Abstract
Single bp mutations in the RET proto-oncogene can cause multiple endocrine neoplasia type 2 syndromes. The conventional approach for genotyping RET mutations is sequencing the exons. A closed-tube RET genotyping assay using a saturating DNA dye, unlabeled probes, and amplicon high-resolution melting analysis was developed. The method required two sequential polymerase chain reaction stages, a primary and secondary assay. The primary assay analyzed RET exons 10, 11, 13, 14, and 16 with a total of seven reactions using eight unlabeled probes. The primary assay genotyped wild-type exons, a common exon 13 polymorphism, and an exon 16 mutation, whereas other RET sequence variation was detected. The primary unlabeled probe data limited the possible genotypes for the detected RET sequence variation, which permitted genotyping in a secondary assay with only two to five reactions. Six probes were designed with the masking technique and masked selected sequence variations to allow unambiguous analysis of other mutations elsewhere under the probe. After this two-stage RET genotyping assay, less than 0.2% of exons tested would require sequencing for genotype. A blinded study generated from five wild type and 29 available RET sequence variation samples was 100% concordant with sequencing. Amplicon high-resolution melting analysis with unlabeled probes and the masking technique is a fast, accurate method for genotyping the >50 RET sequence variations.
Multiple endocrine neoplasia type 2 (MEN2) syndromes, MEN2A, MEN2B, and familial medullary thyroid carcinoma (FMTC), are caused by germline RET mutations in exons 10, 11, and 13 through 16. MEN2 syndromes are autosomal dominant disorders that lead to a high lifetime risk of thyroid cancer. Genetic testing for RET mutations can identify patients at risk of thyroid cancer before cancer progression, when prophylactic thyroidectomy can increase survival rate.
There have been many assays developed to detect and/or genotype RET mutations, such as single-stranded conformation polymorphism analysis, denaturing gradient gel electrophoresis, temperature gradient capillary electrophoresis, restriction enzyme digestion of polymerase chain reaction (PCR) products, pyrosequencing, fluorescently labeled hybridization probes, and microarrays.1,2,3,4,5,6,7,8,9,10 However, most of these methods require additional post-PCR processing of the amplicon to detect mutations. Some of these methods have reported 95% or less sensitivity for mutation detection or have been tested with only a small number of RET mutation samples. The methods that analyze limited regions of RET sequence may target only common mutations and miss rare mutations. Furthermore, the presence of nonpathogenic polymorphisms may also lead to aberrant results.9,11 Because of these limitations, RET exon sequencing remains the gold standard.
Sequencing is a time-consuming and expensive open-tube assay that requires additional processing of PCR products. In contrast, mutation scanning by high-resolution melting analysis is a rapid and cost-effective closed-tube assay that does not require post-PCR processing of the amplicon.12 By using a saturating double-stranded DNA-binding dye, LCGreen Plus, amplicon high-resolution melting analysis can detect sequence variation within a PCR amplicon.12,13,14 A systematic study of high-resolution melting detection of heterozygous point mutations within a PCR amplicon found a sensitivity and specificity of 100% for amplicons <400 bp in size.14 Recently, we demonstrated mutation scanning of RET exons by high-resolution melting analysis.15 Although this assay detected 100% of the mutations in a blinded study, complete genotyping was difficult because some unique mutations had similar melting curves. In these cases, sequencing was required after the initial mutation scanning assay. Alternatively, the addition of hybridization probes may allow the majority of RET mutations to be genotyped without sequencing.
Fluorescently labeled hybridization probe analysis is a rapid, closed-tube assay that can genotype sequence variation by probe/allele melting temperatures (Tm) but requires expensive oligonucleotide modifications.5 Unlabeled probes can also genotype mutations by Tm in a closed-tube assay, using LCGreen Plus.16 Because the detection is with a double-stranded DNA-binding dye, unlabeled probe and amplicon data can be analyzed from one melting curve. Amplicon mutation scanning can be complemented with unlabeled probe genotyping analysis, as recently demonstrated by Zhou et al.17 However, all RET mutations within an exon cannot be genotyped with one probe, because unique mutations under the probe can have similar or identical melting temperatures.18
A two-stage RET genotyping assay was developed for complete closed-tube genotyping, which was more cost effective and less time consuming than sequencing. The first PCR stage (primary assay) used amplicon melting to scan for any sequence variation within the RET exons, whereas the unlabeled probe analysis located the region of variation and identified possible genotypes. A second PCR stage (secondary assay) with a limited number of mutation-specific probes was required for unambiguous genotyping of the >50 RET sequence variations. Mutation identification was simplified by using probes designed with the masking technique.18 The masking technique simplifies probe melting interpretation when the presence of multiple possible sequence variations under the probe complicates analysis. Selected sequence variation can be masked by incorporating mismatches (deletion, unmatched nucleotide, or universal base) into hybridization probes at the polymorphic location. This creates an artificial mismatch with all possible alleles at the masked polymorphic location. Both the wild-type and masked variant alleles have a single mismatch with the probe and a similar Tm, whereas mutant alleles have an additional mismatch elsewhere under the probe and are identified by a lower Tm than alleles that vary only at the masked position.
Materials and Methods
Samples
The deidentified genomic DNA samples of wild-type RET genotype or with RET sequence variation used in this report were described previously.15,18 RET sequence variations that alter RET protein function to cause MEN2 syndromes are mutations, whereas RET sequence changes that do not cause MEN2 syndromes are polymorphisms (>1% population frequency).2,19,20,21,22,23 Rare RET sequence variations of unknown significance or that do not cause MEN2 syndromes are “sequence variants.” In this report, RET sequence variation is listed by the codon number, the wild-type codon DNA sequence followed by the codon sequence change, with the mutant nucleotide in bold [eg, 618 (TGC→TAC)], whereas polymorphism or sequence variant nucleotide change is underlined. All of the RET sequence variation samples tested had a single nucleotide change and were heterozygous unless otherwise stated. RET mutant genotypes as well as polymorphism and sequence variant genotypes are listed in Table 1.3,20,24
Table 1.
RET Proto-Oncogene Sequence Variation
| RET exon | Codon | Codon sequence change* | Amino acid change | References |
|---|---|---|---|---|
| 10 | 603 | AAA → CAA | K603Q | 20 |
| 609 | TGC → AGC† | C609S | 3 | |
| TGC → CGC | C609R | 3, 20, 24 | ||
| TGC → GGC | C609G | 3, 20 | ||
| • TGC → TAC | C609Y | 3, 20, 24 | ||
| • TGC → TCC | C609S | 3, 20 | ||
| TGC → TTC | C609F | 3 | ||
| TGC → TGG† | C609W | 3, 20, 38 | ||
| 611 | TGC → AGC | C611S | 3, 20 | |
| • TGC → CGC | C611R | 3, 20 | ||
| TGC → GGC | C611G | 3, 20, 24 | ||
| • TGC → TAC | C611Y | 3, 20, 24 | ||
| TGC → TCC† | C611S | 3 | ||
| • TGC → TTC | C611F | 3, 20 | ||
| TGC → TGG | C611W | 3, 20, 24 | ||
| 618 | TGC → AGC | C618S | 3, 20, 24 | |
| • TGC → CGC | C618R | 3, 20, 24 | ||
| • TGC → GGC | C618G | 3, 20, 24 | ||
| • TGC → TAC | C618Y | 3, 20, 24 | ||
| • TGC → TCC | C618S | 3, 20, 24 | ||
| • TGC → TTC | C618F | 3, 20, 24 | ||
| TGC → TGG | C618W | 3 | ||
| 620 | • TGC → AGC | C620S | 3, 20 | |
| TGC → CGC | C620R | 3, 20, 24 | ||
| TGC → GGC | C620G | 3, 20, 24 | ||
| • TGC → TAC | C620Y | 3, 20, 24 | ||
| • TGC → TCC | C620S | 3, 20, 24 | ||
| • TGC → TTC | C620F | 3, 20, 24 | ||
| • TGC → TGG | C620W | 3, 20 | ||
| 11 | 630 | TGC → CGC | C630R | 3 |
| TGC → TAC | C630Y | 3, 20 | ||
| TGC → TTC | C630F | 3, 20, 24 | ||
| TGC → TCC | C630S | 3, 20 | ||
| 631 | • GAC → GAT† | D631D | 3 | |
| GAC → TAC† | D631Y | 3 | ||
| GAC → GAA† | D631E | 3, 34 | ||
| GAC → GTC† | D631V | 3, 34 | ||
| 634 | • TGC → AGC | C634S | 3, 20, 24 | |
| • TGC → CGC | C634R | 3, 20, 24 | ||
| • TGC → GGC | C634G | 3, 20, 24 | ||
| • TGC → TAC | C634Y | 3, 20, 24 | ||
| • TGC → TCC | C634S | 3, 20, 24 | ||
| • TGC → TTC | C634F | 3, 20, 24 | ||
| • TGC → TGG | C634W | 3, 20, 24 | ||
| 640 | GCC → GGC | A640G | 20 | |
| 13 | 768 | • GAG → GAC | E768D | 3, 20, 24 |
| GAG → GAT | E768D | 3, 20 | ||
| 769 | • CTT → CTG‡ | L769L | 2, 19, 20 | |
| 778 | GTC → ATC | V778I | 20 | |
| 781 | CAG → CGG | Q781R | 20 | |
| 790 | TTG → TTT | L790F | 20, 24, 28 | |
| TTG → TTC | L790F | 20, 24, 28 | ||
| 791 | TAT → TTT | Y791F | 24, 27, 28 | |
| 14 | 804 | • GTG → ATG | V804M | 3, 20, 24 |
| • GTG → TTG | V804L | 3, 20, 24 | ||
| GTG → CTG | V804L | 3 | ||
| 806 | TAC → TGC§ | Y806C | 24, 35 | |
| 844 | CGG → CTG | R844L | 20, 24 | |
| 852 | ATC → ATG† | I852M | 20, 29 | |
| 16 | 912 | CGG → CCG | R912P | 30 |
| 918 | • ATG → ACG | M918T | 3, 20, 24 | |
| 922 | TCC → TAC† | S922Y | 20, 31 |
Sequence variation is heterozygous unless otherwise stated in the text. Codon DNA sequence is listed with the nucleotide of change in bold for mutations and underlined for polymorphisms/sequence variants. •, available RET sequence variation samples.
Rare sequence variants of unknown significance or not causative of MEN2 syndromes; see references. Three variants in codons 609 or 611 are within RET pathogenic codons yet are not associated with MEN2 syndromes. Variants at codons 922 and 852 were previously found in MTC patients, yet may not be causative of MEN2 syndromes.
Benign polymorphism, 0.26 allelic frequency.
806 is only pathogenic with 804 mutation on same allele.
Primers and Probes
Oligos were synthesized at Integrated DNA Technology (Coralville, IA). Primers were designed using Primer 3.25 Primers and probes were chosen to detect all known RET mutations while excluding common polymorphisms from analysis (Table 2).2,19,20 The unlabeled probes had either a 3′-phosphate or the 3′-amino modifier (Integrated DNA Technology) incorporated to prevent extension during PCR. Probes were designed to analyze the mutation hotspots within each exon.
Table 2.
Primers and Probes
| RET exon | Primers* | Amplicon (bp) | Codon of variation† | Probes‡ | Probe sequence§ |
|---|---|---|---|---|---|
| 10 | 0.275 μmol/L, 1F:9R | 146 | 609 | WT10A | GGCTATGGCACCTGCAACTGCTTCCCTGAG |
| GGGCAGCATTGTTGGGGGAC | 611 | L10A 611mask¶ | GGCTATGGCACCTGCAAC---TTCCCTGAG | ||
| TGGTGGTCCCGGCCGCCA | MS 609/611 TAC∥ | GGCTATGGCACCTACAACTACTTCCCTGAG | |||
| 618 | WT10B | GGAGAAGTGCTTCTGCGAGCCCGAAGACATC | |||
| 620 | L10B 620mask¶ | GGAGAAGTGCTTC---GAGCCCGAAGACATC | |||
| MS 618/620 TAC∥ | GGAGAAGTACTTCTACGAGCCCGAAGACATC | ||||
| 11 | 0.55 μmol/L, 5F:1R | 109 | 630 | WT11 | TGCGATCACCGTGCGGCACAGCTCGTCGCAC |
| TGCCAAGCCTCACACCAC | 631 | L11 634mask¶ | TGCGATCACCGTGCG---CAGCTCGTCGCAC | ||
| GACAGCAGCACCGAGAC | 634 | MS 630/634 TAC∥ | TGCGATCACCGTGCGGTACAGCTCGTCGTAC | ||
| 13 | 1.1 μmol/L, 1F:9R | 187 | WT13 | CCCGAGTGAGCTTCGAGACCTGCTGTCAGA | |
| CTGCATTTCAGAGAACGCCTC | 768 | Mask 1bp 769** | CCCGAGTGAGCT-CGAGACCTGCTGTCAGA | ||
| GAACAGGGCTGTATGGAGC | 769 | MS 768 GAC + mask** | CCCGAGTGACCT-CGAGACCTGCTGTCAGA | ||
| MS 768 GAT + mask** | CCCGAGTGATCT-CGAGACCTGCTGTCAGA | ||||
| 14 | 0.55 μmol/L, 1F:5R | 235 | WT14A | CTCCTCCTCATCGTGGAGTACGCCAAA | |
| CAGGGCCCCCTCTCTCCGC | 804 | MS 804 TTG | CTCCTCCTCATCTTGGAGTACGCCAAA | ||
| TCTCGGCCAGATACTGCAT | 806 | MS 804 ATG | CTCCTCCTCATCATGGAGTACGCCAAA | ||
| MS 804 CTG | CTCCTCCTCATCCTGGAGTACGCCAAA | ||||
| 844 | WT14B | GAGCGGGCCCTCACCATGGGCGACCTCATCTCA | |||
| 852 | MS 844 CTG | GAGCTGGCCCTCACCATGGGCGACCTCATCTCA | |||
| MS 852 ATG | GAGCGGGCCCTCACCATGGGCGACCTCATGTCA | ||||
| 16 | 0.55 μmol/L, 1F:9R | 154 | 918 | MS 918 ACG†† | TCCAGTTAAATGGACGGCAATTGAATCCCT |
| ATAGGGCCTGGGCTTCTC | 922 | ||||
| ACACATCACTTTGCGTGGTG |
Final total primer concentration and forward (F) to reverse (R) primer ratio is displayed above the primer sequences. Primer sequences are listed 5′ to 3′, with the forward primer above the reverse primer.
Codon of variation under each probe set. The underlined codons contain a polymorphism or sequence variant, whereas the other codons contain mutations.
Mask, masking deletion of one or three nucleotides. Wild-type probe sequence (WT exon #) is used in the primary assay for each exon, except for exon 16. If two wild-type probes are used for one exon, they are labeled as A and B probes.
Probe sequences are listed 5′ to 3′. RET exons 10, 13, 14, and 16 have forward probes, whereas exon 11 has reverse probes. The known mutation locations are highlighted in bold, and the possible polymorphism or sequence variant locations are underlined.
Location (L) probes have a three nucleotide masking deletion (−−−) of one pathogenic codon from the wild-type probe sequence.
Example for the MS probes used for genotyping the mutations in exons 10 and 11. The same mutation-specific sequence is at both pathogenic codon positions within the probe. These probes have the mutation-specific sequence in the name, with the changed sequence in bold. There are seven possible mutant sequence changes from the ‘TGC’ wild-type cysteine codon sequence to another amino acid, therefore seven mutation-specific probes (AGC, CGC, GGC, TAC, TCC, TTC, TGG) for a total of 21 MS probes for exon 10 and 11 mutations. The MS probe sets used to genotype each mutation are diagrammed in the figures and Table 3 as well as in Supplemental Tables 1 to 5.
Exon 13 used masking probes in the primary and secondary assays. The ‘Mask 1bp 769’ primary probe and the exon 13 mutation-specific probes have a single nucleotide deletion (−) at the polymorphism position within codon 769 (deletion of the underlined ‘T’ in the wild-type probe sequence).
Exon 16 used a codon 918(ATG→ACG) mutation-specific probe for the primary assay.
Four types of unlabeled probes were used in the RET genotyping assay (Table 2): wild-type (WT) probes, mutation-specific (MS) probes, masking probes that masked a single polymorphic nucleotide, and location (L) probes that masked three polymorphic nucleotides (one codon).18 For example, the exon 13 masking probe (Mask 1bp 769) had a single nucleotide deletion at the polymorphic nucleotide position (the “T” position in WT13 probe; Table 2). The exon 10 and 11 location probes had a three-nucleotide (codon) deletion from the wild-type probe sequence, masking one pathogenic codon to identify the codon position of a mutation. The mutation-specific probes for exon 10 and 11 had the same sequence change at both pathogenic codon positions. For example, in exon 10, the MS 618/620 TAC probe has the “TAC” sequence at both codons 618 and 620 as displayed in Table 2.
PCR
Sample DNA was amplified by asymmetric rapid-cycle PCR as described previously,18 except that 55 cycles of PCR were used with primer concentrations given in Table 2. The exon 14 reaction included two probes: 0.5 μmol/L WT14A probe and 0.25 μmol/L WT14B probe. A wild-type control was run with each probe for the primary and secondary assay reactions. For exon 13, the WT13 primary assay reaction used two additional controls: heterozygous and homozygous codon 769 samples. The exon 10 primary assay reactions used a 620(TGC→AGC) mutant sample as a positive control.
High-Resolution Melting
Analysis was performed on a high-resolution melting instrument, the HR-1 (Idaho Technology, Salt Lake City, UT). The LightCycler capillaries were transferred into the HR-1 and heated at 0.3°C/second. In the primary assay, amplicon and unlabeled probe melting data for the RET exons were acquired between 60°C and 95°C. All amplicon and probe data were analyzed in the primary assay for each exon, except for exon 14. Because all reported exon 14 mutations were under the unlabeled probes, only probe data were analyzed. For the secondary assay, only unlabeled probe melting was analyzed with data acquired between 60°C and 89°C.
The high-resolution melting data were analyzed with custom software written in LabView (National Instruments, Austin, TX) as described previously.12,15 In brief, the melting curve data for the amplicon was normalized, temperature-shifted, and then converted to a derivative plot or fluorescence difference plot. Depending on the data plot used for amplicon analysis, sequence variation was detected by a visual shift from the wild-type controls due to a unique melting curve shape, a broader derivative melting peak, a uniquely shaped derivative melting peak (shoulder or two peaks), a shift in melting temperature, and/or fluorescence difference. The amplicon melting curve, derivative plot, and fluorescence difference plot could all be used to detect RET sequence variation. The unlabeled probe melting data were directly converted to a derivative plot. Exon 14 probe data were normalized, and the exponential background was removed before analysis.26
Tms for the unlabeled probe data were estimated at one-half the area of the derivative melting curve peaks. ΔTms for heterozygous samples were defined as the wild-type allele Tm minus the variant allele Tm (mutation, polymorphism, or sequence variant allele). Note that the ΔTm value will be negative when the variant allele Tm is higher than the wild-type allele Tm (eg, with a complementary mutation-specific probe). When probe data for heterozygous samples resulted in shoulder or plateau shaped peaks, the peak area was divided into ¼, ½, and ¼ regions with the wild-type and variant allele Tms taken at the boundaries of these regions. When only a single peak was apparent because of homozygous sequence variation (no wild-type allele) or the variant allele Tm was nearly identical to the wild-type allele Tm (no distinguishable wild-type allele), then the wild-type control sample Tm was used for the wild-type allele Tm in the ΔTm calculation. Wild-type samples also resulted in a single peak, so the sample’s single allele Tm was subtracted from the wild-type control Tm to calculate ΔTm.
The primary probe ΔTm values for each RET sequence variation determined the mutation-specific probe set used in the secondary genotyping assay. In cases in which the RET mutations were not available (Table 1), predicted ΔTms were estimated using nearest-neighbor parameters as previously described.13 The empirically determined and predicted ΔTm values for RET sequence variations with the primary probes established the ΔTm ranges expected to group mutations for mutation-specific probe selection. Supplemental Tables 1 through 5 (see http://jmd.amjpathol.org) display the ΔTm ranges and mutation grouping with the primary probes, as well as the ΔTm values for each available RET sequence variation with the primary probes and the selected secondary probes. The “Tm guidelines” were placed on a derivative plot at temperatures calculated by subtracting the primary assay ΔTm range values from the control wild-type allele Tm, as shown for exons 10 and 11 primary probe data. These Tm guidelines are convenient for quick, visual grouping of mutant samples to select the secondary assay mutation-specific probes. However, calculating the ΔTm value for each sample can better control for systematic Tm shifts caused by sample-specific differences (altered salt concentration, different sample purification methods, or temperature inaccuracy).
Blinded Study
Twenty-nine variant RET samples (the available mutations, polymorphisms, and sequence variants as listed in Table 1) and five wild-type samples were used to generate the blinded study. Ten samples each for exons 13, 14, and 16; 20 samples for exon 11; and 28 samples for exon 10 were analyzed (Supplemental Table 6; http://jmd.amjpathol.org). For each exon, the number of wild-type and variant samples was unknown. For the blinded study amplicon analysis, the derivative melting curve data were used to visually identify sequence variation by unique derivative melting peak shape and Tm shift from a wild-type control, although RET sequence variation was detectable in all three data plots: melting curve, derivative melting curve, and fluorescence difference. The amplicon derivative melting data from the primary assays using the WT10A and WT13 probes were used to scan for mutations in exons 10 and 13, respectively.
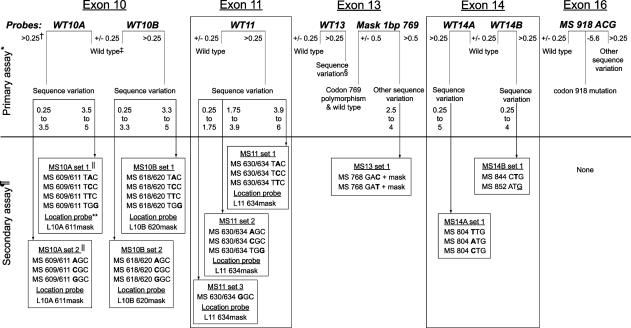
The ΔTm values were calculated from the probe data for each sample in the primary assay (and secondary assay, if needed). The ΔTm data charts were used to determine genotype or to select mutation-specific probes for the secondary assay (Supplemental Tables 1 through 5; http://jmd.amjpathol.org). Table 3 (a simplified version of Supplemental Tables 1 through 5; http://jmd.amjpathol.org) presents the testing schematic for the RET genotyping assay and can also be used to select the secondary probe sets. The ability of the Tm guidelines to select mutation-specific probes for exons 10 and 11 was compared with using the ΔTm calculated for each sample. After the primary assay, samples were labeled as wild-type, exon 13 polymorphism, exon 16 codon 918 mutation, or other RET sequence variations. The primary probe ΔTm for the detected sequence variations determined the set of mutation-specific probes used in the secondary genotyping assay, where the RET sequence variations were genotyped.
Table 3.
Testing Schematic for the RET Genotyping Assay
* Eight probes were used in seven PCR reactions (exon 14 probes are multiplexed) in the primary assay for five RET exons. The primary probes are listed for each RET exon: WT, wild type; MS, mutation-specific. The primary assay uses probe and amplicon data to genotype wild type, exon 13 codon 769(CTT→CTG) polymorphism and exon 16 codon 918(ATG→ACG) mutation. All other sequence variations detected with a primary probe will be tested with a secondary probe set selected by the ΔTm from the primary assay data (∼7% of tested exons). There are two exceptions where samples will instead be sequenced (rare): 1) Exon 16 probe detected sequence variation other than the codon 918 mutation, or 2) the primary probe detected sequence variation has a ΔTm outside the ranges given for secondary probe selection. This could indicate two or more sequence variations detected by the primary probe or novel sequence variation was detected.
† The ΔTm values (°C) listed for the primary assay are used to determine sample genotype or to select the one probe set used in the secondary assay. Wild-type samples were within 0.25°C of the control wild type.
‡ Wild-type sequence under the probe. Analyze amplicon data to call wild-type genotype or to detect rare sequence variation within the amplicon but not under the probe (amplicon melting curve is shifted even though probe data indicated the wild type). Any rare sequence variation detected by only the amplicon melting data would be sequenced for genotype (<0.2% of tested exons).
§ Sequence variation due to mutations or the common codon 769 polymorphism. The Mask 1bp 769 probe differentiates the polymorphism from the other sequence variation detected by the WT13 probe.
¶ Test only the exon(s) where sequence variation was detected with the primary assay probe.
∥ Only one secondary assay probe set will be used to genotype sequence variation detected by the primary assay probe. Probe set was selected by the ΔTm of a sample with the primary probe, either visually using Tm guidelines (Figure 2a, Supplemental Figures 1a and 2a; http://jmd.amjpathol.org) or by calculated ΔTm values (Supplemental Tables 1–5; http://jmd.amjpathol.org). Only sequence the sample if sequence variation is not genotyped by the selected probe set (0.2% of tested exons), for example, the exon 11 (GAC→GAT) polymorphism.
** The location (L) probes: L10A 611mask, L10B 620mask, and L11 634mask, are used with any sequence variation detected with the primary assay probes WT10A, WT10B, and WT11, respectively.
Results
Assay Design
Genotyping of RET exons 10, 11, 13, 14, and 16 was achieved in a two-stage, closed-tube protocol consisting of primary and secondary assays (Table 3). Because a double-stranded DNA dye was used, both unlabeled probe and amplicon melting data were generated in each reaction. The primary assay used eight probes in seven PCR reactions to scan for sequence variation and identify some genotypes (wild-type exons, the exon 13 codon 769 polymorphism, and the exon 16 codon 918 mutation). Only the exons that have sequence variation detected but not genotyped by the primary probe data were tested in the secondary assay with one selected probe set. Samples with rare sequence variation detected by the amplicon data only (wild-type sequence under the probe) or rare sequence variation not genotyped by the mutation-specific probes in the secondary assay required sequencing to determine genotype (∼0.2% of exons tested).
Exons 10 and 11: Multiple Mutation Hotspots
The majority of mutations for MEN2A and FMTC are in exons 10 (codons 609, 611, 618, and 620) and 11 (codons 630 and 634) and are single nucleotide changes from the wild-type “TGC” codon DNA sequence for cysteine. There are >40 sequence variations reported in exons 10 and 11 (Table 1).3,20
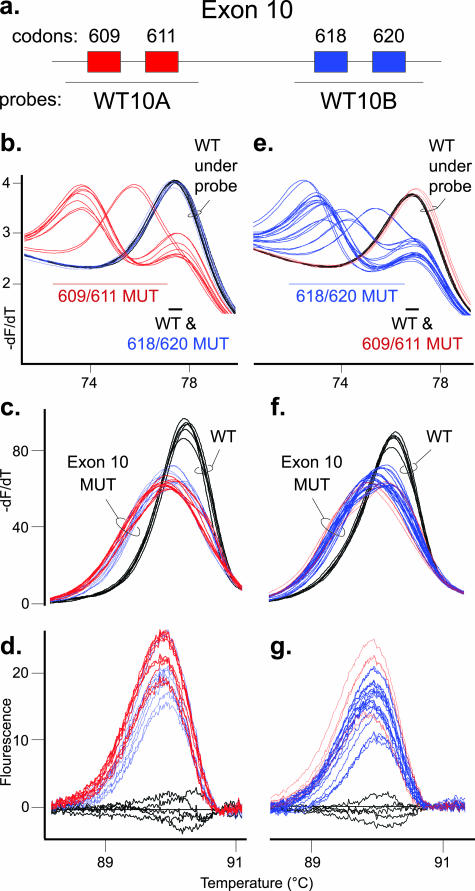
The primary assay for RET exon 10 used two wild-type probes in two separate reactions. The WT10A probe covers codons 609 and 611, whereas the WT10B probe covers codons 618 and 620 (Figure 1a). All codon 618 and 620 mutant samples have wild-type sequence under the WT10A probe and have the same Tm as the control wild-type samples (Figure 1b). Each of the five codon 609 and 611 mutant samples had a mutant allele that melted at a lower Tm than the wild-type allele. Conversely, with the WT10B probe, codon 609 and 611 mutant samples had the same Tm as a wild-type sample, whereas each of the 10 codon 618 and 620 mutant samples had a mutant allele that melted at a lower Tm than the wild-type allele (Figure 1e). All RET sequence variations in exon 10 were detected by amplicon high-resolution melting analysis using normalized melting curves, derivative plots, or fluorescence difference data (Figure 1, c, d, f, and g; data not shown). These data demonstrate how mutations not under the unlabeled probes can still be detected using the amplicon melting data (eg, compare blue mutation traces in Figure 1, b versus c).
Figure 1.
Primary assay for detection of sequence variation in RET exon 10. a: Diagram of RET exon 10 with codons 609 and 611 boxed in red and codons 618 and 620 boxed in blue. The primary assay for exon 10 used two unlabeled probes of WT sequence. As displayed, the WT10A probe detects codon 609 and 611 mutations, whereas the WT10B probe detects codon 618 and 620 mutations. In each graph, three wild-type controls in duplicate (WT, black traces) are shown with two codon 609 and three codon 611 mutant samples in duplicate (red traces) as well as five codon 618 and five codon 620 mutations (blue traces). The three left panels are unlabeled probe and amplicon high-resolution melting data from the WT10A probe reaction, and the three right panels are data from the WT10B probe reaction. For each probe, thick trace lines were used with mutations detected by the probe data, and thin trace lines were used with mutations detected by amplicon data only. b: Derivative melting curve plot for the WT10A probe data. The alleles are divided into two Tm ranges, and predicted codon mutant alleles in each range (609/611 MUT or 618/620 MUT), along with the WT allele, are listed. c: Amplicon derivative melting curve data from the WT10A probe primary assay with the WT and exon 10 mutation (exon 10 MUT) samples labeled. d: Difference plot of same amplicon data. e: Derivative melting curve plot for the WT10B probe data. The alleles are divided into two Tm ranges, and predicted codon mutant alleles in each range, along with the wild-type allele, are listed. f: Amplicon derivative melting curve data from the WT10B probe primary assay with wild-type and exon 10 mutant samples labeled. g: Difference plot of same amplicon data.
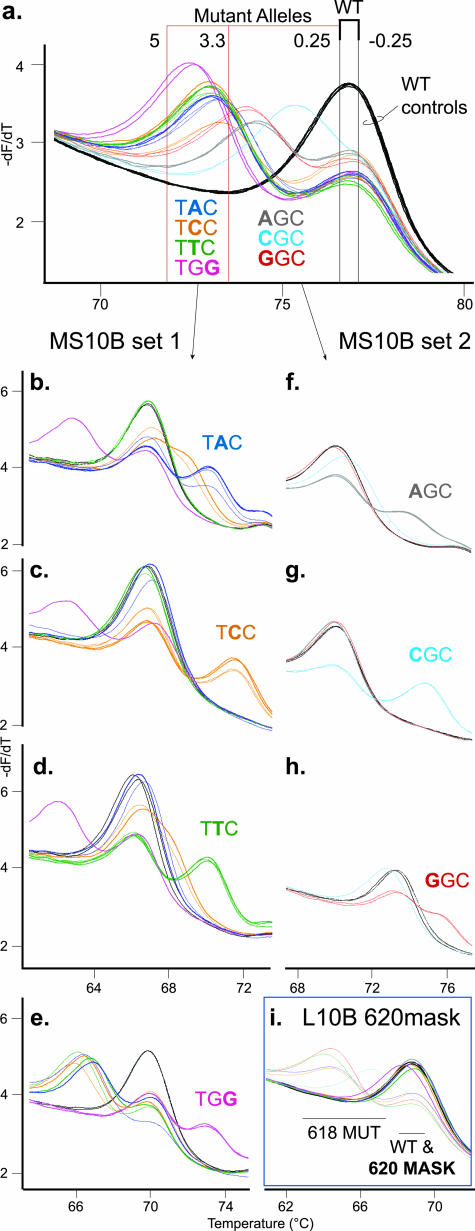
The wild-type probes used in the primary assay identified wild-type sequence under each probe, as indicated by a sample Tm within ±0.25°C of the control wild-type allele Tm (Table 3; Supplemental Table 1; http://jmd.amjpathol.org). Samples with RET sequence variation had an allele with a lower Tm than the wild-type allele and a ΔTm value of >0.25°C. To limit the number of mutation-specific probes required for genotyping in the secondary assay, the ΔTm ranges were used to group RET sequence variation with similar ΔTm values. The ΔTm ranges were used to generate the Tm guidelines for quick, visual selection of secondary assay probes. For example, the WT10B probe data in Figure 2a shows 13 samples visually divided by Tm guidelines into a wild-type and two mutation groups. The first mutation group, consisting of TAC, TCC, TTC, and TGG mutations at either codon 618 or codon 620, had ΔTm values within the 3.3°C to 5°C ΔTm range (Figure 2a; Table 3; Supplemental Table 1; http://jmd.amjpathol.org). Samples in this group required the four probes in MS10B probe set 1 for the secondary genotyping assay (Figure 2, b–e). The second mutation group, consisting of AGC, CGC, and GGC mutations at either codon 618 or codon 620, had ΔTm values within the 0.25°C to 3.3°C ΔTm range. This mutation group required the three probes in MS10B probe set 2 for the secondary assay (Figure 2, f–h). A similar method was used for the WT10A probe data with the codon 609 and 611 mutations (Table 3; Supplemental Figure 1; Supplemental Table 2; http://jmd.amjpathol.org). The exon 10 primary assay used the ΔTm values from two exon 10 wild-type probes to limit the possible genotypes of an unknown mutation from 28 to ≤8 genotypes.
Figure 2.
Genotyping mutations in RET exon 10 at codons 618 and 620. a: Derivative melting curve plot for the primary WT10B probe data from Figure 1e. Three wild-type controls in duplicate (WT, black traces) are shown with 10 heterozygous mutant samples in duplicate, five at codon 618 (thin traces) and five at codon 620 (thick traces). The colors used for mutant codon DNA sequences and traces are consistent throughout the figure. Graph traces for both the codon 618 and 620 mutation sequences are gray for AGC, light blue for CGC, red for GGC, dark blue for TAC, orange for TCC, green for TTC, and pink for TGG. The ΔTm range values used to generate the Tm guidelines are listed next to each Tm guideline. Wild-type allele Tms are within the black Tm guidelines, ±0.25°C of control wild-type sample. The mutant alleles detected by the primary probe were divided into two ΔTm ranges by the red Tm guidelines. The mutation sequences that are predicted to be grouped into each ΔTm range are listed below the traces. The secondary assay data for the MS probe sets, MS10B set 1 or MS10B set 2, are displayed for the two mutation groups. b–e: The mutation group with ΔTm values of 3.3°C to 5°C was tested with the four probes in MS10B set 1: MS 618/620 TAC (b), MS 618/620 TCC (c), MS 618/620 TTC (d), and MS 618/620 TGG (e). f–h: The mutation group with ΔTm values of 0.25°C to 3.3°C was tested with the three probes in MS10B set 2: MS 618/620 AGC (f), MS 618/620 CGC (g), and MS 618/620 GGC (h). On each mutation-specific probe graph (b–h), the codon mutation DNA sequence that complements the MS probe is listed. i: All mutant alleles (primary WT10B probe data, ΔTm of 0.25°C to 5°C) will also use the location probe L10B 620mask to locate codon position of the detected mutations. Two derivative melting temperature ranges are indicated, with the codon 618 mutant alleles (618 MUT) labeled as well as the WT allele and masked codon 620 mutant alleles (620 MASK).
To reduce the number of probes needed to genotype mutations in the secondary assay, each exon 10 mutation-specific probe had the same sequence change at both pathogenic codon positions (Table 2). A mutant allele that was complementary at one pathogenic codon resulted in only one mismatch with the probe and a 2°C to 5°C higher Tm than the wild-type allele with two mismatches (Figure 2, b–h). If the mutation was at a different position, the mutant allele had three mismatches with the probe and melted at a lower Tm than wild type (eg, TGG mutant in Figure 2, b–d, pink traces). If the mutation was at the same position as the probe sequence change but was not complementary to the probe, then the mutant allele Tm was close to the wild-type allele Tm because both alleles had two mismatches with the probe. These mutant alleles were “masked” from analysis with a derivative melting curve that was similar, if not identical, to the wild-type allele melting curve (eg, masked AGC and GGC mutant alleles in Figure 2g, red and gray traces).18 Occasionally, a mutant allele that should appear masked melted higher than expected with a noncomplementary mutation-specific probe. For example, the codon 618(TGC→TCC) and 620(TGC→TCC) mutations were more stable (ΔTm −1.7°C to −2.0°C) than the wild-type allele with the MS 618/620 TAC and TTC probes (Figure 2, b and d, orange traces). Even so, both heterozygous TCC mutations were easily identified by the higher Tm with the complementary MS 618/620 TCC probe (ΔTm −4.7°C) (Figure 2c, orange traces).
The same mutant sequence change at either pathogenic codon under the probe yielded very similar derivative melting curves for the primary probe data, regardless of mutation location (Figures 1and 2a; Supplemental Figure 1a; http://jmd.amjpathol.org).15 For example, the codon 618(TGC→TAC) and 620(TGC→TAC) mutations were indistinguishable from each other, by either the primary unlabeled probe or amplicon melting data. In addition, the secondary assay mutation-specific probes were designed to only determine mutation sequence, not to identify the codon position of the mutation. A location probe was included in the secondary assay for any detected exon 10 sequence variation to identify the codon of mutation. The “L10B 620 mask” location probe, designed with a masking, three nucleotide (codon) deletion of the wild-type probe sequence over the codon 620 position (Table 2), was used to differentiate the codon 618 and 620 mutations. The codon 620 mutant alleles were masked by a similar Tm as the wild-type allele, whereas the codon 618 mutant alleles had a lower Tm than wild-type (Figure 2i). The location probes allowed the exon 10 mutation-specific probes to be designed with the same sequence change at both analyzed codons, thereby reducing the number of mutation-specific probes required in the secondary assay by one-half. Any exon 10 mutation detected by the primary assay was genotyped with only one location probe and three to four mutation-specific probes.
Exon 11 was analyzed by similar methods as exon 10 (Table 3; Supplemental Figure 2; Supplemental Table 3; http://jmd.amjpathol.org), and all of the tested RET sequence variations within exon 11 were detected by amplicon high-resolution melting analysis (data not shown). Exon 11 used one wild-type probe for the primary assay over pathogenic codons 630 and 634. Each of the eight exon 11 sequence variations tested had an allele that melted at a lower Tm than wild-type (Supplemental Figure 2a; http://jmd.amjpathol.org). These samples were divided into three mutation groups by primary probe ΔTm values to select the mutation-specific probes for the secondary assay. The mutation-specific probes had the same sequence change at both codon 630 and 634 positions (Table 2). In the secondary assay, mutations at either codon 630 or codon 634 were genotyped with one location probe and one to three mutation-specific probes (Supplemental Figure 2, b–i; http://jmd.amjpathol.org). Although the codon 631(GAC→GAT) sample was detected in the primary assay, this sequence variant was outside of the pathogenic codons analyzed in the secondary assay and melted at a lower Tm than wild type due to three mismatches with each mutation-specific probe (Supplemental Figure 2, e–g). The rare codon 631 sequence variant was not genotyped by the selected mutation-specific probes and would require sequencing for identification.
Exon 13: Common Polymorphism near Mutations
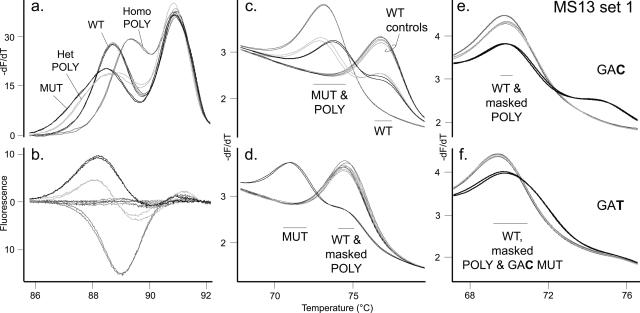
Exon 13 contains seven reported pathogenic mutations (Table 1) and a common polymorphism of 0.26 allelic frequency at codon 769(CTT→CTG).2,19,20 High-resolution amplicon melting revealed two distinct melting domains in the wild-type sequence (Figure 3a). Both heterozygous and homozygous polymorphisms were distinguished from the codon 768(GAG→GAC) mutation by amplicon melting alone (Figure 3, a and b). However, only one of seven reported mutations in exon 13 was available for study.15,20,27,28 Five rare mutations in exon 13 are within the amplicon but not under the exon 13 primary probes. Because these other mutations may be difficult to distinguish from the common polymorphism by amplicon melting analysis data, it is important to identify the presence of the codon 769 polymorphism by using the probe data. Two unlabeled probes, WT13 and Mask 1bp 769, in two separate reactions were included in the primary assay to identify the common polymorphism and clearly detect the two reported mutations under the probe (Tables 2and 3; Supplemental Table 4; http://jmd.amjpathol.org). One probe is wild type and detects any sequence variation under the probe (Figure 3c). The second probe masks the common codon 769 polymorphism with a single nucleotide deletion of the wild-type probe sequence at the polymorphism position.18 With this masking probe, the polymorphism allele has nearly identical Tm as the wild-type allele, whereas the 768(GAG→GAC) mutant allele melts ∼3.6°C lower than the wild-type allele (Figure 3d). If a mutation under the probe was detected, the secondary assay identified the known mutant genotypes at codon 768 (Figure 3, e and f). The mutation-specific probes were also designed with the single nucleotide deletion to mask the codon 769 polymorphism so that the mutations would be clearly genotyped irrespective of whether the polymorphism was present. Note that the mutant 768(GAG→GAC) allele was masked by the MS 768 GAT + mask probe because the sequence change in the probe was at the same position as the mutation but not complementary (Figure 3f). This mutant had the same number and position of mismatches with the probe as the wild-type allele and had a similar, but not identical, Tm as wild type.
Figure 3.
Genotyping sequence variation in RET exon 13. a: Amplicon derivative melting curve data generated in the WT13 probe primary assay. Two wild-type controls in duplicate (WT, thin black traces) are shown with a heterozygous codon 768(GAG→GAC) mutation sample in duplicate (MUT, thick black traces) as well as a heterozygous codon 769(CTT→CTG) polymorphism sample (Het POLY, light gray traces) and homozygous codon 769 polymorphism sample (Homo POLY, gray traces) in duplicate. The colors and trace thickness used for each genotype are consistent throughout the figure. b: Difference plot of the same amplicon data. c: Derivative melting curve plot for the primary WT13 probe data. Two Tm ranges are indicated, with the mutant (MUT) and polymorphism (POLY) alleles labeled as well as the WT allele. d: Derivative melting curve plot for primary “Mask 1bp 769” probe data (the codon 769 polymorphism masking probe). Two derivative melting temperature ranges are indicated, with the mutant allele (MUT) labeled as well as the WT allele and masked polymorphism allele (masked POLY). e and f: The sample genotypes were tested with the two probes in MS13 set 1: MS 768 GAC + mask (e) and MS 768 GAT + mask (f). These two mutation-specific probes also have the codon 769 polymorphism position masked by deletion. The mutant allele complementary to the mutation-specific probe is labeled (GAC or GAT), as well as the WT allele, masked polymorphism allele (masked POLY), and masked codon 768(GAG→GAC) mutant allele (“masked GAC MUT” in f). The codon 768 (GAG→GAT) mutation was not available for testing.
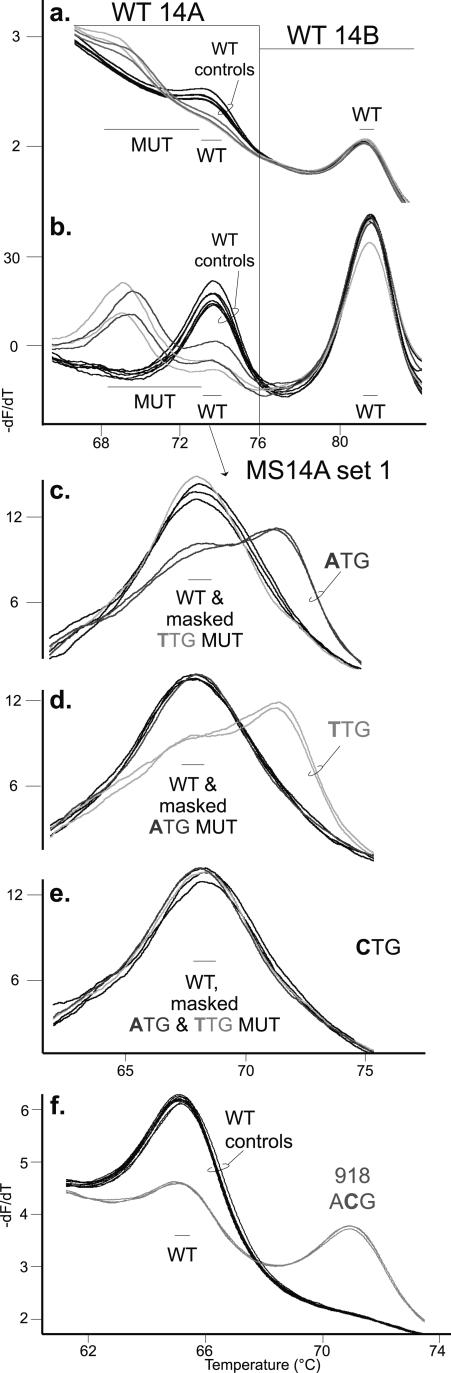
Exon 14: Multiplexed Probe Analysis
For exon 14 in the primary assay, two wild-type probes were multiplexed in one reaction to detect all reported exon 14 mutations while eliminating analysis of the codon 836 (p.S836S) polymorphism (Figure 4, a and b; Table 3; Supplemental Table 5; http://jmd.amjpathol.org). The WT14A probe detects codon 804 and 806 mutations between 65°C and 76°C, whereas the WT14B probe detects sequence variation at codons 844 and 852 between 76°C and 85°C. For sequence variation detected by the primary WT14A probe, the secondary genotyping assay used three mutation-specific probes (MS14A set 1; Figure 4, c–e). For example, the codon 804(GTG→ATG) mutant allele was genotyped by a higher Tm than the wild-type allele with the complementary MS 804 ATG probe (Figure 4c) and masked by both noncomplementary MS 804 TTG and CTG probes (Figure 4, d and e). All of the tested codon 804 mutant alleles expected to be masked by noncomplementary, mutation-specific probes had identical Tm as the wild-type allele. The two mutation-specific probes, MS 844 CTG and MS 852 ATG, were designed to detect the known sequence variation under the primary WT14B probe (data not shown).29
Figure 4.
Genotyping mutations within exon 14 and exon 16. a: Derivative melting curve plot of the primary assay probe data generated by two wild-type exon 14 probes in one reaction. The WT14A probe data are in the temperature range of 65°C to 76°C, and the WT14B probe data are in the temperature range of 76°C to 85°C. Three wild-type controls in duplicate (black traces) are shown with two heterozygous mutation samples, at codon 804(GTG→ATG) (dark gray traces) and codon 804(GTG→TTG) (light gray traces), in duplicate. The codon 804(GTG→CTG) mutation was not available. The colors used for each genotype are consistent throughout the figure. Derivative melting temperature ranges are indicated, with the mutant alleles (MUT) labeled as well as the WT allele. b: Same data as in a with additional normalization and exponential background removal as described in Materials and Methods. c–e: Graphs were also analyzed as in b. Mutant samples detected by the WT14A probe were tested in the secondary assay with the three mutation-specific probes in MS14A set 1: MS 804 ATG (c), MS 804 TTG (d), and MS 804 CTG (e). The complementary codon mutation DNA sequence was labeled as well as the WT allele and masked mutant alleles (masked “codon DNA sequence” MUT). f: Derivative melting curve plot for the exon 16 codon 918 mutation-specific probe (MS 918 ACG) primary assay data. Three wild-type controls in duplicate (black traces) are shown with a heterozygous mutation sample 918(ATG→ACG) in duplicate (gray traces). The mutant allele (918 ACG) is labeled as well as the WT allele.
Exon 16: Mutation-Specific Primary Probe
Since ∼95% of MEN2B cases are caused by the RET exon 16 codon 918(ATG→ACG) mutation, a mutation-specific probe was used in the primary assay for exon 16. The codon 918 mutant allele had a higher Tm than the wild-type allele (ΔTm −5.6°C; Figure 4f), and the amplicon data demonstrated a shift from the wild-type control (data not shown). The sequence variant at codon 922, which was under the probe, and the rare codon 912 mutation, which was within the amplicon, were unavailable for testing.20,30,31
Blinded Study
A blinded combination of five wild-type samples and 29 available RET sequence variation samples were tested by exon in the RET genotyping assay (Supplemental Table 6; http://jmd.amjpathol.org). All 59 RET sequence variations were distinguished from the 19 wild-type exons genotyped in the primary assay. In addition, the heterozygous and homozygous exon 13 polymorphism samples, as well as the exon 16 codon 918(ATG→ACG) mutation, were genotyped in the primary assay. The probe and amplicon data results were always consistent. Identical results were obtained for exons 10 and 11 whether the secondary assay mutation-specific probe sets were selected by the Tm guidelines (eg, Figure 2a) or by calculating the ΔTm for each sample and then using the ΔTm data charts (Supplemental Tables 1 through 3; http://jmd.amjpathol.org). For each mutation, only two to five probes were needed for genotyping, and no sequencing was performed. The secondary assay genotyped all of the RET mutations detected in the primary assay. Although the exon 11 codon 631 sequence variant samples were detected in the primary assay, the mutation-specific probes in the secondary assay were not designed to genotype this variant, and only these samples required sequencing for genotype.
Discussion
Although sequencing is the standard for RET genotyping, mutation scanning is more cost effective because most exons tested are wild type. Patients with apparent sporadic medullary thyroid carcinoma are tested for RET mutations, whereas only 10 to 25% actually have a germline RET mutation. Furthermore, because MEN2 is a rare autosomal dominant disorder, MEN2 patients with a RET mutation in one exon will be wild type (or carry polymorphisms) in the other RET exons.21,28 Relatives of MEN2 patients are commonly tested for RET mutations and may have a RET mutation in one exon or be wild type in all exons tested. Assuming 33% of patients tested for MEN2 mutations actually have a RET germline mutation in only one of the five exons tested, and then 93% of exons tested will be wild type or nonpathogenic polymorphism genotype.
Mutation scanning by amplicon high-resolution melting analysis can detect 100% of RET sequence variation.15 Although high-resolution amplicon melting can distinguish unique heterozygotes,33 many RET genotypes could not be distinguished because the different sequence variations had similar amplicon melting curves.15 The addition of unlabeled probes allows analysis of both amplicon and probe from the same melting curve data.17 However, some RET mutations were still not distinguished when unlabeled probes of wild-type sequence were included in the reaction. For example, the RET mutation “TAC” can be found at codon 618 or 620. Either mutation resulted in nearly identical probe melting, because the wild-type probe covered both mutations, and the nearest neighbors were identical.
To completely genotype RET mutations with a minimal number of unlabeled probes and reactions, a sequential two-stage method was developed that consisted of a primary and secondary assay. The primary assay detected mutations anywhere between the primers by amplicon melting and used unlabeled probes to analyze RET mutation hotspots. Most probes for the primary assay were of wild-type sequence. An exception was the exon 16 mutation-specific probe that complemented the common MEN2B mutation at codon 918 (∼95% of MEN2B cases).32 In addition, a primary exon 13 probe was designed to clearly differentiate the common codon 769(CTT→CTG) polymorphism from the codon 768 mutations using the masking technique.18 The majority of RET exons (93%) were genotyped as wild type or the common exon 13 polymorphism in the primary assay, using eight probes in seven PCR reactions. This primary assay also genotyped the exon 16 codon 918(ATG→ACG) mutation.
The percentage of RET mutations detected by the primary probes was estimated using the published codon mutation frequencies for each MEN2 syndrome24 and the average incidence of each MEN2 syndrome: MEN2A, 75%; FMTC, 20%; and MEN2B, 5%.32 The primary probes for exons 10, 11, 13, 14, and 16 will detect ∼98% of reported MEN2 mutations. The majority of MEN2A and FMTC mutations are in exon 11 at codon 634, and the most common mutation in MEN2A patients is codon 634 (TGC→CGC) at ∼50% frequency.24,32 There are also rare mutations (Table 1) outside of the probe analyzed region, and these will be detected by the amplicon melting analysis only and then sequenced.14 The secondary assay was used to genotype any RET sequence variation detected by the probes in the primary assay. This second stage of PCR required two to four mutation-specific probes, selected by primary assay probe data, to genotype each mutant sample. A location probe was also required for exon 10 and 11 mutations to identify the codon of the mutation, because the mutation-specific probes for exons 10 and 11 had the same mutant nucleotide change at each pathogenic codon in the probe. Less than 0.2% of RET exons tested would need sequencing due to rare sequence variation detected by amplicon melting analysis only, or the primary probe-detected sequence variant was not genotyped by the selected secondary assay probes.
In general, mutation analysis with the mutation-specific probes produced one of three results. First, when the mutant sequence complemented the mutation-specific probe, then the mutant allele melted at a higher Tm than the wild-type allele (ΔTm of −2°C to −5°C). Second, when the mutation was at the same position but did not complement the mutation-specific probe sequence, then the mutant allele was masked by a similar stability and Tm as the wild-type allele (Tm± 0.5°C of the wild-type allele Tm).18 In this case, both the noncomplementary mutant and wild-type alleles have a mismatch with the probe at the same position, but the exact nucleotides forming the mismatch are different. How close the masked allele Tm is to the wild-type allele Tm depends on the mismatched sequence, number of other mismatches with the probe, and the surrounding complementary nucleotides. For example, the A::G mismatched mutant allele melted 2°C higher than the A::C mismatched wild-type allele (Figure 2b, TCC mutant and MS 618/620 TAC probe), whereas the two mismatches C::T and C::C had identical Tms (Figure 2c, TAC mutant and MS 618/620 TCC probe). This result held true whether these mutant codon sequence changes were at RET codon 618, 620, or 609 (611 unavailable for testing). Third, when the mutation was at a different position than the mutant-specific probe sequence change, the mutant allele had an additional mismatch with the probe and melted at a lower Tm than the wild-type allele (ΔTm 0.5°C to 5°C). If this last result was obtained with all mutation-specific probes within the selected probe set for the secondary assay, then the sequence variation was not at a known mutation location under the probe and was not genotyped. Any sample not genotyped in the secondary assay would be sequenced (eg, the exon 11 codon 631 sequence variants).
Six probes were designed using the masking technique to simplify analysis of complicated regions. The masking technique uses artificial mismatches at selected sequence variation positions to allow clear analysis of other mutations elsewhere under the probe.18 The exon 13 masking probe (Mask 1bp 769) in the primary assay and the two exon 13 mutation-specific probes had a single nucleotide deletion at the common polymorphism position of codon 769 (CTT→CTG). This allowed clear detection and genotyping of the two mutations at codon 768 (GAG→GAC and GAT). The three location probes for exons 10 and 11 each used a three-nucleotide (codon) deletion to mask one pathogenic codon position. Location probes were used to identify the mutation codon position under the probe and to decrease the number of mutation-specific probes required in the secondary assay by one-half.
It is possible that known RET mutations are in cis or in trans with other sequence variation of known or unknown significance, such as in exon 14 (codons 804 and 806 mutations) or in exon 13 (codon 791 mutation with the codon 769 polymorphism).27,34,35 For probe data, if the two nucleotide changes are on the same allele, a Tm lower than either mutation alone would be expected. If the nucleotide changes are on opposite alleles, both alleles would have a lower Tm than wild type, and the sample may appear to be homozygous for mutation. In both of these cases, the samples would be sequenced, because either the variant allele was not within the ΔTm ranges predicted for RET mutations or the secondary test would not identify the genotype. If the second nucleotide change is not under the probe, but within the amplicon, a lower Tm shift or a unique amplicon derivative curve shape from the RET mutation genotyped from probe data would be expected, and the sample would be sequenced. This situation is probable for exon 13 (eg, codon 791 mutation with the codon 769 polymorphism).27 When the exon 13 amplicon data are analyzed for rare mutations not analyzed by the exon 13 primary probes, the presence of the common codon 769(CTT→CTG) polymorphism, as determined by the probe data, needs to be taken into consideration. If the sample is wild-type sequence as determined by the WT13 probe data, the sample should only be sequenced for rare mutations if the amplicon trace is shifted from the wild-type control. If the WT13 and Mask 1bp 769 probe data indicate the common codon 769 polymorphism (heterozygous or homozygous), the sample should only be sequenced if the amplicon curve is shifted from the codon 769 polymorphism control melting curves. Although extremely rare, RET mutations can be homozygous. If so, both alleles would complement the correct mutation-specific probe.
Table 1 lists the reported MEN2 mutations, polymorphisms, and sequence variants in exons 10, 11, 13, 14, and 16 that are analyzed by the RET genotyping assay. The extremely rare MEN2 mutations in exon 15, exon 8, and the recently reported exon 536 were not included and would have to genotyped by sequencing these exons. The exon 11 codon 631 sequence variants listed in Table 1 are expected to have the same result as the tested codon 631(GAC→GAT) sample: detection in the primary assay, not genotyped in the secondary assay, and then sequenced for genotype. Two common RET polymorphisms found in patients as well as the general population were not analyzed because they are not causative of the MEN2 syndromes: exon 14 p.S836S (3 to 10% allelic frequency) and exon 11 p.G691S (17 to 28% allelic frequency).21,22 The exon 11 codon 691 polymorphism was excluded by primer selection, whereas the exon 14 polymorphism was eliminated from analysis by probe selection. Two probes were used in one reaction to detect all of the known exon 14 mutations surrounding the 836 polymorphism but without detecting the polymorphism. The exon 14 amplicon melting data for the codon 836 polymorphism was similar in Tm and derivative melting curve shape to a mutation and could not be distinguished.15 Therefore the amplicon data for exon 14 was not analyzed, because the codon 836 polymorphism would then be detected but not distinguished from mutations, creating unnecessary sequencing steps.
There are other reported sequence variations within the RET proto-oncogene that are causative of Hirschsprung disease (HSCR) instead of MEN2 syndromes,20 and there is always the possibility of novel mutations. Novel or HSCR mutations detected with the primary assay probe would not be genotyped with the MEN2 mutation-specific probes in the secondary assay and would be sequenced. Novel or HSCR mutations detected by the amplicon melting data only would also be sequenced for genotype. These occurrences should be extremely uncommon, because MEN2 and HSCR have different clinical presentations.32,37 Even so, there are reports of exon 10 codon 618(TGC→CGC) and codon 620(TGC→CGC) mutations causing both MEN2 and HSCR. In addition, the three sequence variants in exon 10 codon 609(TGC→AGC and TGG) and codon 611(TGC→TCC) are associated with HSCR, not MEN2.3,19,20,38,39 With the developed RET genotyping assay, any sequence variation within the targeted exons will be genotyped, and appropriate treatment can be determined based on the genotype.
In summary, the RET genotyping assay has two PCR stages: a primary and secondary assay. The primary assay was designed so that wild-type exons are genotyped, polymorphisms are identified (or eliminated from analysis), rare mutations are detected by amplicon melting data, and common mutations are detected by both unlabeled probe and amplicon melting data, all without sequencing. The secondary assay genotypes RET mutations with a limited number of mutation-specific probes. This two-stage assay detected and genotyped all of the tested RET wild-type, mutation, polymorphism, and sequence variant samples in a blinded study with 100% accuracy in less than one-half the time required for sequencing the five RET exons. The RET genotyping assay can be used in its entirety for unknown or suspected RET mutations, by individual exon, or by using one mutation-specific probe (and location probe, if needed) when the RET mutation within the family is known.
Supplementary Material
Acknowledgments
We thank Dr. Karl Voelkerding and Maria Erali for helpful discussions and reviewing the manuscript and Lisa Collins for editing.
Footnotes
Aspects of high-resolution melting analysis are licensed from the University of Utah to Idaho Technology.
Supplemental material for this article can be found on http://jmd.amjpathol.org.
C.T.W. holds equity interest in Idaho Technology.
References
- Kambouris M, Jackson CE, Feldman GL. Diagnosis of multiple endocrine neoplasia [MEN] 2A, 2B and familial medullary thyroid cancer [FMTC] by multiplex PCR and heteroduplex analyses of RET proto-oncogene mutations. Hum Mutat. 1996;8:64–70. doi: 10.1002/(SICI)1098-1004(1996)8:1<64::AID-HUMU9>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Ceccherini I, Hofstra RM, Luo Y, Stulp RP, Barone V, Stelwagen T, Bocciardi R, Nijveen H, Bolino A, Seri M. DNA polymorphisms and conditions for SSCP analysis of the 20 exons of the ret proto-oncogene. Oncogene. 1994;9:3025–3029. [PubMed] [Google Scholar]
- Kruckeberg KE, Thibodeau SN. Pyrosequencing technology as a method for the diagnosis of multiple endocrine neoplasia type 2. Clin Chem. 2004;50:522–529. doi: 10.1373/clinchem.2003.027128. [DOI] [PubMed] [Google Scholar]
- de la Fuente M, Quinteiro C, Dominguez F, Loidi L. LightCycler PCR assay for genotyping codon 634 mutations in the RET protooncogene. Clin Chem. 2001;47:1131–1132. [PubMed] [Google Scholar]
- Ruiz A, Antinolo G, Marcos I, Borrego S. Novel technique for scanning of codon 634 of the RET protooncogene with fluorescence resonance energy transfer and real-time PCR in patients with medullary thyroid carcinoma. Clin Chem. 2001;47:1939–1944. [PubMed] [Google Scholar]
- Xue F, Yu H, Maurer LH, Memoli VA, Nutile-McMenemy N, Schuster MK, Bowden DW, Mao J, Noll WW. Germline RET mutations in MEN 2A and FMTC and their detection by simple DNA diagnostic tests. Hum Mol Genet. 1994;3:635–638. doi: 10.1093/hmg/3.4.635. [DOI] [PubMed] [Google Scholar]
- Kim IJ, Kang HC, Park JH, Ku JL, Lee JS, Kwon HJ, Yoon KA, Heo SC, Yang HY, Cho BY, Kim SY, Oh SK, Youn YK, Park DJ, Lee MS, Lee KW, Park JG. RET oligonucleotide microarray for the detection of RET mutations in multiple endocrine neoplasia type 2 syndromes. Clin Cancer Res. 2002;8:457–463. [PubMed] [Google Scholar]
- Siegelman M, Mohabeer A, Fahey TJ, III, Tomlinson G, Mayambala C, Jafari S, Noll WW, Thibodeau SN, Dawson DB. Rapid, nonradioactive screening for mutations in exons 10, 11, and 16 of the RET protooncogene associated with inherited medullary thyroid carcinoma. Clin Chem. 1997;43:453–457. [PubMed] [Google Scholar]
- Blank RD, Sklar CA, Martin ML. Denaturing gradient gel electrophoresis to diagnose multiple endocrine neoplasia type 2. Clin Chem. 1996;42:598–603. [PubMed] [Google Scholar]
- Lundquist P, Thorland E, Highsmith W, Thibodeau S, Dawson B. Evaluation of the SpectruMedix RevealTM mutation discovery system (Abstract G22). J Mol Diagn. 2003;5:260. [Google Scholar]
- Orban TI, Csokay B, Olah E. Sequence alterations can mask each other’s presence during screening with SSCP or heteroduplex analysis: bRCA genes as examples. Biotechniques. 2000;29:94–98. doi: 10.2144/00291st04. [DOI] [PubMed] [Google Scholar]
- Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 2003;49:853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, Wittwer C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem. 2004;50:1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem. 2004;50:1748–1754. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- Margraf RL, Mao R, Highsmith WE, Holtegaard LM, Wittwer CT. Mutation scanning of the RET protooncogene using high-resolution melting analysis. Clin Chem. 2006;52:138–141. doi: 10.1373/clinchem.2005.052951. [DOI] [PubMed] [Google Scholar]
- Zhou L, Myers AN, Vandersteen JG, Wang L, Wittwer CT. Closed-tube genotyping with unlabeled oligonucleotide probes and a saturating DNA dye. Clin Chem. 2004;50:1328–1335. doi: 10.1373/clinchem.2004.034322. [DOI] [PubMed] [Google Scholar]
- Zhou L, Wang L, Palais R, Pryor R, Wittwer CT. High-resolution DNA melting analysis for simultaneous mutation scanning and genotyping in solution. Clin Chem. 2005;51:1770–1777. doi: 10.1373/clinchem.2005.054924. [DOI] [PubMed] [Google Scholar]
- Margraf RL, Mao R, Wittwer CT. Masking selected sequence variation by incorporating mismatches into melting analysis probes. Hum Mutat. 2006;27:269–278. doi: 10.1002/humu.20290. [DOI] [PubMed] [Google Scholar]
- Eng C, Mulligan LM. Mutations of the RET proto-oncogene in the multiple endocrine neoplasia type 2 syndromes, related sporadic tumours, and hirschsprung disease. Hum Mutat. 1997;9:97–109. doi: 10.1002/(SICI)1098-1004(1997)9:2<97::AID-HUMU1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- Ruiz A, Antinolo G, Fernandez RM, Eng C, Marcos I, Borrego S. Germline sequence variant S836S in the RET proto-oncogene is associated with low level predisposition to sporadic medullary thyroid carcinoma in the Spanish population. Clin Endocrinol (Oxf) 2001;55:399–402. doi: 10.1046/j.1365-2265.2001.01328.x. [DOI] [PubMed] [Google Scholar]
- Elisei R, Cosci B, Romei C, Bottici V, Sculli M, Lari R, Barale R, Pacini F, Pinchera A. RET exon 11 (G691S) polymorphism is significantly more frequent in sporadic medullary thyroid carcinoma than in the general population. J Clin Endocrinol Metab. 2004;89:3579–3584. doi: 10.1210/jc.2003-031898. [DOI] [PubMed] [Google Scholar]
- Borrego S, Saez ME, Ruiz A, Gimm O, Lopez-Alonso M, Antinolo G, Eng C. Specific polymorphisms in the RET proto-oncogene are over-represented in patients with Hirschsprung disease and may represent loci modifying phenotypic expression. J Med Genet. 1999;36:771–774. doi: 10.1136/jmg.36.10.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansford JR, Mulligan LM. Multiple endocrine neoplasia type 2 and RET: from neoplasia to neurogenesis. J Med Genet. 2000;37:817–827. doi: 10.1136/jmg.37.11.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Seitz O, Marx A (Eds): Methods in Molecular Biology–Molecular Beacons–Signaling Nucleic Acid Probes. Totowa, NJ, Humana Press, (in press) [DOI] [PubMed] [Google Scholar]
- Baumgartner-Parzer SM, Lang R, Wagner L, Heinze G, Niederle B, Kaserer K, Waldhausl W, Vierhapper H. Polymorphisms in exon 13 and intron 14 of the RET proto-oncogene: genetic modifiers of medullary thyroid carcinoma ? J Clin Endocrinol Metab. 2005;90:6232–6236. doi: 10.1210/jc.2005-1278. [DOI] [PubMed] [Google Scholar]
- Berndt I, Reuter M, Saller B, Frank-Raue K, Groth P, Grussendorf M, Raue F, Ritter MM, Hoppner W. A new hot spot for mutations in the ret protooncogene causing familial medullary thyroid carcinoma and multiple endocrine neoplasia type 2A. J Clin Endocrinol Metab. 1998;83:770–774. doi: 10.1210/jcem.83.3.4619. [DOI] [PubMed] [Google Scholar]
- Demeester R, Parma J, Cochaux P, Vassart G, Abramowicz MJ. A rare variant, I852M, of the RET proto-oncogene in a patient with medullary thyroid carcinoma at age 20 years. Hum Mutat. 2001;17:354. doi: 10.1002/humu.42. [DOI] [PubMed] [Google Scholar]
- Jimenez C, Dang GT, Schultz PN, El-Naggar A, Shapiro S, Barnes EA, Evans DB, Vassilopoulou-Sellin R, Gagel RF, Cote GJ, Hoff AO. A novel point mutation of the RET protooncogene involving the second intracellular tyrosine kinase domain in a family with medullary thyroid carcinoma. J Clin Endocrinol Metab. 2004;89:3521–3526. doi: 10.1210/jc.2004-0073. [DOI] [PubMed] [Google Scholar]
- Kouvaraki MA, Shapiro SE, Perrier ND, Cote GJ, Gagel RF, Hoff AO, Sherman SI, Lee JE, Evans DB. RET proto-oncogene: a review and update of genotype-phenotype correlations in hereditary medullary thyroid cancer and associated endocrine tumors. Thyroid. 2005;15:531–544. doi: 10.1089/thy.2005.15.531. [DOI] [PubMed] [Google Scholar]
- Wiesner G, Snow-Bailey K: Multiple Endocrine Neoplasia Type 2. GeneReviews at GeneTests: Medical Genetics Information Resource. University of Washington, Seattle. Updated March 7, 2005; accessed February 2004. Available from http://www.geneclinics.org/servlet/access?db=geneclinics&site=gt&id=8888892&key=2paoNNdpDmir&gry=&fcn=y&fw=ZVFB&filename=/profiles/men2/index.html [Google Scholar]
- Graham R, Liew M, Meadows C, Lyon E, Wittwer CT. Distinguishing different DNA heterozygotes by high-resolution melting. Clin Chem. 2005;51:1295–1298. doi: 10.1373/clinchem.2005.051516. [DOI] [PubMed] [Google Scholar]
- Ahmed SA, Snow-Bailey K, Highsmith WE, Sun W, Fenwick RG, Mao R. Nine novel germline gene variants in the RET proto-oncogene identified in twelve unrelated cases. J Mol Diagn. 2005;7:283–288. doi: 10.1016/S1525-1578(10)60556-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi A, Futami H, Hai N, Yokozawa T, Kuma K, Aoki N, Kosugi S, Sugano K, Yamaguchi K. Two germline missense mutations at codons 804 and 806 of the RET proto-oncogene in the same allele in a patient with multiple endocrine neoplasia type 2B without codon 918 mutation. Jpn J Cancer Res. 1999;90:1–5. doi: 10.1111/j.1349-7006.1999.tb00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorakova S, Vaclavikova E, Duskova J, Vlcek P, Ryska A, Bendlova B. Exon 5 of the RET proto-oncogene: a newly detected risk exon for familial medullary thyroid carcinoma, a novel germ-line mutation Gly321Arg. J Endocrinol Invest. 2005;28:905–909. doi: 10.1007/BF03345322. [DOI] [PubMed] [Google Scholar]
- Parisi M: Hirschsprung Disease Overview. GeneReviews at GeneTests: Medical Genetics Information Resource. University of Washington, Seattle. Updated July 28, 2004; accessed April 2006. Available from http://www.geneclinics.org/servlet/access?db=geneclinics&site=gt&id=8888892&key=2fNQelv81Dr2o&gry=&fcn=y&fw=kx5p&filename=/profiles/hirschsprung-ov/index.html [Google Scholar]
- Mulligan LM, Eng C, Attie T, Lyonnet S, Marsh DJ, Hyland VJ, Robinson BG, Frilling A, Verellen-Dumoulin C, Safar A. Diverse phenotypes associated with exon 10 mutations of the RET proto-oncogene. Hum Mol Genet. 1994;3:2163–2167. doi: 10.1093/hmg/3.12.2163. [DOI] [PubMed] [Google Scholar]
- Mulligan LM, Eng C, Healey CS, Clayton D, Kwok JB, Gardner E, Ponder MA, Frilling A, Jackson CE, Lehnert H. Specific mutations of the RET proto-oncogene are related to disease phenotype in MEN 2A and FMTC. Nat Genet. 1994;6:70–74. doi: 10.1038/ng0194-70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.