Abstract
Gastrointestinal stromal tumors (GISTs) frequently harbor mutations in the KIT and PDGFRA genes, the presence and type of which correlate with the response to the kinase inhibitor imatinib mesylate. Because most GIST mutations are deletions/insertions, we used a microfluidic apparatus to detect these size variations in polymerase chain reaction-amplified DNA. This approach, termed microfluidic deletion/insertion analysis (MIDIA), identified mutations in 30 of 50 DNA samples from paraffin-embedded CD117-positive GISTs (60%), comprising 25 deletions and five insertions. Sequencing of 14 MIDIA-positive samples confirmed the deletions/insertions, including two 3-bp alterations. Sequencing of all 20 MIDIA-negative samples also showed highly consistent results with MIDIA because 10 cases were wild type and eight displayed a single base substitution in which detection by MIDIA was not expected. Sequencing also revealed a 3-bp deletion undetected by MIDIA, thus establishing the resolution limit of MIDIA at deletions/insertions ≥3 bp. Denaturing high-pressure liquid chromatography analysis confirmed all mutations detected by MIDIA and sequencing. We propose MIDIA as the first step in mutational screening of GIST because it allowed the detection of 75% of mutated cases (94% of deletions/insertions) in less than 30 minutes after polymerase chain reaction amplification and at a lower cost compared with denaturing high-pressure liquid chromatography and sequencing, which might then be used only for MIDIA-negative cases.
Gastrointestinal stromal tumors (GISTs) are infrequent neoplasms1,2 in which the putative cell of origin is the interstitial cell of Cajal.3,4 They typically show immunohistochemical positivity for CD34 and KIT (CD117).5 KIT is a 145-kd transmembrane tyrosine kinase receptor, whose ligand is the stem cell factor. In the vast majority of GISTs, KIT protein shows abnormal functional properties attributable to gene mutations6 that sustain tumor growth. PDGFRA mutations are an alternative mechanism of GIST tumorigenesis.7,8 KIT and PDGFRA are the molecular targets of imatinib mesylate,9 a low-toxicity compound that inhibits their kinase activity and has been approved for clinical use in unresectable, recurrent, or metastatic GISTs. Interestingly, the type of mutation also predicts response to this molecular therapy.9
In order of frequency, KIT mutations in GIST cluster in exons 11,6 9,10 13,10 and 17,11 whereas those in PDGFRA are found in exons 18 and 12.7,8 The relative frequency of different mutations varies among different reports, but there is consensus that KIT exon 11 is the most frequently involved because approximately two-thirds of GISTs harbor sequence abnormalities in this DNA segment.12 Deletions and insertions account for the large majority of these abnormalities.12
Various methods have been used for mutational detection in GISTs, including direct polymerase chain reaction (PCR) sequencing, PCR-single strand conformation polymorphism,13 length analysis of polymerase chain reaction products,14 and denaturing high-pressure liquid chromatography (DHPLC).15 Direct PCR sequencing is by far the most widely used and is a relatively high-cost technique. PCR-single strand conformation polymorphism may have higher sensitivity, although its discriminative capacity is affected by several variables that render optimization rather difficult. These include gel composition and thickness, electrophoresis conditions and temperature, and type of mutation in relation to the sequence context. Length analysis of polymerase chain reaction products has a good sensitivity for deletions/insertions but requires the use of labeled primers and an automated capillary electrophoresis apparatus, both increasing the expense of the technique. DHPLC is possibly the most efficient mutation screening method; however, it requires high-cost instrumentation and experienced operators because its sensitivity is affected by sequence context and temperature, often requiring repeated analyses at different temperatures for the screening of sequences carrying different types of mutations.
Because most kinase mutations in GISTs are size-altering changes, we considered using the accurate length discrimination of microfluidic chip-based electrophoresis as an initial screening approach, which we termed microfluidic deletion/insertion analysis (MIDIA). This may be performed using a novel and relatively low-cost microfluidic apparatus (Agilent 2100 Bioanalyzer and DNA500 chip; Agilent Technologies, Waldbronn, Germany) that reduces the separation time and sample consumption compared with gel electrophoresis. The chip accommodates 12 sample wells and a well for an external standard (sizing ladder). Glass microchannels create interconnected networks among these wells. Once the wells and channels are filled with a sieving polymer and fluorescence dye, the chip becomes an integrated electrical circuit. DNA is electrophoretically driven by a voltage gradient, dye molecules intercalate into DNA, and these complexes are detected by laser-induced fluorescence. The chip preparation and loading takes only a few minutes and the electrophoretic run of 12 samples and size ladder is completed in less than 30 minutes. Sizing and quantitative information are then converted into gel-like images (bands) and electropherograms (peaks), interpretation of which requires little experience.16
Materials and Methods
Study Cases
Fifty formalin-fixed, paraffin-embedded CD117-positive GISTs from the files of the Department of Pathology, University Hospital of Verona, Italy, were included in the study (Table 1). These included 36 spindle, eight epithelioid, and six mixed GISTs. The primary tumor site was stomach (n = 23), duodenum (n = 8), small intestine (n = 17), colon (n = 1), and rectum (n = 1). Three patients (SP33, SP34, SP35) were affected by a neurofibromatosis type 1 syndrome. Eleven samples of nonneoplastic gastric mucosa were used as controls.
Table 1.
Gastrointestinal Stromal Tumor Series and Results of MIDIA and Sequencing Analysis
| Case | Site | Histology | Prognostic category* | Gene | MIDIA† | Sequencing‡ |
|---|---|---|---|---|---|---|
| SP1 | Stomach | Spindle | Probably benign | KIT exon 11 | DEL | DEL D579 |
| SP2 | Stomach | Spindle | Probably benign | KIT exon 11 | DEL | Not done |
| SP3 | Stomach | Spindle | Probably benign | KIT exon 11 | DEL | Not done |
| SP4 | Stomach | Spindle | Probably benign | KIT exon 11 | DEL | Not done |
| SP5 | Stomach | Spindle | Uncertain or LMP | KIT exon 11 | DEL | DEL Q556_V559 |
| SP6 | Stomach | Spindle | Uncertain or LMP | KIT exon 11 | DEL | Not done |
| SP7 | Stomach | Spindle | Uncertain or LMP | KIT exon 11 | DEL | DEL K550_P551 |
| SP8 | Stomach | Spindle | Probably malignant | KIT exon 11 | DEL | Not done |
| SP9 | Stomach | Spindle | Probably malignant | KIT exon 11 | DEL | Not done |
| SP10 | Stomach | Spindle | Probably malignant | KIT exon 11 | DEL | Not done |
| SP11 | Stomach | Spindle | Malignant | KIT exon 11 | DEL | DEL K558_E562 |
| SP12 | Stomach | Spindle | Malignant | KIT exon 11 | DEL | DEL K550_K558 |
| SP13 | Duodenum | Spindle | Probably malignant | KIT exon 11 | DEL | Not done |
| SP14 | Small intestine | Spindle | Probably malignant | KIT exon 11 | DEL | DEL W557_N564 |
| SP15 | Small intestine | Spindle | Probably malignant | KIT exon 11 | DEL | K550L, DEL P551_E554 |
| SP16 | Small intestine | Spindle | Malignant | KIT exon 11 | DEL | Not done |
| SP17 | Small intestine | Spindle | Probably malignant | KIT exon 11 | DEL | Not done |
| SP18 | Rectum | Spindle | Probably malignant | KIT exon 11 | DEL | Not done |
| SP19 | Liver¶ | Spindle | Malignant | KIT exon 11 | DEL | Not done |
| SP20 | Omentum¶ | Spindle | Malignant | KIT exon 11 | DEL | Not done |
| SP21 | Liver¶ | Spindle | Malignant | KIT exon 11 | DEL | Not done |
| SP22 | Peritoneum¶ | Spindle | Malignant | KIT exon 11 | DEL | DEL V555_V559 |
| SP23 | Stomach | Spindle | Probably benign | KIT exon 11 | WT | W557R |
| SP24 | Stomach | Spindle | Uncertain or LMP | KIT exon 11 | WT | V560E |
| SP25 | Stomach | Spindle | Probably malignant | KIT exon 11 | WT | K558I, DEL V559 |
| SP26 | Small intestine | Spindle | Probably malignant | KIT exon 11 | WT | V559D |
| SP27 | Small intestine | Spindle | Probably malignant | KIT exon 11 | WT | K558I, DEL V559 |
| SP28 | Small intestine¶ | Spindle | Malignant | KIT exon 11 | WT | L576P |
| SP29 | Omentum¶ | Spindle | Malignant | KIT exon 11 | INS | INS P559 |
| SP30 | Duodenum | Spindle | Probably malignant | KIT exon 9 | INS | DUPL A502-Y503 |
| SP31 | Small intestine | Spindle | Uncertain or LMP | KIT and PDGFRA | WT | WT |
| SP32 | Stomach | Spindle | Probably malignant | KIT and PDGFRA | WT | WT |
| SP33§ | Peritoneum¶ | Spindle | Malignant | KIT and PDGFRA | WT | WT |
| SP34§ | Small intestine | Spindle | Uncertain or LMP | KIT and PDGFRA | WT | WT |
| SP35§ | Small intestine | Spindle | Probably malignant | KIT and PDGFRA | WT | WT |
| SP36 | Small intestine | Spindle | Probably malignant | KIT and PDGFRA | WT | WT |
| EP1 | Duodenum | Epithelioid | Probably malignant | KIT exon 11 | INS | INS P559 |
| EP2 | Stomach|| | Epithelioid | Malignant | KIT exon 11 | WT | V559D |
| EP3 | Duodenum | Epithelioid | Uncertain or LMP | KIT exon 9 | INS | DUPL A502-Y503 |
| EP4 | Stomach|| | Epithelioid | Malignant | PDGFRA exon 18 | WT | D842V |
| EP5 | Stomach | Epithelioid | Probably malignant | PDGFRA exon 12 | DEL | DEL S566_E571, E571R |
| EP6 | Duodenum|| | Epithelioid | Malignant | KIT exon 11 | DEL | Not done |
| EP7 | Stomach | Epithelioid | Probably malignant | KIT and PDGFRA | WT | WT |
| EP8 | Stomach | Epithelioid | Probably malignant | KIT and PDGFRA | WT | WT |
| MIX1 | Small intestine | Mixed | Probably malignant | KIT exon 11 | DEL | Not done |
| MIX2 | Small intestine | Mixed | Probably benign | KIT exon 11 | WT | V559D |
| MIX3 | Stomach | Mixed | Uncertain or LMP | KIT exon 11 | WT | V559D |
| MIX4 | Duodenum | Mixed | Probably malignant | KIT exon 9 | INS | DUPL A502-Y503 |
| MIX5 | Colon|| | Mixed | Malignant | KIT and PDGFRA | WT | WT |
| MIX6 | Small intestine | Mixed | Probably malignant | KIT and PDGFRA | WT | WT |
Prognostic category according to Miettinen et al.25
MIDIA, microfluidic deletion/insertion analysis.
Mutations were numbered according to GenBank sequences X06182 for KIT and NM006206 for PDGFRA.
SP33, SP34, and SP35 are from patients affected by neurofibromatosis type 1 syndrome.
Relapse or metastasis originating from: SP19, small intestine synchronous; SP20, small intestine 2 years before; SP21, small intestine 6 years before; SP22, stomach 20 years before; SP28, duodenum 4 years before; SP29, small intestine synchronous; SP33, duodenum synchronous.
Cases with metastatic disease to peritoneum and/or omentum.
Control of Tumor Cell Content and Nucleic Acid Purification
DNA was extracted from five 5-μm paraffin sections after microscopic examination of a section contiguous to that used. Manual microdissection was applied in selected cases to assure the prevalence of neoplastic over normal cells.17 Tissue sections were deparaffinized and digested with proteinase K at a final concentration of 1 μg/μl, in a total volume of 400 μl of the same buffer used for PCR reactions, for 2 hours at 55°C or overnight at 37°C. The concentration of DNA was assessed with a Qubit fluorometer (Invitrogen, Eugene, OR), and 100 ng of DNA (generally corresponding to 1.5 to 2 μl of crude lysate) were used for PCR amplification. For case SP12, RNA was extracted from frozen tissue using RNAeasy spin columns (Qiagen, Hilden, Germany). cDNA was synthesized using AMV reverse-transcriptase and random hexamer primers (First Strand cDNA synthesis kit; Roche, Mannheim, Germany).
Primers and PCR Amplification of KIT and PDGFR
The primers used for KIT and PDGFRA amplification were designed using specific software tools (DNAsis MAX from Hitachisoft, Hamburg, Germany or Primer3 from http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi/), and their sequences are reported in Table 2. These primers were chosen among a set of several different primer pairs that were tested because they allowed the efficient amplification of the entire coding region for each exon using DNA from paraffin-embedded tissues. In particular, the 246-bp exon 11 amplimer includes the entire exon and the 3′ end of intron 10, in which mutations have been described. The reaction was performed in a final volume of 50 μl, containing 1× HotMaster buffer (Eppendorf-5 Prime, Inc., Boulder, CO), 5 pmol of each primer, 200 μmol/L each dNTP, and 1.25 U of HotMaster Taq polymerase (Eppendorf). Cycling conditions were 90 seconds at 94°C, followed by 40 cycles of 30 seconds at 94°C, 20 seconds at 55°C, 20 seconds at 65°C; followed by a final elongation of 5 minutes at 65°C.
Table 2.
Primers Used for PCR Amplification of the KIT and PDGFRA Genes
| Name | Forward primer | Reverse primer | Amplicon length |
|---|---|---|---|
| KIT exon 9 | 5′-TTCCTAGAGTAAGCCAGGGC-3′ | 5′-CCTAAACATCCCCTTAAATTGG-3′ | 273 bp |
| KIT exon 11 | 5′-TGTTCTCTCTCCAGAGTGCTCTAA-3′ | 5′-ACCCAAAAAGGTGACATGGA-3′ | 246 bp |
| KIT exon 13 | 5′-TGCCAGTTGTGCTTTTTGCTA-3′ | 5′-GCTTTACCTCCAATGGTGCAG-3′ | 161 bp |
| KIT exon 17 | 5′-ATGGTTTTCTTTTCTCCTCC-3′ | 5′-TACATTATGAAAGTCACAGG-3′ | 243 bp |
| PDGFR exon 12 | 5′-CTCTGGTGCACTGGGACTTT-3′ | 5′-GGAGGTTACCCCATGGAACT-3′ | 212 bp |
| PDGFR exon 14 | 5′-GAGAACAGGAAGATGGTAGCTCA-3′ | 5′-TTCACAACCACATGTGTCCA-3′ | 230 bp |
| PDGFR exon 18 | 5′-CATTTCTTCCTTTTCCATGCA-3′ | 5′-TGTGGGAAGTGTGGACGTAC-3′ | 165 bp |
MIDIA
One μl of each PCR sample was analyzed in the microfluidic apparatus (Agilent 2100 Bioanalyzer) using the DNA 500 chip (Agilent Technologies) following the manufacturer’s instructions. Data were analyzed using the bundle software package (2100 Expert) that automatically provides peak sizing and quantification. A wild-type sample was run in parallel as a control. The presence of an additional peak with lower or higher molecular weight was considered as a deletion or insertion, respectively.
DNA Sequencing
Sequence analysis was done on all MIDIA-negative samples and in a subset of 14 MIDIA-positive samples. PCR products were purified by centrifugal devices (Centricon 100; Millipore, Bedford, MA) and reamplified using dye-terminator chemistry (BigDye v3.1; Applied Biosystems, Foster City, CA) and the same forward and reverse primers used for PCR amplification. Samples were run on a capillary automated sequencer (310 or 3130xl; Applied Biosystems) and analyzed by specific DNA-analysis software (DNAsis MAX; Hitachisoft, Tokyo, Japan).
KIT Reverse Transcriptase (RT)-PCR and Sequencing
In the only sample with a 5′ deletion in KIT exon 11 for which frozen tissue was available (SP12), a 1300-bp fragment of KIT cDNA spanning exons 8 to 17 was amplified using primers KIT exon 8F, 5′-GACGTCAATGCTGCCATAGCAT-3′, and KIT exon 17R, 5′-AGTCGAGCGTTTCCTTTAACCA-3′. The cycling conditions were 2 minutes at 94°C, 38 cycles of 15 seconds at 94°C, 30 seconds at 56°C, 1 minute at 65°C; followed by a terminal elongation of 7 minutes at 65°C. The primer used for sequencing KIT exon 11 in this case was 5′-CCACACCCTGTTCACTCCTT-3′.
DHPLC
Mutational analysis using DHPLC was performed for KIT exons 9 and 11 and for PDGFRA exons 12 and 18, which were PCR-amplified with the same sets of primers used for MIDIA. A positive control and negative control (water) were included in each assay. Amplicons were then subjected to denaturation at 95°C for 3 minutes, followed by tapered cooling at 1°C/minute decrements to a final temperature of 25°C. This treatment allows the formation of mismatched double strands whenever a mutated sequence is present along with the wild type. Thirty to 60 μg of PCR product was injected into a Wave nucleic acid fragment analysis system (Transgenomic, La Jolla, CA) containing a DNA-Sep column composed of alkylated nonporous (styrene divinyl benzene) particles. The melting temperatures, estimated using the Wave Maker software (Transgenomic), for optimal resolution were 58°C for KIT exon 11, 60°C for KIT exon 9 and PDGFRA exon 12, and 62°C for PDGFRA exon 18.
Results
The results of the mutational analysis of 50 GISTs are summarized in Table 1. Forty cases (80%) showed a mutation in KIT or PDGFRA detected by either MIDIA or direct PCR sequencing. Mutations were detected in 83% (30 of 36) of spindle, 75% (six of eight) of epithelioid, and 67% (four of six) of mixed variants of GIST. Deletions and insertions accounted for 80% of all mutations (32 of 40). Mutations showed a bias for known hot spots, with 35 mutations involving KIT exon 11 (87.5%), three KIT exon 9 (7.5%), one PDGFRA exon 12 (2.5%), and one PDGFRA exon 18 (2.5%). Notably, 3 of the 10 samples showing wild-type KIT and PDGFRA sequences were from patients affected by neurofibromatosis 1 syndrome (cases SP33, SP34, and SP35 in Table 1), which was an expected result.18,19,20
MIDIA Recognized 75% of Mutated Cases (94% of Deletions/Insertions)
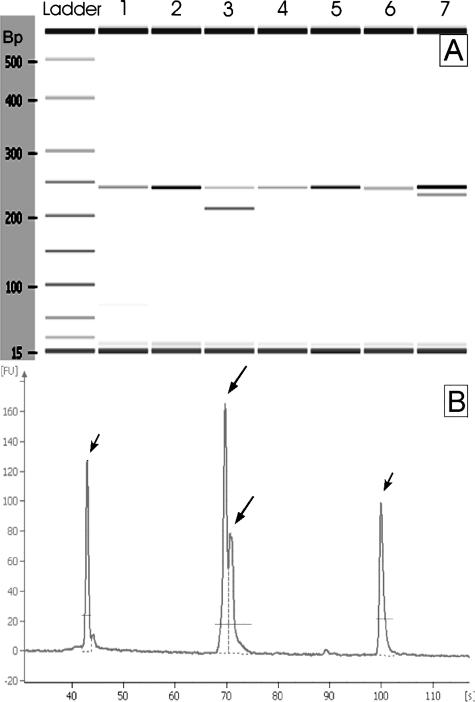
MIDIA identified deletions/insertions in 30 of the 50 cases (60%) (Figure 1). These represented 75% of all mutations and 94% of the deletions/insertions. In particular, the 30 mutations detected by MIDIA included 25 cases (50%) that showed an additional lower molecular weight peak, consistent with a deletion (24 cases in KIT exon 11, and one in PDGFRA exon 12), and five cases (10%) that showed an additional higher molecular weight peak, consistent with an insertion (two in KIT exon 11 and three in KIT exon 9). All of the deletions/insertions were heterozygous mutations, with the exception of case SP11 that had a hemi/homozygous deletion, as assessed by the presence of only one peak shorter than expected. Twenty cases (40%) only showed the wild-type peak by MIDIA analysis. Moreover, 11 DNAs from normal gastric mucosa used as negative controls also showed only the wild-type peak at MIDIA analysis.
Figure 1.
Example of MIDIA analysis. A: Pseudo-gel image, samples 3 and 7 harbor a KIT deletion. B: Electropherogram of the smallest KIT alteration detected by MIDIA, a 3-bp insertion of sample 25. Short arrows: 15-bp and 600-bp markers; long arrows: PCR product peaks.
Control of False-Positive and -Negative Results
To check for false-positive results, a subset of 14 MIDIA-positive samples were sequenced. None of these resulted in wild type, whereas all showed an alteration fully consistent with MIDIA results (Table 1). To check for false-negative results, all 20 GIST samples that were wild type by MIDIA were sequenced. Of these, eight showed a single base substitution (Table 1), whose detection by MIDIA analysis was not expected. However, two samples showed the same 3-bp deletion (K558I-DEL V559, cases SP25 and SP27) that was not detected by the MIDIA analysis. In summary, MIDIA detected all insertions/deletions larger than 3 bp and two of three size-altering mutations of 3 bp, thus establishing the resolution limit of MIDIA at deletions/insertions ≥3 bp.
Sensitivity of MIDIA
The sensitivity of MIDIA was assessed by dilution experiments using DNA from case SP11 harboring a homozygous deletion in KIT exon 11 and DNA from a wild-type sample. The two samples were mixed so that the percentage of DNA from neoplastic cells was 1, 5, 10, 20, 35, and 50%. The samples were amplified for KIT exon 11, and PCR products were run on a chip. A shifted band was clearly detected when mutated DNA accounted for 10% of total DNA.
Primers for Exon 11 Analysis Identified a New KIT Mutation
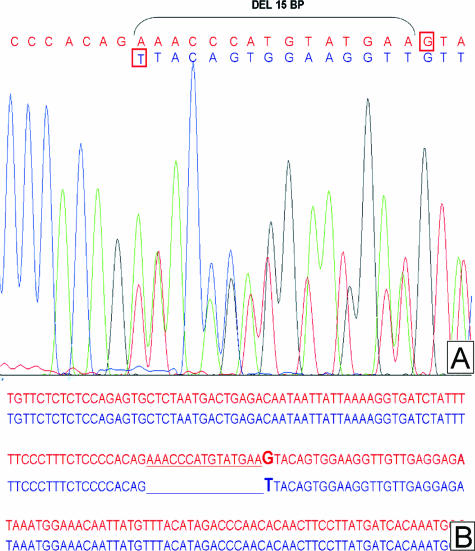
Because KIT mutations at the boundary between intron 10 and exon 11 have been described, particular care was taken to design primers for the efficient analysis of the entire exon and its 5′ adjacent intron 10 sequence. The chosen 246-bp amplimer allowed the MIDIA detection of two such mutations. In fact, sequencing analysis of these cases showed that the deletion was localized at the KIT intron 10-exon 11 boundary. The first case (SP12) showed a deletion of the most 3′ 5 bp of intron 10 and 25 bp of exon 11; cDNA sequencing of this case showed a 27-bp deletion in the coding sequence, consistent with a splicing aberration, as previously reported.12,21,22 The second case (SP15) presented a 15-bp deletion of the most 5′ region of exon 11 together with a G to T mutation of the first nondeleted base (Figure 2). To our knowledge, this is the first report of this type of mutation in KIT.
Figure 2.
A: Chromatogram view of the new combined deletion and point mutation at 5′ end of KIT exon 11. Wild-type sequence in red, mutated sequence in blue; G to T point mutation squared in red. B: Sequence alignment of wild-type (red) and mutated (blue) sequences.
Mutational Analysis by DHPLC
The results of DHPLC analysis of KIT exons 9 and 11 and PDGFRA exons 12 and 18 matched those obtained by MIDIA for deletion/insertion changes, including the newly detected deletion; all point mutations detected by sequencing were also confirmed.
Cost and Time Assessment
A direct comparison of the costs and labor involved in direct sequencing, DHPLC and MIDIA is reported in Table 3. This analysis showed that MIDIA is the fastest and most inexpensive approach for the screening of size-altering mutations. The results from MIDIA are obtained in 18 minutes, whereas sequencing analysis and DHPLC require ∼5 hours and 3 hours, respectively. The cost of the chip-based analysis is lower than that of DHPLC; the cost of the former is estimated at 1.5 €/sample and that of the latter at 2.4 €/sample. However, because DHPLC is a high-throughput device, it will incur higher costs in the analysis of single cases; moreover, repeated analyses at more than one temperature are also necessary to avoid false-negative results. The cost of direct sequencing is estimated at ∼13 €/sample, which is approximately an order of magnitude higher than that of MIDIA.
Table 3.
Comparison of Cost and Time Requirements of MIDIA, PCR Sequencing, and DHPLC Analysis (per Sample)
| PCR sequencing | MIDIA | DHPLC | |
|---|---|---|---|
| Cost analysis | |||
| Sample preparation | 6.63 €* | 0 € | 0 € |
| Instrument consumables | 4 € | 1.2 € | 1.83 € |
| Plastic material | 2 € | 0.30 € | 0.60 € |
| Total cost | 13 € | 1.5 € | 2.4 € |
| Time requirements | |||
| Sample/instrument preparation | 4 hours* | 13 minutes | 15 minutes |
| Run time | 1 hour | 2.5 minutes | 2 hours, 7 minutes |
| Results analysis | 30 minutes | 2 minutes | 5 minutes |
| Total time | 5 hours | 18 minutes | 2 hours, 45 minutes |
PCR purification, sequencing reaction, and purification of the same.
Discussion
Despite their relative rarity, there is great interest in GISTs because they represent a paradigm of successful molecular therapy. The basis of such therapy is the knowledge of the precise molecular alteration driving tumor cell growth. For GISTs this alteration is known, namely KIT tyrosine kinase-activating mutation6 or, more rarely, a similar abnormality of PDGFRA.7,8 Specific mutations correlate with the response to imatinib mesylate, which inhibits the kinase activity of both KIT and PDGFRA.9,23
Because deletions and insertions account for nearly 70% of all of the GIST kinase mutations,15 we assessed whether a chip-based microfluidic electrophoresis could be used as an initial screening methodology for the detection of size altering mutations in GISTs. We called this approach MIDIA. Our results show that MIDIA can reliably detect the vast majority of GIST-specific kinase mutations in less than 3 hours starting from PCR and with very little hands-on operator time. In fact, MIDIA identified size-altering mutations in 30 of 50 cases, representing 75% of all mutated cases and 94% of those harboring deletions/insertions. These latter included five insertions and 25 deletions, one of which was hemi/homozygous. Importantly, all of the mutations detected by MIDIA are among the good responders to kinase inhibition therapy. The mutations not recognized by MIDIA included eight point mutations and only one size alteration, which were detected by PCR sequencing of MIDIA-negative cases.
The discriminative power of MIDIA seems good because it was able to pick up differences as small as 3 bp between the two amplified alleles in two of three different alterations. Any deletion or insertion longer than 3 bp was always detected by MIDIA. Its sensitivity was also high because 94% of deletion/insertion mutations were detected, and dilution experiments assessed that a shifted band was clearly detected when mutated DNA accounted for 10% of total DNA. The specificity was excellent because no false-positive cases were detected, as assessed by direct PCR sequencing and DHPLC analysis.
MIDIA performed as efficiently as DHPLC in detecting deletion/insertion mutations. The high efficiency of DHPLC in the detection of sequence alterations was also seen for the analysis of KIT exons 9 and 11, and PDGFRA exons 12 and 18 in the present series of GISTs. At variance with MIDIA, DHPLC offers the advantage of discovering any sequence alteration, including point mutations, although requiring repeated analyses at more than one temperature to avoid false-negative reports. The interpretation of chromatograms also warrants expertise that is especially necessary when analyzing PCR products generated with DNA derived from formalin-fixed, paraffin-embedded tissues.
Because most KIT mutations cluster in exon 11 and alterations occurring at the boundary between intron 10 and exon 11 have been also described, particular care was taken to design primers for the efficient analysis of this region. The 246-bp amplimer used here was chosen after using different sets of primers amplifying either the entire region or overlapping segments of it. The 246-bp amplimer was the best performer because it allowed the detection of all deletions/insertions larger than 3 bp within both the exon itself and its 5′ intron 10 region. Moreover, chip electrophoresis using the same amplimer resolved two of three size-altering mutations involving 3 bp. The latter (K558I-DEL V559) remained undetected also using a set of shorter amplimers, the shortest being 112 bp.
The use of the above-mentioned amplimer permitted the detection of two deletions at the KIT intron 10-exon 11 boundary. The first was identical to cases 9 and 10 reported by Corless and colleagues22 and to case 1 by Chen and colleagues.21 The second showed a combined 15-bp deletion of the most 5′ region of exon 11 and a mutation of the first nondeleted nucleotide. To our knowledge, this is the first time a mutation of this kind has been reported for the KIT gene. This mutation differs from the other exon 11 5′ region deletions in that no part of intron 10 is deleted because the deletion starts from the first nucleotide in the exon. The G to T mutation following the deletion was somewhat unexpected because it disrupts the AG/GT consensus for intron/exon boundaries. However, only the AG is strictly conserved for both the major and minor classes of introns.24 The GTA to TTA mutation theoretically substitutes a valine residue with a leucine residue, but because frozen tissue was not available for this case, we could not assess the consequences of this mutation.
Comparison of the cost and labor required by MIDIA, PCR sequencing, and DHPLC suggests that MIDIA may be used as a first step in GIST mutation screening because it allowed the detection of 75% of mutated cases (94% of all deletions/insertions) in less than 30 minutes after PCR amplification and at a lower cost compared with DHPLC and PCR sequencing. These latter procedures may then be used for MIDIA-negative cases.
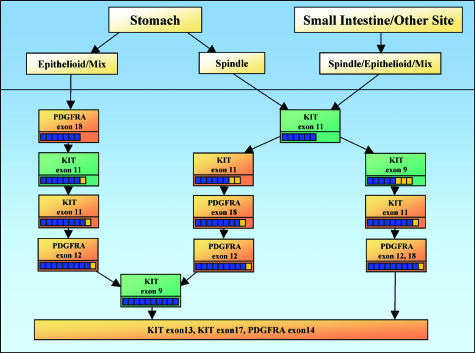
Considering that some authors propose using KIT mutations as a diagnostic criterion (apparently stronger than KIT immunostaining),12 a fast and inexpensive mutational screening approach would be of great value. Based on our results and on the published literature,25,26,27,28 we developed a flow chart that may optimize the process of mutational detection in GISTs. This includes the use of MIDIA and direct sequencing or DHPLC in an order driven by the site of origin of GIST and the morphological distinction between spindle and epithelioid variants (Figure 3). The workflow begins with MIDIA analysis of KIT exons 11 in all spindle-type GISTs of any location, whereas sequencing/DHPLC of PDGFRA exon 18 is warranted as first analysis for epithelioid/mixed type GISTs originating in the stomach.
Figure 3.
Proposed flow chart for mutation detection in GISTs. The suggested flow chart differs according to the site of origin and the histological variant of the GIST. Green boxes refer to MIDIA, orange boxes refer to PCR sequencing or DHPLC analysis. Small squares below the indicated analysis describe the percentages of the resolved cases at each step. For example, among the spindle GIST originating from the stomach, 60% of mutations are detected at the first step by KIT exon 11 MIDIA analysis, a further 20% is detected by KIT exon 11 PCR sequencing/DHPLC, and an additional 20% by PDGFRA exon 12 and 18 DHPLC/sequencing. The remaining mutations are detected by KIT exon 9 MIDIA and by further DHPLC/sequencing analysis as indicated.
Footnotes
Supported by the Fondazione Cassa di Risparmio di Verona (2005), Italy; the Ministero Università e Ricerca (Programmi di Ricerca di Interesse Nazionale 2005) and Ministero Salute, Rome, Italy; the European Community (grant PL018771); and the Associazione Italiana Ricerca Cancro, Milan, Italy (to A.S.).
References
- Nilsson B, Bumming P, Meis-Kindblom JM, Oden A, Dortok A, Gustavsson B, Sablinska K, Kindblom LG. Gastrointestinal stromal tumors: the incidence, prevalence, clinical course, and prognostication in the preimatinib mesylate era—a population-based study in western Sweden. Cancer. 2005;103:821–829. doi: 10.1002/cncr.20862. [DOI] [PubMed] [Google Scholar]
- DeMatteo RP, Lewis JJ, Leung D, Mudan SS, Woodruff JM, Brennan MF. Two hundred gastrointestinal stromal tumors: recurrence patterns and prognostic factors for survival. Ann Surg. 2000;231:51–58. doi: 10.1097/00000658-200001000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindblom LG, Remotti HE, Aldenborg F, Meis-Kindblom JM. Gastrointestinal pacemaker cell tumor (GIPACT): gastrointestinal stromal tumors show phenotypic characteristics of the interstitial cells of Cajal. Am J Pathol. 1998;152:1259–1269. [PMC free article] [PubMed] [Google Scholar]
- Tsujimura T, Makiishi-Shimobayashi C, Lundkvist J, Lendahl U, Nakasho K, Sugihara A, Iwasaki T, Mano M, Yamada N, Yamashita K, Toyosaka A, Terada N. Expression of the intermediate filament nestin in gastrointestinal stromal tumors and interstitial cells of Cajal. Am J Pathol. 2001;158:817–823. doi: 10.1016/S0002-9440(10)64029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blay JY, Bonvalot S, Casali P, Choi H, Debiec-Richter M, Dei Tos AP, Emile JF, Gronchi A, Hogendoorn PC, Joensuu H, Le Cesne A, McClure J, Maurel J, Nupponen N, Ray-Coquard I, Reichardt P, Sciot R, Stroobants S, van Glabbeke M, van Oosterom A, Demetri GD, GIST Consensus Meeting Panelists Consensus meeting for the management of gastrointestinal stromal tumors. Report of the GIST Consensus Conference of 20–21 March 2004, under the auspices of ESMO. Ann Oncol. 2005;16:566–578. doi: 10.1093/annonc/mdi127. [DOI] [PubMed] [Google Scholar]
- Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, Muhammad Tunio G, Matsuzawa Y, Kanakura Y, Shinomura Y, Kitamura Y. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A, Demetri GD, Fletcher CD, Fletcher JA. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- Hirota S, Ohashi A, Nishida T, Isozaki K, Kinoshita K, Shinomura Y, Kitamura Y. Gain-of-function mutations of platelet-derived growth factor receptor alpha gene in gastrointestinal stromal tumors. Gastroenterology. 2003;125:660–667. doi: 10.1016/s0016-5085(03)01046-1. [DOI] [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, McGreevey LS, Chen CJ, Van den Abbeele AD, Druker BJ, Kiese B, Eisenberg B, Roberts PJ, Singer S, Fletcher CD, Silberman S, Dimitrijevic S, Fletcher JA. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- Lux ML, Rubin BP, Biase TL, Chen CJ, Maclure T, Demetri G, Xiao S, Singer S, Fletcher CD, Fletcher JA. KIT extracellular and kinase domain mutations in gastrointestinal stromal tumors. Am J Pathol. 2000;156:791–795. doi: 10.1016/S0002-9440(10)64946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin BP, Singer S, Tsao C, Duensing A, Lux ML, Ruiz R, Hibbard MK, Chen CJ, Xiao S, Tuveson DA, Demetri GD, Fletcher CD, Fletcher JA. KIT activation is a ubiquitous feature of gastrointestinal stromal tumors. Cancer Res. 2001;61:8118–8121. [PubMed] [Google Scholar]
- Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal stromal tumors. J Clin Oncol. 2004;22:3813–3825. doi: 10.1200/JCO.2004.05.140. [DOI] [PubMed] [Google Scholar]
- Sakurai S, Fukasawa T, Chong JM, Tanaka A, Fukayama M. C-kit gene abnormalities in gastrointestinal stromal tumors (tumors of interstitial cells of Cajal). Jpn J Cancer Res. 1999;90:1321–1328. doi: 10.1111/j.1349-7006.1999.tb00715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emile JF, Lemoine A, Bienfait N, Terrier P, Azoulay D, Debuire B. Length analysis of polymerase chain reaction products: a sensitive and reliable technique for the detection of mutations in KIT exon 11 in gastrointestinal stromal tumors. Diagn Mol Pathol. 2002;11:107–112. doi: 10.1097/00019606-200206000-00007. [DOI] [PubMed] [Google Scholar]
- Corless CL, McGreevey L, Haley A, Town A, Heinrich MC. KIT mutations are common in incidental gastrointestinal stromal tumors one centimeter or less in size. Am J Pathol. 2002;160:1567–1572. doi: 10.1016/S0002-9440(10)61103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller O, Hahnenberger K, Dittmann M, Yee H, Dubrow R, Nagle R, Ilsley D. A microfluidic system for high-speed reproducible DNA sizing and quantitation. Electrophoresis. 2000;21:128–134. doi: 10.1002/(SICI)1522-2683(20000101)21:1<128::AID-ELPS128>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Achille A, Biasi MO, Zamboni G, Bogina G, Magalini AR, Pederzoli P, Perucho M, Scarpa A. Chromosome 7q allelic losses in pancreatic carcinoma. Cancer Res. 1996;56:3808–3813. [PubMed] [Google Scholar]
- Andersson J, Sihto H, Meis-Kindblom JM, Joensuu H, Nupponen N, Kindblom LG. NF1-associated gastrointestinal stromal tumors have unique clinical, phenotypic, and genotypic characteristics. Am J Surg Pathol. 2005;29:1170–1176. doi: 10.1097/01.pas.0000159775.77912.15. [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Hirota S, Isozaki K, Ohashi A, Nishida T, Kitamura Y, Shinomura Y, Matsuzawa Y. Absence of c-kit gene mutations in gastrointestinal stromal tumours from neurofibromatosis type 1 patients. J Pathol. 2004;202:80–85. doi: 10.1002/path.1487. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Fetsch JF, Sobin LH, Lasota J. Gastrointestinal stromal tumors in patients with neurofibromatosis 1: a clinicopathologic and molecular genetic study of 45 cases. Am J Surg Pathol. 2006;30:90–96. doi: 10.1097/01.pas.0000176433.81079.bd. [DOI] [PubMed] [Google Scholar]
- Chen LL, Sabripour M, Wu EF, Prieto VG, Fuller GN, Frazier ML. A mutation-created novel intra-exonic pre-mRNA splice site causes constitutive activation of KIT in human gastrointestinal stromal tumors. Oncogene. 2005;24:4271–4280. doi: 10.1038/sj.onc.1208587. [DOI] [PubMed] [Google Scholar]
- Corless CL, McGreevey L, Town A, Schroeder A, Bainbridge T, Harrell P, Fletcher JA, Heinrich MC. KIT gene deletions at the intron 10-exon 11 boundary in GI stromal tumors. J Mol Diagn. 2004;6:366–370. doi: 10.1016/S1525-1578(10)60533-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corless CL, Schroeder A, Griffith D, Town A, McGreevey L, Harrell P, Shiraga S, Bainbridge T, Morich J, Heinrich MC. PDGFRA mutations in gastrointestinal stromal tumors: frequency, spectrum and in vitro sensitivity to imatinib. J Clin Oncol. 2005;23:5357–5364. doi: 10.1200/JCO.2005.14.068. [DOI] [PubMed] [Google Scholar]
- Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Sobin LH, Lasota J. Gastrointestinal stromal tumors of the stomach: a clinicopathologic, immunohistochemical, and molecular genetic study of 1765 cases with long-term follow-up. Am J Surg Pathol. 2005;29:52–68. doi: 10.1097/01.pas.0000146010.92933.de. [DOI] [PubMed] [Google Scholar]
- Wardelmann E, Hrychyk A, Merkelbach-Bruse S, Pauls K, Goldstein J, Hohenberger P, Losen I, Manegold C, Buttner R, Pietsch T. Association of platelet-derived growth factor receptor alpha mutations with gastric primary site and epithelioid or mixed cell morphology in gastrointestinal stromal tumors. J Mol Diagn. 2004;6:197–204. doi: 10.1016/s1525-1578(10)60510-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen M, Makhlouf H, Sobin LH, Lasota J. Gastrointestinal stromal tumors of the jejunum and ileum: a clinicopathologic, immunohistochemical, and molecular genetic study of 906 cases before imatinib with long-term follow-up. Am J Surg Pathol. 2006;30:477–489. doi: 10.1097/00000478-200604000-00008. [DOI] [PubMed] [Google Scholar]
- Penzel R, Aulmann S, Moock M, Schwarzbach M, Rieker RJ, Mechtersheimer G. The location of KIT and PDGFRA gene mutations in gastrointestinal stromal tumours is site and phenotype associated. J Clin Pathol. 2005;58:634–639. doi: 10.1136/jcp.2004.021766. [DOI] [PMC free article] [PubMed] [Google Scholar]