Abstract
An acquired mutation in Janus kinase 2 (JAK2), V617F, has recently been identified in human myeloproliferative disorders. Detection of the mutation is helpful in differential diagnosis, prognosis, and predication of therapeutic response. Because the mutation can be present in a small proportion of granulocytic populations in myeloproliferative disorder patients, a highly sensitive detection method is required. In this study, we systematically optimized the reaction conditions of a published amplification refractory mutation system-polymerase chain reaction research protocol to make it a robust clinical diagnostic test. The modifications led to a clear demonstration of the V617F mutation in a patient who would have been easily missed by the original amplification refractory mutation system-polymerase chain reaction assay. The test detects the V617F mutation not only with a high analytic sensitivity of 0.05 to 0.1% but also with a high diagnostic specificity of 99%. In addition, the assay has the ability to distinguish cases with only mutant alleles from cases with mixed normal and mutant alleles. The assay is fast and easy to perform, and no special equipment other than thermocyclers is required. All these features make the assay readily and broadly applicable in clinical molecular diagnostic laboratories.
An acquired mutation in the JAK2 gene has recently been described in human myeloproliferative disorders.1,2,3,4,5 JAK2 is a cytoplasmic tyrosine kinase that plays an essential role in the signaling pathways of cytokines and growth factors. The mutation 1849 G>T, which leads to amino acid substitution of phenylalanine for a highly conserved valine (V617F), renders JAK2 kinase constitutively active and leads to cell proliferation in the absence of the growth factors.2,4,5
The V617F mutation has been detected in 65 to 97% of patients with polycythemia vera, 23 to 57% of those with essential thrombocythemia (ET), and 30 to 57% of idiopathic myelofibrosis.1,2,3,4,5 In the vast majority of literature, JAK2mutation is absent in normal individuals, in patients with secondary erythrocytosis and thrombocytosis, or in patients with chronic myelogenous leukemia; detection of the mutation thus aids in differential diagnosis in cases with similar clinicopathological features.6 In addition, studies have found that JAK2 V617F mutation is associated with response to hydroxyurea in patients with ET and may be correlated with poorer survival in patients with idiopathic myelofibrosis.7,8
Several laboratory techniques have been developed for JAK2 V617F genotypic analysis. Because V617F is an acquired mutation that can be present in a small proportion of granulocytic populations in some cases, especially of ET, a highly sensitive detection method is essential.7 Different sensitivity of various assay methods partially accounts for the wide range of mutation frequencies reported in the literature.9 As a matter of fact, re-evaluation of the same cases with a more sensitive technique has increased the detection rate from 73 to 97% in patients with polycythemia vera.1 In addition, a highly sensitive technique is potentially useful to monitor residual disease after treatment. Methodologies reported in the literature include direct sequencing,1,2,4,5 allele-specific polymerase chain reaction (PCR),1,10,11 PCR-restriction fragment length polymorphism,5,7 pyrosequencing,3,12 and amplification refractory mutation system (ARMS)-PCR.3 The advantages and disadvantages of the methods are briefly discussed below, and interested readers are referred to a detailed review by Steensma13 for principles and potential clinical utilities of each assay.
The JAK2 mutation was discovered using direct sequencing after PCR. However, the method only detects the mutation present in more than 20 to 30% of total DNA. Compared with sequencing, allele-specific PCR detects the mutation with a much higher sensitivity at 1 to ∼3%. However, the allele-specific PCR assay does not amplify the normal alleles, so cases that carry only mutant alleles show the same pattern as those that carry both the mutant and the normal alleles as a result of either heterozygosity or mixed clonality. With a sensitivity of ∼4% for mutation detection, PCR-restriction fragment length polymorphism using restriction enzyme BsaXI has the ability to show both mutant and normal allele in a sample. However, the assay may cause misinterpretation because incomplete BsaXI digestion would generate a mutant plus normal pattern in patients with only wild-type alleles. In addition, a restriction digestion step increases the cost, time, and labor of the test. Pyrosequencing is an emerging technique based on nucleotide extension by DNA polymerase; it is able to detect V617F in ∼5% of DNA, and the level of the mutant allele can be quantified. However, requirement of expensive equipment and special reagents makes it less ideal to serve as a routine clinical test. ARMS-PCR is a procedure originally developed for the analysis of single nucleotide polymorphisms, and the assay has been successfully adopted to analyze the JAK2 genotypes.3 This technique enables simultaneous amplification of the mutant and normal alleles plus a DNA quality control with just two pairs of primers in a single PCR tube (Figure 1). The test is therefore a simple, fast, and inexpensive procedure that does not entail any special equipment other than a thermocycler. However, the method is relatively insensitive in detecting low levels of the mutation, possibly because of primer competitions among different PCR reactions. An ET case that was almost missed by the assay prompted us to optimize the reaction conditions systematically for clinical use. The modified assay detects mutation with a highly reproducible analytic sensitivity of 0.05 to 0.1%, making it suitable for the detection of low levels of JAK2 mutation. The test will be potentially useful to follow minimal residual disease when targeted therapies against JAK2 kinase become available.
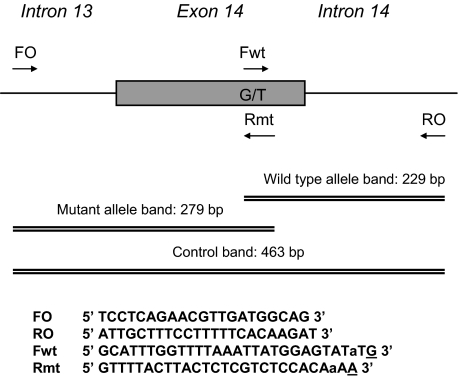
Figure 1.
Schematic diagram of the ARMS-PCR assay. Two forward and two reverse primers are used in different combinations to generate three potential PCR products. Primers FO and RO flank the exon 14 of the JAK2 gene, resulting in a band of 463 bp to control for DNA quality and quantity. Primers Fwt and RO amplify a wild-type allele, generating a band of 229 bp, and primers FO and Rmt generate a band of 279 bp from the mutant allele. Primer sequences are shown below the diagram. The intended mismatches in Fwt and Rmt are shown in lowercase, and genotype-specific nucleotides are underlined.
Materials and Methods
Cells and Patient Samples
The HEL cell line that carries homozygous mutant T alleles was obtained from Drs. Shahin Rafii and David Jin at Weill Medical College of Cornell University and was originally purchased from (American Type Culture Collection, Rockville, MD). Leukocytes from a healthy donor were purchased from the New York Blood Center (New York, NY). Eighteen DNA samples for validation were obtained from patients with myeloproliferative disorders who were referred to Salisbury District Hospital for routine analysis. Informed consents were obtained, and personal identifiers were removed to protect patient confidentiality for the purposes of this study. One hundred seventeen residual DNA samples from patients who were tested for other diseases were used to study the assay specificity and were obtained from the DNA bank at Weill Medical College of Cornell University in an anonymous manner.
DNA Extraction
DNA from HEL cells and patient samples was extracted using the QIAamp DNA mini kit or EZ1 system (Qiagen, Valencia, CA) following the manufacturer’s instructions. DNA was quantified using spectrophotometric measurements.
ARMS-PCR
Principle of the ARMS-PCR assay is illustrated, and the primer sequences are shown in Figure 1 (modified from Jones et al3). Mismatches are included to maximize discrimination of the wild-type and mutant alleles. Amplifications were performed using HotStart Taq polymerase (Qiagen). PCR conditions of both modified and the original method are shown in Table 1, and reactions were conducted in T3000 and Tgradient thermocyclers (Biometra, Goettingen, Germany). PCR products were resolved on 2% agarose gels for 45 minutes at 80 V.
Table 1.
Comparison of PCR Conditions of the Original and Modified ARMS PCR Assays
| Condition types | Original | Modified |
|---|---|---|
| Reaction conditions | ||
| dNTP | 200 μmol/L | 120 μmol/L |
| Primer FO | 1.0 μmol/L | 0.4 μmol/L |
| Primer RO | 1.0 μmol/L | 0.3 μmol/L |
| Primer Fwt | 0.5 μmol/L | 0.5 μmol/L |
| Primer Rmt | 0.5 μmol/L | 1.0 μmol/L |
| DNA polymerase | AmpliTaq Gold, 1 U | HotStart, 1 U |
| Template DNA | 25 ng | 100 ng |
| Total volume | 25 μl | 25 μl |
| Cycling conditions | ||
| Denaturing | 94°C, 1 minute | 94°C, 30 seconds |
| Annealing | 60°C, 1 minute | 58°C, 45 seconds |
| Extension | 72°C, 1 minute | 72°C, 45 seconds |
| Cycle number | 30 | 40 |
Pyrosequencing
Quantitation of the percentage of the V617F alleles was performed by pyrosequencing as described.14
Results and Discussion
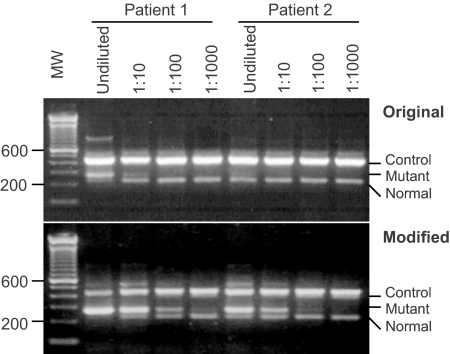
Design of the ARMS-PCR assay is shown in Figure 1. With the original reaction conditions, a faint band of the mutant allele was barely detected in an undiluted DNA specimen from a patient with clinical and bone marrow features consistent with ET (Figure 2, top, patient 2). Given the low levels of the V617F mutation observed in ET patients, further investigation was performed using a single pair of mutant-specific primers (FO and Rmt in Figure 1). The presence of the V617F mutation was clearly demonstrated (data not shown and see below), suggesting that the analytic sensitivity of the original ARMS-PCR was suboptimal. We suspect that the low sensitivity was caused by competing PCR reactions occurring in the same reaction tube. In particular, amplification of the control DNA fragment seemed to out-compete the reactions of the mutant and/or normal alleles for primers (Figure 2, top; note the relative intensity of the three bands). Several changes were made to improve systematically the analytic sensitivity of the assay. The differences between the original ARMS-PCR and modified version are listed in Table 1. The major changes include the following. 1) The concentration of the mutant-specific primer (Rmt) was raised, and concentrations of common primers (FO and RO) were lowered to favor amplification from the mutant allele. 2) The annealing temperature was lowered from 60 to 58°C to favor the binding of both Fwt and Rmt primers that contain mismatches to the templates (Figure 1). The best temperature was determined to be 58°C in the temperature range of 55 to 63°C tested with a gradient thermocycler (data not shown). 3) The number of cycles was increased from 30 to 40 cycles, significantly enhancing the yields of all three PCR products (data not shown). Together, these changes resulted in a more robust amplification of the mutant allele and a less competing reaction from the control, as evidenced by the relative intensities of the corresponding bands on agarose gel electrophoresis (Figure 2, bottom). Figure 2 shows a side-by-side comparison of the modified and the original ARMS-PCR in detection of the JAK2 mutation in two patient samples. The intensities of mutant products in both cases were significantly enhanced by the modified procedure. In patient 1, a band from the mutant allele can be seen when the patient DNA was diluted up to 1000-fold. In patient 2, the mutant allele was barely detectable in an undiluted DNA specimen by the original procedure but yielded a strong band that can be visualized even after 100-fold dilution. Taken together, the modifications enhanced the analytic sensitivity by approximately two orders of magnitude.
Figure 2.
The modified ARMS-PCR has a higher sensitivity than the original method. DNA samples from two patients were diluted with DNA from peripheral blood mononuclear cell at ratios of 1:10, 1:100, and 1:1000. The positions of the DNA control, mutant, and normal alleles are indicated. MW, 100-bp molecular weight markers.
We verified modified ARMS-PCR results by the pyrosequencing technique in 18 clinical specimens in a blinded manner. As shown in Table 2, the modified ARMS obtained results in 100% concordance with those obtained by pyrosequencing. Note that the lower detection limit of pyrosequencing quantitative assessment is 5%. Any peaks of the mutant T allele that were less than 5% were considered background noises rather than signals and were thus interpreted as normal.
Table 2.
Validation of the Modified ARMS-PCR Assay by Pyrosequencing
| Specimens | Modified ARMS | Pyrosequencing (% of mutant T allele*) |
|---|---|---|
| 1 | Normal | 0 |
| 2 | Normal | 0 |
| 3 | Normal | 4.3 |
| 4 | Mutant only | 95 |
| 5 | Normal | 2.6 |
| 6 | Normal | 2.0 |
| 7 | Normal | 2.1 |
| 8 | Mutant + normal | 23.7 |
| 9 | Mutant + normal | 34.3 |
| 10 | Mutant + normal | 39.8 |
| 11 | Mutant + normal | 58.5 |
| 12 | Normal | 1.0 |
| 13 | Normal | 0.9 |
| 14 | Mutant + normal | 70.1 |
| 15 | Mutant + normal | 27.7 |
| 16 | Normal | 1.6 |
| 17 | Normal | 1.0 |
| 18 | Mutant + normal | 77.6 |
Less than 5% of T by pyrosequencing is scored as normal.
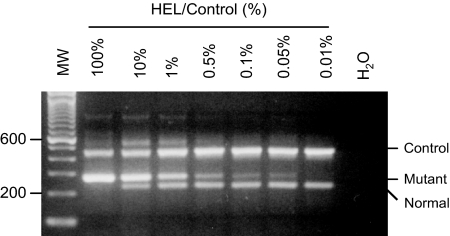
To determine the analytic sensitivity of the assay, the HEL cell line that harbors homozygous V617F was serially diluted in peripheral blood mononuclear cells from a normal individual. As shown in Figure 3, a sensitivity of 0.05% was reached. To determine whether such sensitivity can be routinely reproduced, multiple runs were performed by several operators. Fresh serial dilutions were made and assayed in each run. Table 3 shows 0.1% was achieved in all 14 runs by four different operators, and 0.05% was achieved in 9 of 10 independent runs and 0.025% in four of eight independent runs. We conclude that the modified ARMS-PCR is highly sensitive and highly reproducible.
Figure 3.
Analytic sensitivity of the ARMS-PCR assay. Genomic DNA from HEL cells was diluted with DNA from human peripheral blood mononuclear cell at indicated percentages. The positions of the DNA control, mutant, and normal alleles are indicated. MW, 100-bp molecular weight markers.
Table 3.
Sensitivity and Reproducibility of the ARMS-PCR Assay
| Staff member | HEL/control (%)
|
|||
|---|---|---|---|---|
| 0.1% | 0.05% | 0.025% | 0.0125% | |
| A | 6 of 6* | 1 of 2 | 0 of 2 | 0 of 2 |
| B | 2 of 2 | 2 of 2 | 1 of 2 | 0 of 2 |
| C | 2 of 2 | 2 of 2 | 2 of 2 | 1 of 2 |
| D | 4 of 4 | 4 of 4 | 1 of 2 | 0 of 2 |
| Total | 14 of 14 | 9 of 10 | 4 of 8 | 1 of 8 |
x of y means that out of y number of test runs, x numbers of runs reached sensitivity indicated.
To ensure that the increased sensitivity does not give rise to false-positive results, samples from patients with nonmyeloproliferative disorders were tested for the JAK2 mutation. Of 117 DNA specimens, 116 were normal and one carried the V617F mutation, suggesting the diagnostic specificity of the assay is greater than 99%. At the present time, the significance of this positive finding in a patient who did not have MPD is unclear. On one hand, it is possible that the patient had MPD in its early stage that had not yet been detected by any other clinical or laboratory evaluations. The vast majority of the literature does not report finding the V617F mutation in normal individuals or patients with reactive cytosis. However, given the sensitive nature of our ARMS-PCR, it would not be surprising that cases of early MPD with a small evolving clone of tumor cells are revealed. On the other hand, it is equally possible that the result is false because of the high sensitivity of the ARMS-PCR assay. Indeed, low levels of the mutant allele have been identified in 5 of 52 normal volunteers by an extremely sensitive assay that combines locked nucleic acid with molecular beacon technology. With its ability to enrich the PCR products of the mutant allele, the method carries a sensitivity of 0.01%.15 Apparently, the molecular beacon assay might be overly sensitive because it is unlikely that 10% of the population will eventually develop MPD. MPDs are considered rare diseases (a few in 100,000), and epidemiology studies are currently ongoing to determine the actual incidence and prevalence of MPDs in the United States. Nevertheless, our ARMS-PCR assay does not seem to give rise to unusually high false-positive rate. With a sensitivity of 0.05 to 0.1%, the diagnostic specificity remains greater than 99%, which is generally acceptable for a clinical test.
In summary, by changing the PCR reaction conditions, we developed an ARMS-PCR assay for clinical testing of the JAK2 V617F mutation. During the preparation of this article, Vannucchi and colleagues16 published a report describing a JAK2 ARMS-RT-PCR assay with a sensitivity of ∼1%. The RT-PCR is coupled with capillary electrophoresis and is able to determine normal and mutant allele in a relatively quantitative manner. However, the method has yet to be validated against an independent direct quantitative method such as pyrosequencing. In addition, the analyte is RNA, which is much more labile than DNA, and the step of reverse transcription is required to convert RNA to cDNA. Currently, without a clearly demonstrated clinical utility of V617F levels, the DNA-based assay described here is probably more cost efficient in a clinical setting. We show that the test reaches a high sensitivity in the range of 0.05 to 0.1%, which is among the highest reported in the literature.17 The high sensitivity is beneficial in detecting the JAK2 mutation in patients with a small proportion of the malignant cells as we have shown in Figure 2. Further, we have demonstrated that this high sensitivity is not accompanied by any significant compromise in the diagnostic specificity of the assay. In addition, the ARMS-PCR assay shows different patterns for normal, mutant, or mixed genotypes, minimizing the chance of misinterpretation. With simply one tube of PCR reactions and obviation for special equipment and reagents, ARMS-PCR represents a time- and labor-saving as well as cost-efficient method that is readily and broadly applicable in clinical molecular diagnostic laboratories.
References
- Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, Scott MA, Erber WN, Green AR. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, Garcon L, Raslova H, Berger R, Bennaceur-Griscelli A, Villeval JL, Constantinescu SN, Casadevall N, Vainchenker W. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- Jones AV, Kreil S, Zoi K, Waghorn K, Curtis C, Zhang L, Score J, Seear R, Chase AJ, Grand FH, White H, Zoi C, Loukopoulos D, Terpos E, Vervessou EC, Schultheis B, Emig M, Ernst T, Lengfelder E, Hehlmann R, Hochhaus A, Oscier D, Silver RT, Reiter A, Cross NC. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood. 2005;106:2162–2168. doi: 10.1182/blood-2005-03-1320. [DOI] [PubMed] [Google Scholar]
- Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, Adelsperger J, Koo S, Lee JC, Gabriel S, Mercher T, D’Andrea A, Frohling S, Dohner K, Marynen P, Vandenberghe P, Mesa RA, Tefferi A, Griffin JD, Eck MJ, Sellers WR, Meyerson M, Golub TR, Lee SJ, Gilliland DG. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Vainchenker W, Constantinescu SN. A unique activating mutation in JAK2 (V617F) is at the origin of polycythemia vera and allows a new classification of myeloproliferative diseases. Hematology (Am Soc Hematol Educ Program) 2005:195–200. doi: 10.1182/asheducation-2005.1.195. [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Scott LM, Buck G, Wheatley K, East CL, Marsden JT, Duffy A, Boyd EM, Bench AJ, Scott MA, Vassiliou GS, Milligan DW, Smith SR, Erber WN, Bareford D, Wilkins BS, Reilly JT, Harrison CN, Green AR. Definition of subtypes of essential thrombocythaemia and relation to polycythaemia vera based on JAK2 V617F mutation status: a prospective study. Lancet. 2005;366:1945–1953. doi: 10.1016/S0140-6736(05)67785-9. [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Griesshammer M, Dohner K, Dohner H, Kusec R, Hasselbalch HC, Larsen TS, Pallisgaard N, Giraudier S, Le Bousse-Kerdiles MC, Desterke C, Guerton B, Dupriez B, Bordessoule D, Fenaux P, Kiladjian JJ, Viallard JF, Briere J, Harrison CN, Green AR, Reilly JT. V617F mutation in JAK2 is associated with poorer survival in idiopathic myelofibrosis. Blood. 2006;107:2098–2100. doi: 10.1182/blood-2005-08-3395. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Pardanani A. Mutation screening for JAK2V617F: when to order the test and how to interpret the results. Leuk Res. 2006;30:739–744. doi: 10.1016/j.leukres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- James C, Delhommeau F, Marzac C, Teyssandier I, Couedic JP, Giraudier S, Roy L, Saulnier P, Lacroix L, Maury S, Tulliez M, Vainchenker W, Ugo V, Casadevall N. Detection of JAK2 V617F as a first intention diagnostic test for erythrocytosis. Leukemia. 2006;20:350–353. doi: 10.1038/sj.leu.2404069. [DOI] [PubMed] [Google Scholar]
- McClure R, Mai M, Lasho T. Validation of two clinically useful assays for evaluation of JAK2 V617F mutation in chronic myeloproliferative disorders. Leukemia. 2006;20:168–171. doi: 10.1038/sj.leu.2404007. [DOI] [PubMed] [Google Scholar]
- Jelinek J, Oki Y, Gharibyan V, Bueso-Ramos C, Prchal JT, Verstovsek S, Beran M, Estey E, Kantarjian HM, Issa JP. JAK2 mutation 1849G>T is rare in acute leukemias but can be found in CMML, Philadelphia chromosome-negative CML, and megakaryocytic leukemia. Blood. 2005;106:3370–3373. doi: 10.1182/blood-2005-05-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensma DP. JAK2 V617F in myeloid disorders: molecular diagnostic techniques and their clinical utility: a paper from the 2005 William Beaumont Hospital Symposium on Molecular Pathology. J Mol Diagn. 2006;8:397–411. doi: 10.2353/jmoldx.2006.060007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AV, Silver RT, Waghorn K, Curtis C, Kreil S, Zoi K, Hochhaus A, Oscier D, Metzgeroth G, Lengfelder E, Reiter A, Chase AJ, Cross NC. Minimal molecular response in polycythemia vera patients treated with imatinib or interferon alpha. Blood. 2006;107:3339–3341. doi: 10.1182/blood-2005-09-3917. [DOI] [PubMed] [Google Scholar]
- Sidon P, El Housni H, Dessars B, Heimann P. The JAK2V617F mutation is detectable at very low level in peripheral blood of healthy donors. Leukemia. 2006;20:1622. doi: 10.1038/sj.leu.2404292. [DOI] [PubMed] [Google Scholar]
- Vannucchi AM, Pancrazzi A, Bogani C, Antonioli E, Guglielmelli P. A quantitative assay for JAK2(V617F) mutation in myeloproliferative disorders by ARMS-PCR and capillary electrophoresis. Leukemia. 2006;20:1055–1060. doi: 10.1038/sj.leu.2404209. [DOI] [PubMed] [Google Scholar]
- Greiner TC. Diagnostic assays for the JAK2 V617F mutation in chronic myeloproliferative disorders. Am J Clin Pathol. 2006;125:651–653. doi: 10.1309/NXXT-GRCX-D0TM-A3C2. [DOI] [PubMed] [Google Scholar]