Abstract
Germline mutations in the tumor suppressor gene APC are the underlying cause of familial adenomatous polyposis, an autosomal-dominant cancer predisposition syndrome of the colorectum. Here, we describe a complex pathogenic rearrangement in the APC gene that was detected during deletion screening and transmitted throughout at least three generations. The rearrangement consists of a deletion of 604 bp in intron 4 that impairs the binding site of the reverse primer for exon 4 and of an insertion of 119 bp in exon 4 that interferes with the binding site of the multiplex ligation-dependent probe amplification (MLPA) probes for exon 4. The insertion is composed of three duplicated sequences derived from exon 4, intron 3, and intron 4, all in inverse direction. By transcript analysis, we found that the mutation results in complete skipping of exon 4 and that it leads to a frameshift. The rearrangement would not have been identified had it occurred outside the MLPA hybridization site. Our findings demonstrate that part of the pathogenic mutations remain undetected by routine methods. Moreover, MLPA and RNA analysis alone would have led to an incorrect interpretation of a genomic deletion of exon 4.
Germline mutations in the tumor suppressor gene APC are known to cause familial adenomatous polyposis (FAP) (MIM *175100, http://www.ncbi.nlm.nih.gov), an early onset autosomal-dominant cancer predisposition syndrome typically characterized by the presence of more than 100 adenomatous polyps in the entire colorectum.1,2 In the attenuated phenotype (AFAP), the number of colorectal adenomas is often less than 100, and diagnosis of both polyposis and colorectal cancer is at a later age than in typical FAP.3
Traditional diagnostic methods can identify APC mutations in 50 to 90% of patients with typical FAP and in 20 to 30% of patients with AFAP, depending on the patients examined and the methods used (Solomon C, Burt RW: APC-Associated Polyposis Conditions. GeneReviews: http://www.geneclinics.org., accessed October 21, 2005).4,5,6,7 Recently, large genomic deletions encompassing one to several exons or even the entire APC gene have been identified by use of multiplex ligation-dependent probe amplification (MLPA) in ∼7 to 12% of all patients with typical FAP.8,9,10 Biallelic mutations in the base excision repair gene MUTYH have been identified in a subset of patients with an attenuated or atypical course of the disease.11,12,13 Evidence for the existence of unidentified APC mutations came from other methods including mRNA inactivation studies7,14 and monoallelic mutation analysis.15,16 Complex genomic rearrangements encompassing the APC gene have been rarely described. Here, we report on a novel complex intragenic mutation involving intron 3, exon 4, and intron 4 of the APC gene that was detected in a FAP family by use of MLPA.
Materials and Methods
Patients
The female index patient was diagnosed with familial adenomatous polyposis (FAP) at the age of 44 years because of rectal bleeding and abdominal pain. Multiple tubular and tubulo-villous adenomas showing low-grade dysplasia were present in the entire colorectum. Fundic gland polyps were seen in the stomach; no duodenal polyps were found. No congenital hypertrophy of the retinal pigment epithelium could be detected. A colonoscopy on her 69-year-old mother revealed an adenocarcinoma at the right colon flexure and, in addition, multiple tubulo-villous adenomas with low-grade dysplasia. In the 18-year-old daughter of the index patient, ∼30 small tubular adenomas without dysplasia were apparent throughout the entire colorectum.
Mutation Analysis
Routine screening for APC mutations was performed on genomic DNA by use of the protein truncation test for mutations in exon 15, denaturing high-performance liquid chromatography for mutations in exons 1 to 14 and the first 500 bp of exon 15, and MLPA for the presence of large genomic deletions (SALSA P043 APC exon deletion test kit; MRC Holland, Amsterdam, The Netherlands), as described.8,17
Long-Range Polymerase Chain Reaction (PCR) on Genomic DNA
Long-range PCR was used to characterize the presumed deletion uncovered by MLPA. With the Expand High Fidelity PCR system (Roche Diagnostics, Mannheim, Germany), different primers located 5′ and 3′ to exon 4 were applied (Table 1). PCR was performed according to the manufacturer’s recommendations. PCR products were separated on a 1% agarose gel and visualized by ethidium bromide staining on a UV imaging system (Bio-Rad Laboratories, Münich, Germany). To determine the breakpoints of the rearrangement, long-range PCR products containing the assumed deletion were excised from the gel, purified by QIAquick PCR purification kit (Qiagen, Hilden, Germany), and sequenced on an ABI Prism 3100 automated sequencer (Applied Biosystems, Darmstadt, Germany) using the cycle-sequencing procedure and the BigDye terminator kit version 1.1 (Applied Biosystems).
Table 1.
Primer Sequences Used for Characterization of the Rearrangement on mRNA or DNA Level
| Primer no. | Exon/intron | Primer sequences | Location of primers GenBank AC008575.7 |
|---|---|---|---|
| 834 | Intron 2F | 5′-TGGTTAAAATGTAAACCTAATATTTC-3′ | 56460_56485 |
| 2119 | Exon 3F | 5′-GGAAGCAGAGAAAGTACTGG-3′ | 56723_56742 |
| 2121 | Intron 3F | 5′-CACTGTGAGTCAGTAGTATGC-3′ | 60475_60495 |
| 2122 | Intron 3F | 5′-CACCTTGGACCTGGTAGGTC-3′ | 62276_62295 |
| 1595 | Intron 3F | 5′-ACAACTGATGTAAGTATTGCTC-3′ | 64928_64949 |
| 2173 | Exon 4F | 5′-AGGTCATTGCTTCTTGCTGATC-3′ | 65006_65027 |
| 1040 | Exon 4F | 5′-ACGCTCAACTTCAGAATCTCAC-3′ | 65061_65082 |
| 2164 | Intron 4R | 5′-CCTAGGTACTTTAAAATATCAAG-3′ | 65180_65158 |
| 1596 | Intron 4R | 5′-TGAATTTTAATGGATTACCTAGGT-3′ | 65197_65174 |
| 2123 | Intron 4R | 5′-GTTACCATTGCTAGCAACATCC-3′ | 66366_66345 |
| 2120 | Exon 5R | 5′-GCAACTCTGATTTGCCTTGC-3′ | 70233_70214 |
| 921 | Intron 5R | 5′-TGTAATTCATTTTATTCCTAATAGCTC-3′ | 70344_70318 |
APC Transcript Analysis
Fresh venous blood samples (2.5 ml) were collected into PAXgene blood RNA tubes (PreAnalytiX; Qiagen) containing RNA stabilizing solution. Total RNA was extracted by use of the PAXgene blood RNA kit (Qiagen) according to the manufacturer’s protocol. First strand cDNA was synthesized from 2 to 3 μg of total RNA by random hexamer-primed reverse transcription with the Superscript first strand system for reverse transcriptase (RT)-PCR (Invitrogen GmbH, Karlsruhe, Germany) according to the manufacturer’s protocol. RT-PCR fragments were obtained according to a standard PCR protocol by use of a forward primer localized in exon 3 and a reverse primer in exon 5. RT-PCR products were separated on a 1% agarose gel and visualized with ethidium bromide. Individual bands were excised from the gel and eluted by use of the High Pure PCR product purification kit (Roche Diagnostics). Eluted products were reamplified with the same pair of primers and sequenced as described above.
Results
No point mutation in the APC gene was identified by protein truncation test or denaturing high-performance liquid chromatography screening in the patient. However, search for large deletions by MLPA indicated a heterozygous deletion of exon 4. This deletion appeared not to be an artifact because of a variant in the binding site of MLPA probes for exon 4 because no mutation was detected by sequencing of the PCR product obtained with primers used in routine mutation analysis of exon 4 (primer no. 1595F and 1596R; Table 1).
A hint for a small deletion was obtained by long-range PCR using primers localized in intron 2 (834F) and intron 5 (921R). The region involved was narrowed down by long-range PCR with different primers localized in intron 3 and intron 4 or in exon 4, respectively. With primers in exon 4 (1040F) and intron 4 (2123R), an aberrant product of ∼700 bp was obtained in addition to the normal product of 1305 bp in the patient but not in controls (not shown). To our surprise, sequencing of the 700-bp fragment revealed a deletion of 604 bp localized exclusively in intron 4 (deleted nucleotides g.65185_65788, GenBank AC008575.7) (not shown). This 604-bp deletion is not a common polymorphism because it was not present in 100 normal unrelated controls. The extent of the deletion was in line with the pattern obtained on agarose gels but could not explain the heterozygous deletion of exon 4 detected by MLPA.
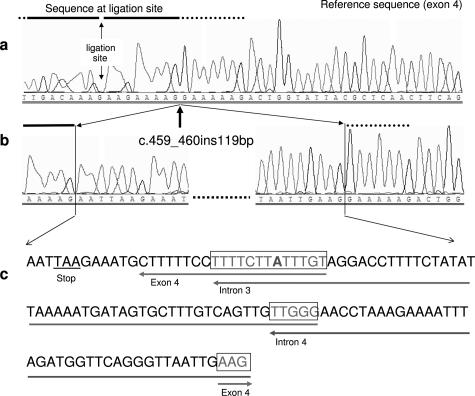
We found that the 5′ breakpoint of the deletion disrupted the sequence of the reverse primer (1596R) of exon 4. Sequencing of a PCR product obtained with primers 1595F and a newly designed reverse primer located more 5′ (2164R) that was not impaired by the deletion, revealed an insertion of 119 bp (insertion site g.65044_65045; designed as c.459_460ins119). This insertion occurred within the hybridization sequence of MLPA probes for exon 4 (Figure 1). The inserted nucleotides 13 to 119 were composed of three different fragments of the APC gene with small overlapping sequences at the ends of each fragment (http://www.ncbi.nlm.nih.gov/BLAST/). Nucleotides 13 to 33 of the inserted 119-bp sequence originated from exon 4 (nucleotides g.65051_65031), nucleotides 21 to 80 from intron 3 (nucleotides g.63194_63135), and nucleotides 76 to 119 derived from intron 4 (nucleotides g. 67389_67346) (Figures 1and 2). All fragments were inserted in an inverse direction. No homology was found for the first 12 bp of the 119-bp insertion. Nucleotides 4 to 6 formed a premature in-frame termination codon (TAA). A detailed description of the rearrangement is shown in the supplementary online material (Supplemental Figure S1 at http://jmd.amjpathol.org/). The insertion in exon 4 was not expressed in mRNA. Instead, a deletion of exon 4 was detected (not shown). Deletion of exon 4 of the APC gene is predicted to result in a frameshift.
Figure 1.
Partial sequencing pattern of exon 4 and the inserted fragments. a: Normal sequence of APC exon 4. The sequence at the ligation site of MLPA probes for exon 4 (according to MRC Holland) is marked by a horizontal bar. b: Partial sequence of the mutant allele showing the breakpoint in exon 4 (arrow) and the sequence of the 5′ and 3′ end of the 119-bp insertion. c: Complete sequence of the 119-bp insertion in exon 4. The origin of the first 12 bp is unknown. The corresponding genomic origin of the other fragments is marked by horizontal arrows. The framed regions indicate the overlapping sequences at the ends of each fragment [one mismatched nucleotide (A) is shown in bold].
Figure 2.
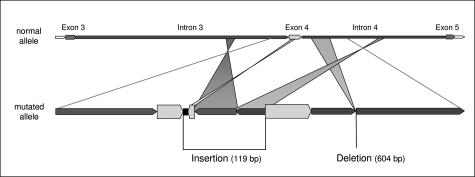
Diagram of the APC rearrangement in the family. The insertion of 119 bp is composed of a sequence of 12 bp of unknown origin (black box) and of three fragments derived from the APC gene, each in inverse direction: 21 bp originated from exon 4, 60 bp from intron 3, and 44 bp from intron 4 (with overlaps of 13 and 5 bp, respectively).
Both the deletion and the insertion were localized within the same allele, as shown by sequencing of the complete aberrant band obtained with primers at the 5′ end of exon 4 (2173F) and in intron 4 (2123R). The region in intron 3 and intron 4 where the inserted sequences originated were not deleted in the patient’s DNA. The rearrangement segregated in a stable form throughout three generations because it was identified in all three affected family members.
Discussion
In a three-generation FAP family, we characterized a complex genomic rearrangement of the APC gene, composed of a 119-bp insertion in exon 4 and a 604-bp deletion in intron 4, that was uncovered by coincidence because the insertion had occurred close to the ligation site of MLPA probes for exon 4. By transcript analysis, we could demonstrate that the mutation results in skipping of exon 4. The attenuated course of the polyposis disease in the family and the absence of congenital hypertrophy of the retinal pigment epithelium are in accordance with the known genotype-phenotype correlation.18,19,20,21
Different mechanisms have been proposed to contribute to the origin of gross genomic alterations including unequal crossing over by retrotransposon-mediated homologous recombination and replication slippage.22,23 Alu-driven recombination seems to be a frequent cause of large deletions in APC and other (tumor suppressor) genes.24,25,26,27 Complex rearrangements other than indels are difficult to explain in terms of their underlying mutational mechanisms, most of the assumed causes are highly hypothetical.28 Complex alterations encompassing exon 14 of APC, also resulting in inappropriate splicing, have been described by Su and colleagues25 and were supposed to be Alu mediated. This mechanism is unlikely in our case because all six intronic breakpoints are localized outside repetitive sequences. The composition of three duplicated and inverted insertions (and one deletion) involving four different intragenic sites (genomic distance between the two inserted fragments: 4152 bp) makes also simple (serial) replication slippage implausible. Nevertheless, the presence of short homologous sequence repeats at the ends of the inserted fragments (13 bp containing one mismatch, 5 bp, and 3 bp, respectively; Supplemental Figure S1 at http://jmd.amjpathol.org/) is striking. The length of the repeats is probably insufficient to mediate unequal crossing over by homologous recombination.28 In contrast, they may have facilitated the formation of more complex secondary genomic structures such as loops within the mutated allele, that serve as templates for the sequences between the shared short sequences, possibly in a multistep process. Chen and colleagues23,28 provide the explanation of untemplated DNA incorporation as an attempt to repair broken DNA strands. It comprises the capture of DNA oligonucleotides, which could be promoted by short sequence complementarity.
Our findings highlight the difficulties in the identification and correct interpretation of some specific mutation types or sites. As in our patient, a variant in the primer sequence may prohibit the amplification of one allele and mask a mutation in the respective exon. The alteration described here would not have been noticed at all had the insertion occurred outside the hybridization sequence of MLPA probes. Moreover, combined results of MLPA and RT-PCR would have led to the wrong result of a true deletion of exon 4 instead of an insertion. On the other hand, deletions localized completely within introns are also shown to be pathogenic by affecting splice sites.25
Gross deletions are a common cause of typical FAP.8,9,10,25,29 The identification of the mutations described here and by others25 suggests that complex rearrangements might be more frequent than suspected, but fail to be detected by routine diagnostic procedures. Additional methods for mutation detection should be applied in FAP families without identified mutation in APC or MUTYH.
Supplementary Material
Footnotes
Supported by the German Cancer Aid (Deutsche Krebshilfe e.V. Bonn, project no. 106244).
Supplemental material for this article can be found on http://jmd.amjpathol.org.
Databases: Reference sequence: mRNA sequence NM_000038.2 GI:21626462; Genomic sequence: gi 14971161 gb AC008575.7 (NCBI); Online Mendelian Inheritance of Man no. 175100.
References
- Bülow S. Clinical features in familial polyposis coli. Results of the Danish Polyposis Register. Dis Colon Rectum. 1986;29:102–107. doi: 10.1007/BF02555389. [DOI] [PubMed] [Google Scholar]
- Fearnhead NS, Britton MP, Bodmer WF. The ABC of APC. Hum Mol Genet. 2001;10:721–733. doi: 10.1093/hmg/10.7.721. [DOI] [PubMed] [Google Scholar]
- Knudsen AL, Bisgaard ML, Bulow S. Attenuated familial adenomatous polyposis (AFAP): a review of the literature. Fam Cancer. 2003;2:43–55. doi: 10.1023/a:1023286520725. [DOI] [PubMed] [Google Scholar]
- Friedl W, Aretz S. Familial adenomatous polyposis—experience from a study on 1164 German unrelated polyposis patients. Hered Cancer. 2005;3:95–114. doi: 10.1186/1897-4287-3-3-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis YL, Morton DG, McKeown CM, Macdonald F. Molecular analysis of the APC gene in 205 families: extended genotype-phenotype correlations in FAP and evidence for the role of APC amino acid changes in colorectal cancer predisposition. J Med Genet. 1999;36:14–20. [PMC free article] [PubMed] [Google Scholar]
- van der Luijt RB, Khan PM, Vasen HF, Tops CM, van Leeuwen- Cornelisse IS, Wijnen JT, van der Klift HM, Plug RJ, Griffioen G, Fodde R. Molecular analysis of the APC gene in 105 Dutch kindreds with familial adenomatous polyposis: 67 germline mutations identified by DGGE, PTT, and Southern analysis. Hum Mutat. 1997;9:7–16. doi: 10.1002/(SICI)1098-1004(1997)9:1<7::AID-HUMU2>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Powell SM, Petersen GM, Krush AJ, Booker S, Jen J, Giardiello FM, Hamilton SR, Vogelstein B, Kinzler KW. Molecular diagnosis of familial adenomatous polyposis. N Engl J Med. 1993;329:1982–1987. doi: 10.1056/NEJM199312303292702. [DOI] [PubMed] [Google Scholar]
- Aretz S, Stienen D, Uhlhaas S, Pagenstecher C, Mangold E, Caspari R, Propping P, Friedl W. Large submicroscopic genomic APC deletions are a common cause of typical familial adenomatous polyposis. J Med Genet. 2005;42:185–192. doi: 10.1136/jmg.2004.022822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunyan DJ, Eccles DM, Sillibourne J, Wilkins E, Thomas NS, Shea-Simonds J, Duncan PJ, Curtis CE, Robinson DO, Harvey JF, Cross NC. Dosage analysis of cancer predisposition genes by multiplex ligation-dependent probe amplification. Br J Cancer. 2004;91:1155–1159. doi: 10.1038/sj.bjc.6602121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michils G, Tejpar S, Thoelen R, van Cutsem E, Vermeesch JR, Fryns JP, Legius E, Matthijs G. Large deletions of the APC gene in 15% of mutation-negative patients with classical polyposis (FAP): a Belgian study. Hum Mutat. 2005;25:125–134. doi: 10.1002/humu.20122. [DOI] [PubMed] [Google Scholar]
- Aretz S, Uhlhaas S, Goergens H, Siberg K, Vogel M, Pagenstecher C, Mangold E, Caspari R, Propping P, Friedl W. MUTYH-associated polyposis: 70 of 71 patients with biallelic mutations present with an attenuated or atypical phenotype. Int J Cancer. 2006;119:807–814. doi: 10.1002/ijc.21905. [DOI] [PubMed] [Google Scholar]
- Sampson JR, Dolwani S, Jones S, Eccles D, Ellis A, Evans DG, Frayling I, Jordan S, Maher ER, Mak T, Maynard J, Pigatto F, Shaw J, Cheadle JP. Autosomal recessive colorectal adenomatous polyposis due to inherited mutations of MYH. Lancet. 2003;362:39–41. doi: 10.1016/S0140-6736(03)13805-6. [DOI] [PubMed] [Google Scholar]
- Nielsen M, Franken PF, Reinards TH, Weiss MM, Wagner A, van der Klift H, Kloosterman S, Houwing-Duistermaat JJ, Aalfs CM, Ausems MG, Brocker-Vriends AH, Gomez Garcia EB, Hoogerbrugge N, Menko FH, Sijmons RH, Verhoef S, Kuipers EJ, Morreau H, Breuning MH, Tops CM, Wijnen JT, Vasen HF, Fodde R, Hes FJ. Multiplicity in polyp count and extracolonic manifestations in 40 Dutch patients with MYH associated polyposis coli (MAP). J Med Genet. 2005;42:e54. doi: 10.1136/jmg.2005.033217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkonen ET, Nieminen P, Abdel-Rahman WM, Moisio AL, Jarvela I, Arte S, Jarvinen HJ, Peltomaki P. Adenomatous polyposis families that screen APC mutation-negative by conventional methods are genetically heterogeneous. J Clin Oncol. 2005;23:5651–5659. doi: 10.1200/JCO.2005.14.712. [DOI] [PubMed] [Google Scholar]
- Papadopoulos N, Leach FS, Kinzler KW, Vogelstein B. Monoallelic mutation analysis (MAMA) for identifying germline mutations. Nat Genet. 1995;11:99–102. doi: 10.1038/ng0995-99. [DOI] [PubMed] [Google Scholar]
- Laken SJ, Papadopoulos N, Petersen GM, Gruber SB, Hamilton SR, Giardiello FM, Brensinger JD, Vogelstein B, Kinzler KW. Analysis of masked mutations in familial adenomatous polyposis. Proc Natl Acad Sci USA. 1999;96:2322–2326. doi: 10.1073/pnas.96.5.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl W, Caspari R, Sengteller M, Uhlhaas S, Lamberti C, Jungck M, Kadmon M, Wolf M, Fahnenstich J, Gebert J, Moslein G, Mangold E, Propping P. Can APC mutation analysis contribute to therapeutic decisions in familial adenomatous polyposis? Experience from 680 FAP families. Gut. 2001;48:515–521. doi: 10.1136/gut.48.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aretz S, Uhlhaas S, Sun Y, Pagenstecher C, Mangold E, Caspari R, Moslein G, Schulmann K, Propping P, Friedl W. Familial adenomatous polyposis: aberrant splicing due to missense or silent mutations in the APC gene. Hum Mutat. 2004;24:370–380. doi: 10.1002/humu.20087. [DOI] [PubMed] [Google Scholar]
- Heppner Goss K, Trzepacz C, Tuohy TM, Groden J. Attenuated APC alleles produce functional protein from internal translation initiation. Proc Natl Acad Sci USA. 2002;99:8161–8166. doi: 10.1073/pnas.112072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samowitz WS, Thliveris A, Spirio LN, White R. Alternatively spliced adenomatous polyposis coli (APC) gene transcripts that delete exons mutated in attenuated APC. Cancer Res. 1995;55:3732–3734. [PubMed] [Google Scholar]
- Giardiello FM, Brensinger JD, Luce MC, Petersen GM, Cayouette MC, Krush AJ, Bacon JA, Booker SV, Bufill JA, Hamilton SR. Phenotypic expression of disease in families that have mutations in the 5′ region of the adenomatous polyposis coli gene. Ann Intern Med. 1997;126:514–519. doi: 10.7326/0003-4819-126-7-199704010-00003. [DOI] [PubMed] [Google Scholar]
- Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN. Complex gene rearrangements caused by serial replication slippage. Hum Mutat. 2005;26:125–134. doi: 10.1002/humu.20202. [DOI] [PubMed] [Google Scholar]
- Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN. Intrachromosomal serial replication slippage in trans gives rise to diverse genomic rearrangements involving inversions. Hum Mutat. 2005;26:362–373. doi: 10.1002/humu.20230. [DOI] [PubMed] [Google Scholar]
- Petrij-Bosch A, Peelen T, van Vliet M, van Eijk R, Olmer R, Drusedau M, Hogervorst FB, Hageman S, Arts PJ, Ligtenberg MJ, Meijers-Heijboer H, Klijn JG, Vasen HF, Cornelisse CJ, van ’t Veer LJ, Bakker E, van Ommen GJ, Devilee P. BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients. Nat Genet. 1997;17:341–345. doi: 10.1038/ng1197-341. [DOI] [PubMed] [Google Scholar]
- Su LK, Steinbach G, Sawyer JC, Hindi M, Ward PA, Lynch PM. Genomic rearrangements of the APC tumor-suppressor gene in familial adenomatous polyposis. Hum Genet. 2000;106:101–107. doi: 10.1007/s004399900195. [DOI] [PubMed] [Google Scholar]
- Wang Y, Friedl W, Lamberti C, Jungck M, Mathiak M, Pagenstecher C, Propping P, Mangold E. Hereditary nonpolyposis colorectal cancer: frequent occurrence of large genomic deletions in MSH2 and MLH1 genes. Int J Cancer. 2003;103:636–641. doi: 10.1002/ijc.10869. [DOI] [PubMed] [Google Scholar]
- Cao X, Eu KW, Seow-Choen F, Zhao Y, Cheah PY. Topoisomerase-I- and Alu-mediated genomic deletions of the APC gene in familial adenomatous polyposis. Hum Genet. 2001;108:436–442. doi: 10.1007/s004390100492. [DOI] [PubMed] [Google Scholar]
- Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN. Meta-analysis of gross insertions causing human genetic disease: novel mutational mechanisms and the role of replication slippage. Hum Mutat. 2005;25:207–221. doi: 10.1002/humu.20133. [DOI] [PubMed] [Google Scholar]
- Sieber OM, Lamlum H, Crabtree MD, Rowan AJ, Barclay E, Lipton L, Hodgson S, Thomas HJ, Neale K, Phillips RK, Farrington SM, Dunlop MG, Mueller HJ, Bisgaard ML, Bulow S, Fidalgo P, Albuquerque C, Scarano MI, Bodmer W, Tomlinson IP, Heinimann K. Whole-gene APC deletions cause classical familial adenomatous polyposis, but not attenuated polyposis or “multiple” colorectal adenomas. Proc Natl Acad Sci USA. 2002;99:2954–2958. doi: 10.1073/pnas.042699199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.