim N-G, Kim H, Hyung WJ, Noh SH, Kim J-H, Yun C-O, Liu ET, Kim H. Correlation of KIT and platelet-derived growth factor receptor α mutations with gene activation and expression profiles in gastrointestinal stromal tumors. Oncogene. 2005;24:1066–1074. doi: 10.1038/sj.onc.1208358. [DOI] [PubMed] [Google Scholar]
- Lasota J, Stachura J, Miettinen J. GISTs with PDGFRA exon 14 mutations represent subset of clinically favorable gastric tumors with epithelioid morphology. Lab Invest. 2006;86:94–100. doi: 10.1038/labinvest.3700360. [DOI] [PubMed] [Google Scholar]
- Rubin BP, Singer S, Tsao C, Duensing A, Lux ML, Ruiz R, Hibbard MK, Chen CJ, Xiao S, Tuveson DA, Demetri GD, Fletcher CD, Fletcher JA. KIT activation is a ubiquitous feature of gastrointestinal stromal tumors. Cancer Res. 2001;61:8118–8121. [PubMed] [Google Scholar]
- Wardelmann E, Neidt I, Bierhoff E, Speidel N, Manegold C, Fischer HP, Pfeifer U, Pietsch T. c-kit mutations in gastrointestinal stromal tumors occur preferentially in the spindle rather than in the epithelioid cell variant. Mod Pathol. 2002;15:125–136. doi: 10.1038/modpathol.3880504. [DOI] [PubMed] [Google Scholar]
- Debiec-Rychter M, Dumez H, Judson I, Wasag B, Verweij J, Brown M, Dimitrijevic S, Sciot R, Stul M, Vranck H, Scurr M, Hagemeijer A, Van Glabbeke M, van Oosterom AT. Use of c-KIT/PDGFRA mutational analysis to predict the clinical response to imatinib in patients with advanced gastrointestinal stromal tumours entered on phase I and II studies of the EORTC soft tissue and bone sarcoma group. Eur J Cancer. 2004;40:689–695. doi: 10.1016/j.ejca.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Feng F, Liu X-H, Xie Q, Liu W-Q, Bai C-G, Ma D-L. Expression and mutation of c-kit gene in gastrointestinal stromal tumors. World J Gastroenterol. 2003;9:2548–2551. doi: 10.3748/wjg.v9.i11.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y-Y, Tan Y-S, Sun M-H, Wei Y-K, Xu J-F, Lu S-H, A-Ke-Su S-J, Zhou Y-N, Gao F, Zheng A-H, Zhang T-M, Hou W-Z, Wang J, Du X, Zhu X-Z. C-kit gene mutation in human gastrointestinal stromal tumors. World J Gastroenterol. 2004;10:1310–1314. doi: 10.3748/wjg.v10.i9.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Lee H, Kang Y-K, Choe MS, Ryu M-H, Chang HM, Kim JS, Yook JH, Kim BS, Lee JS. Prognostic significance of c-kit mutation in localized gastrointestinal stromal tumors. Clin Cancer Res. 2004;10:3076–3081. doi: 10.1158/1078-0432.ccr-03-0581. [DOI] [PubMed] [Google Scholar]
- Subramanian S, West RB, Corless CL, Ou W, Rubin BP, Chu KM, Leung SY, Yuen ST, Zhu S, Hernandez-Boussard T, Montgomery K, Nielsen TO, Patel RM, Goldblum JR, Heinrich MC, Fletcher JA, van de Rijn M. Gastrointestinal stromal tumors (GISTs) with KIT and PDGFRA mutations have distinct gene expression profiles. Oncogene. 2004;23:7780–7790. doi: 10.1038/sj.onc.1208056. [DOI] [PubMed] [Google Scholar]
- Wardelmann E, Hrychyk A, Merkelbach-Bruse S, Pauls K, Goldstein J, Hohenberger P, Losen I, Manegold C, Buttner R, Pietsch T. Association of platelet-derived growth factor receptor α mutations with gastric primary site and epithelioid or mixed cell morphology in gastrointestinal stromal tumors. J Mol Diagn. 2004;6:197–204. doi: 10.1016/s1525-1578(10)60510-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Went PT, Dirnhofer S, Bundi M, Mirlacher M, Schraml P, Mangialaio S, Dimitrijevic S, Kononen J, Lugli A, Simon R, Sauter G. Prevalence of KIT expression in human tumors. J Clin Oncol. 2004;22:4514–4522. doi: 10.1200/JCO.2004.10.125. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Besmer P, Guo T, Arkun K, Hom G, Koryotowski B, Leversha MA, Jeffrey PD, Desantis D, Singer S, Brennan MF, Maki RG, DeMatteo RP. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin Cancer Res. 2005;11:4182–4190. doi: 10.1158/1078-0432.CCR-04-2245. [DOI] [PubMed] [Google Scholar]
- Haller F, Gunawan B, von Heydebreck A, Schwager S, Schulten H-J, Wolf-Salgo J, Langer C, Ramadori G, Sultmann H, Fuzesi L. Prognostic role of E2F1 and members of the CDKN2A network in gastrointestinal stromal tumors. Clin Cancer Res. 2005;11:6589–6597. doi: 10.1158/1078-0432.CCR-05-0329. [DOI] [PubMed] [Google Scholar]
- Théou N, Gil S, Devocelle A, Julie C, Lavergne-Slove A, Beauchet A, Callard P, Farinotti R, Le Cesne A, Lemoine A, Faivre-Bonhomme L, Emile J-F. Multidrag resistance proteins in gastrointestinal stromal tumors: site-dependent expression and initial response to imatinib. Clin Cancer Res. 2005;11:7593–7598. doi: 10.1158/1078-0432.CCR-05-0710. [DOI] [PubMed] [Google Scholar]
- Lasota J, Wozniak A, Sarlomo-Rikala M, Rys J, Kordek R, Nassar A, Sobin LH, Miettinen M. Mutations in exons 9 and 13 of KIT gene are rare events of gastrointestinal stromal tumors: a study of 200 cases. Am J Pathol. 2000;157:1091–1095. doi: 10.1016/S0002-9440(10)64623-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YR, Kim H, Kang HJ, Kim NG, Kim JJ, Park KS, Paik YK, Kim HO, Kim H. Overexpression of high mobility group box 1 in gastrointestinal stromal tumors wit KIT mutation. Cancer Res. 2003;63:2188–2193. [PubMed] [Google Scholar]
- Lasota J, Kopczynski J, Sarlomo-Rikala M, Schneider-Stock R, Stachura T, Kordek R, Michal M, Boltze C, Roessner A, Stachura J, Miettinen M. KIT 1530ins6 mutation defines subset of predominantly malignant gastrointestinal stromal tumors (GIST) of intestinal origin. Hum Pathol. 2003;34:1306–1312. doi: 10.1016/s0046-8177(03)00407-6. [DOI] [PubMed] [Google Scholar]
- Lasota J, Dansonka-Mieszkowska A, Sobin LH, Miettinen M. A great majority of GISTs with PDGFRA mutations represents gastric tumors of low or no malignant potential. Lab Invest. 2004;84:874–883. doi: 10.1038/labinvest.3700122. [DOI] [PubMed] [Google Scholar]
- Wasag B, Debiec-Rychter M, Pauwels P, Stul M, Vranckx H, Oosterom AV, Hagemeijer A, Scoit R. Differential expression of KIT/PDGFRA mutant isoforms in epithelioid and mixed variants of gastrointestinal stromal tumors depends predominantly on the tumor site. Mod Pathol. 2004;17:889–894. doi: 10.1038/modpathol.3800136. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Monihan JM, Sarlomo-Rikala M, Kovatich AJ, Carr NJ, Emory TS, Sobin LH. Gastrointestinal stromal tumors/smooth muscle tumors (GISTs) primary in the omentum and mesentery: clinicopathologic and immunohistochemical study of 26 cases. Am J Surg Pathol. 1999;23:1109–1118. doi: 10.1097/00000478-199909000-00015. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Sobin LH, Lasota J. Gastrointestinal stromal tumors (GISTs) of the stomach: a clinicopathologic, immunohistochemical and molecular genetic study of 1756 cases with long-term follow-up. Am J Surg Pathol. 2005;29:52–68. doi: 10.1097/01.pas.0000146010.92933.de. [DOI] [PubMed] [Google Scholar]
- Emmerson P, Maynard J, Jones S, Butler R, Sampson JR, Cheadle JP. Characterizing mutations in samples with low-level mosaicism by collection and analysis of DHPLC fractionated heteroduplexes. Human Mutat. 2003;21:112–115. doi: 10.1002/humu.10159. [DOI] [PubMed] [Google Scholar]
- Metaxa-Mariatou V, Papadopoulos S, Papadopoulos E, Passa O, Georgiadis T, Arapadoni-Dadioti P, Leondara V, Nasioulas G. Molecular analysis of GISTs: evaluation of sequencing and dHPLC. DNA Cell Biol. 2004;23:777–782. doi: 10.1089/dna.2004.23.777. [DOI] [PubMed] [Google Scholar]
- van Oosterom AT, Judson I, Verweij J, Stroobants S, Donato di Paola E, Dimitrijevic S, Martens M, Webb A, Sciot R, Van Glabbeke M, Silberman S, Nielsen OS, European Organisation for Research and Treatment of Cancer Soft Tissue and Bone Sarcoma Group Safety and efficacy of imatinib (STI571) in metastatic gastrointestinal stromal tumours: a phase I study. Lancet. 2001;358:1421–1423. doi: 10.1016/s0140-6736(01)06535-7. [DOI] [PubMed] [Google Scholar]
- Demetri GD, von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, Heinrich MC, Tuveson DA, Singer S, Janicek M, Fletcher JA, Silverman SG, Silberman SL, Capdeville R, Kiese B, Peng B, Dimitrijevic S, Druker BJ, Corless C, Fletcher CD, Joensuu H. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- Van Glabbeke M, Verweij J, Casali PG, Le Cesne A, Hohenberger P, Ray-Coquard I, Schlemmer M, van Oosterom AT, Goldstein D, Sciot R, Hogendoorn PCW, Brown M, Bertulli R, Judson IR. Initial and late resistance to imatinib in advanced gastrointestinal stromal tumors are predicted by different prognostic factors: a European Organization for Research and Treatment of Cancer-Italian Sarcoma Group-Australasian Gastrointestinal Trials Group study. J Clin Oncol. 2005;23:5795–5804. doi: 10.1200/JCO.2005.11.601. [DOI] [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, McGreevey LS, Chen CJ, Van den Abbeele AD, Druker BJ, Kiese B, Eisenberg B, Roberts PJ, Singer S, Fletcher CDM, Silberman S, Dimitrijevic S, Fletcher JA. Kinase mutations in imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- Corless CL, Schroeder A, Griffith D, Town A, McGreevey L, Harrell P, Shiraga S, Bainbridge T, Morich J, Heinrich MC. PDGFRA mutations in gastrointestinal stromal tumors: frequency, spectrum and in vitro sensitivity to imatinib. J Clin Oncol. 2005;23:5357–5364. doi: 10.1200/JCO.2005.14.068. [DOI] [PubMed] [Google Scholar]