Abstract
We report a novel TaqMan assay for JAK2 V617F that measures averaged copies per cell in absolute terms, as opposed to a ratio of mutant to wild-type alleles. Measurements were obtained by comparing the JAK2 V617F signal generated by the test samples to that generated by a set of external plasmid standards containing the sequence of interest. Specificity of the assay was demonstrated above 36 cycles of amplification, and endpoint titration experiments indicated sensitivity down to 0.05% clinical dilutions. The test measured linearly over a wide logarithmic range and exhibited good reproducibility. Combination of this assay with another TaqMan method for determining cell number allowed identification of 14 cases of myeloproliferative disease with greater than two copies per cell. Mutational frequency was 68% among polycythemia vera (n = 44), 59% (n = 37) among essential thrombocythemia and 46% (n = 13) among idiopathic myelofibrosis. Levels of the mutation were significantly higher in polycythemia vera compared with essential thrombocythemia (P = 0.0005) and correlated with the following jointly significant variables at diagnosis: PRV-1, hemoglobin, white cell count, neutrophil count, and red cell count, using multiple regression analyses (P = 0.015). This method should be useful for assessing the relationship of gene dose to phenotype and possibly for monitoring therapy.
Classification and management of myeloproliferative diseases is currently hampered by a paucity of etiopathogenic understanding and a lack of informative disease indicators. The occurrence of the JAK2 V617F mutation has been strongly linked with cases of myeloproliferative disease and promises improved molecular-based diagnosis of these disorders.1 However, it is still unclear why all patients do not carry the mutation and why there is phenotypic pleiotropy among those who do. An effect of mutational dose on disease phenotype has been indicated,2 and it is possible that this accounts for some of the biological spectrum at presentation. Other variations in disease phenotype may potentially be explained by the presence of activating mutations in other receptor tyrosine kinases such as the MPL thrombopoietin receptor.3 To test whether detection and quantitation of the JAK2 mutation is useful for the management of myeloproliferative disease, we have developed a TaqMan-based assay that reports the absolute number of JAK2 V617F per cell averaged in a population of cells. Here, we describe the performance characteristics of the assay and report JAK2 V617F copy number per cell among controls (n = 18) and among individuals referred for hematological assessment (n = 147) with possible myeloproliferative disease.
Materials and Methods
Patients
Individuals (n = 147) who had been referred for clinical assessment with suspected or previously diagnosed myeloproliferative disorders to the Department of Hematology at Pathwest in Western Australia were enrolled in the study. The study included 38 individuals with polycythemia vera (PV), 37 individuals with essential thrombocythemia (ET), and six with idiopathic myelofibrosis (IMF), diagnosed according to the World Health Organization criteria.4 Samples from deidentified healthy controls (n = 18) were obtained from the Red Cross Blood Bank.
Granulocyte Preparation, DNA and RNA Extraction, and cDNA Synthesis
Blood sampling was performed after informed consent either at diagnosis or during the follow-up. Granulocytes were separated by differential centrifugation using a density gradient (Polymorphprep; Axis Shield, Oslo, Norway), starting from 30 ml of peripheral blood collected in ethylenediaminetetraacetic acid within 24 hours from sampling. When necessary, contaminating red cells were removed by hypotonic lysis. Cell pellets, which contained >98% of granulocytes by visual inspection of cytospins, were extracted for DNA or RNA using the QIAmp DNA Blood Mini kit (Qiagen, Valencia, CA) and RNeasy Mini kit (Qiagen), respectively, both according to the manufacturer’s protocol. Concentrations were determined using NanoDrop technology (Biolab, Auckland, New Zealand). cDNA was reverse transcribed starting from 100 ng of RNA with oligo dT primers using Omniscript reverse transcriptase (Qiagen) according to the recommended protocol. For three patients with high granulocytic mutational load, we also measured mutational load from CD14+ or CD3+ cells using immunomagnetic beads (Invitrogen, Perth, Australia).
Quantitative Determination of JAK2 V617F by TaqMan
A polymerase chain reaction (PCR) assay using TaqMan (fluorescence-based, real-time PCR) and minor groove binding probes was designed for the specific and quantitative determination of DNA copy number of JAK2 V617F in clinical samples. The 7700 Sequence Detection System (ABI, Melbourne, Australia) amplifies and monitors an increase in fluorescence generated by binding of the allele-specific probe during the PCR cycle. In the intact probe, the quencher absorbs fluorescence emitted by the reporter. The 5′ nuclease activity of the polymerase degrades the hybridization probe during replication, thereby releasing the reporter and producing an increase in fluorescent emission. The point at which the fluorescent signal rises above the background, or the crossing threshold (Ct), is directly proportional to the input DNA and is thus a measure of the starting template. The test uses a forward primer that has been designed with deliberate mismatches to specifically amplify the mutant sequence.
Twenty nanograms of DNA was added to MicroAmp Reaction Plate (ABI) tubes containing 20 μl of reaction mix. The working master mix contained 5 pmol of mutant-specific forward primer 5′-AGCATTTGGTTTTAAATTATGGAGTATaTT-3′ containing a deliberate mismatch and 5 pmol of non-allele-specific reverse primer 5′-CAAAAACAGATGCTCTGAGAAAGG-3′, 2× Taqman Universal Master mix [containing heat-activated AmpliTaq Gold DNA polymerase, AmpErase uracil-N-glycosylase for carryover prevention, dNTPs with dUTP, Passive Reference dye (Rox), and buffer components; ABI], and 5 pmol of a 5-carboxyfluorescein (FAM) fluorescently labeled probe specific for the reverse strand of the mutant allele, 5′-fam-ctccacagaaacatactc-3′.
JAK2 V617F amplifications were performed using the following cycling conditions. One cycle at 50°C for 2 minutes of incubation was performed for activation of AmpErase uracil-N-glycosylase, followed by a 10-minute incubation at 95°C for amplification of AmpliTaq Gold, then 40 cycles of denaturation at 95°C (15 seconds), and an annealing/extension step at 60°C for 1 minute. Reactions were kept on hold at 4°C. All samples and standards were run in triplicate.
Copy number for JAK2 V617F was calculated in absolute units by comparing the signal generated by the test samples to that generated by a set of external plasmid standards containing the sequence of interest. The stock plasmid standard was created by ligating a PCR product containing the JAK2 V617F sequence into a plasmid vector system (Zero Blunt Topo; Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. The amount of plasmid DNA was determined by spectrophotometric analyses of the insert-containing plasmid DNA at A260 (1 optical density = 50 μg/ml plasmid DNA), and the copy number per milliliter was determined based on molecular weight. Dilutions of the plasmid ranged from 10 to 300,000 copies per reaction, and quantitative determination for clinical samples was determined by reading from this standard curve.
Quantitative Determination of Cell Number by TaqMan
The number of cells per reaction was determined in a separate real-time PCR reaction using a previously validated assay for measuring the number of copies of a single-copy region of the human growth hormone gene (HGH).5,6 Quantitative amplification of this highly conserved region of the nuclear-encoded human growth hormone gene used primer sets and probes designed using Primer Express software (ABI) according to primer design guidelines. The reaction used forward primer 5′-TATCCCAAAGGACAGAAGTATTCATT-3′ and reverse primer 5′-TTGTGTTTCCCTCCCTGTTGGA-3′ to amplify a 141-nucleotide product. The fluorophore-labeled probe used was VIC-ACCTCCCTCTGTTTCTCAGAGTCTATTCCGACA-quencher TAMRA-3′.
Amplification of the HGH product was performed separately to the JAK2 V617F assay using another aliquot of the same DNA and using cycling conditions identical to that of the JAK2 V617F assay. Each 20-μl reaction contained 20 ng of DNA, and the final concentration of each component was as follows: 2.5 pmol of probe, 40 pmol of forward primer, 40 pmol of reverse primer, and 2× Taqman Universal Master mix (ABI). All samples and standards were run in duplicate. Determination of the HGH copy number was similar to that of the JAK2 V617F assay, in that known dilutions of a plasmid containing the HGH sequence of interest were measured to generate a standard curve, from which quantitative determination of HGH copy number in clinical samples could be made. The number of cells per reaction was calculated by dividing HGH copy number by 2, based on every nucleated cell carrying two copies of the HGH sequence.
Quantitative Determination of JAK2 V617F per Cell
Determination of JAK2 V617F per cell was achieved by normalizing JAK2 V617F per PCR reaction (20 ng of input DNA) to cell number per PCR reaction (20 ng of input DNA). Because the measured fluorescence in the quantitative PCR assay is directly proportional to the (base 10) logarithm of the initial DNA content in the sample, results were expressed and analyzed in logarithmic form. Hence, calculations of JAK2 V617F/cell from the JAK2 V617F and nuclear quantitative PCR assay results used the formula log10JAK2 V617F DNA/cell = log10JAK2 V617F DNA − log10nDNA − log100.5. Final results were reported in logarithmic form as copies of JAK2 V617F per cell (or per 20 ng of input DNA).
Detection of JAK2 V617F by Sequence Analysis
To confirm the specificity of the real-time quantitative PCR for the G to T mutation, paired samples (n = 17) of granulocyte and lymphocyte populations were analyzed by sequencing reaction. Amplifications for 40 cycles used Platinum Taq polymerase (Invitrogen), 2 mmol/L MgSO4, and 10 pmol each of sense and antisense primers (forward primer 5′-TGTAAAACGACGGCCAGTATCTATGTCATGCTGAAAGTAGGAGAAAG-3′ and reverse primer 5′-CAGGAAACAGCTATGACCCTGAATAGTCCTACAGTGTTTTCAGTTTCA-3′). PCR sequencing reactions used Big Dye terminator chemistry and M13 reverse primer and forward primer 5′-ttccttagtctttctttgaagc-3′. Products were resolved on an ABI Prism 3730 48 capillary sequencer. Typing for the mutation in JAK2 exon 12 was made with alignment of test sequence to a reference sequence in GenBank (accession NM_00497215) using Conexio software (Perth, Australia).
Dilution Method for Determining Sensitivity of JAK2 V617F Assay
DNA from an individual with polycythemia vera carrying 1.8 copies of the JAK2 mutation per cell as determined by our assay was diluted with DNA from a healthy control to provide mixtures of 100, 95, 80, 50, 20, 5, 1, 0.5, and 0.05%. Taqman JAK2 V617F measurement of duplicate dilutions was performed and used to assess the sensitivity of the assay to detect dilutions of the mutation.
Dilution Method for Determining Specificity of JAK2 V617F Assay
DNA (sample A) from a healthy control carrying 0.007 copies of the JAK2 mutation per cell as determined by our assay was diluted with DNA (sample B) from another healthy control carrying 0.006 copies of the JAK2 mutation per cell to provide mixtures of 100, 75, 25, and 0%. Similar dilutions of sample A were generated with distilled water instead of DNA. TaqMan JAK2 V617F measurement of triplicate dilutions was performed and used to assess the specificity of the assay.
Quantitative Determination of PRV-1 Overexpression by TaqMan
Neutrophil PRV-1 expression levels were quantified similarly to the method described by Klippel et al7 using TaqMan and glyceraldehyde-3-phosphate dehydrogenase as the housekeeping gene. PRV-1 was amplified from cDNA using 20 pmol of forward primer 5′-CTGCGTGGCCCAACCTT-3′ and reverse primer 5′-GCAGAGAAGATCCCGATTTGTCT-3′ and detected using 3.5 pmol of a FAM-labeled probe 5′-CCAGCTTCTTGTTGAAC-3′ in a 20-μl reaction with 2× Universal PCR Master Mix (ABI) using real-time quantitative PCR on the ABI 7700 SDS. Amplification was performed in triplicate samples in a two-step cycle (denaturation, 95°C for 15 seconds, annealing/extension at 62°C for 60 seconds) for 40 cycles. Data collected during cycling were analyzed with the FAM threshold set at 0.05.
Each glyceraldehyde-3-phosphate dehydrogenase reaction used another aliquot of the same cDNA and consisted of 20-μl reactions with 2× Universal MasterMix (ABI) and 20× glyceraldehyde-3-phosphate dehydrogenase premix control reagent (ABI). Reactions were performed in triplicate, as for the PRV-1 assay. Data collected during cycling were analyzed with the VIC threshold set at 0.05. A mean Ct value for each triplicate measurement was calculated. The PRV-1 Ct ratio for each sample was calculated from the mean value of triplicate PRV-1 Ct determinations divided by the mean glyceraldehyde-3-phosphate dehydrogenase Cts.
Results
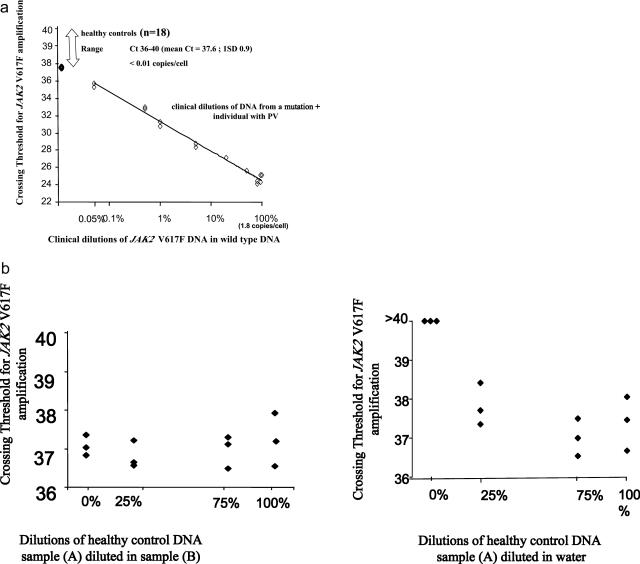
The JAK2 V617F assay was measured linearly over five log10 dilutions of the plasmid standard and demonstrated sensitivity to 0.05% clinical dilution (Figure 1a). Fluorescent detection of 20 ng of undiluted input template DNA from controls (n = 18) was detectable only after 35 cycles of amplification. To test whether fluorescence occurring after 35 cycles was an artifact of the assay arising from nonspecific priming, we diluted DNA from a healthy control (sample A) in water and also in DNA from another control (sample B). As shown in Figure 1b, triplicate measures of different dilutions showed a poor relationship with mutational load, suggesting that nonspecific amplification occurs after 36 cycles of PCR.
Figure 1.
a: Specificity versus sensitivity. The assay demonstrated sensitivity to 0.05% clinical dilution. Determining the specificity of the assay is hampered by the fact that it is not known whether healthy controls carry low levels of the mutation. However, we can say that amplification of the mutation in controls occurred only after the number of cycles required to detect a 0.05% dilution of the positive sample. b: Measurement of JAK2 V617F copies per reaction in mixtures of DNA from two healthy controls demonstrated a poor relationship between crossing threshold values and dilutions of DNA in water (right) and in DNA from other control sample (left), suggesting nonspecific amplification.
An optimally efficient PCR reaction yields a slope of −3.3 on the standard curve, with the y intercept values (theoretical value for detection of one template molecule) between 36.1 and 45.99. Assessment of our assay performance over 15 runs showed that the efficiency of amplification was 98%, with a slope of −3.29 (mean value; range, −3.16 to −3.46). Average y intercept was 40.4 with a SD of 0.6. The correlation coefficient (r2 value) of standard curves was 0.97 overall, with a range of 0.94 to 0.99. Precision (coefficient of variation of replicate values of the crossing threshold) was relatively constant across dilutions (ranging from 0.8 to 2.2% and not associated with concentration) for clinical samples and for the plasmid stock. Intra-assay precision (averaged over runs; n = 15), as assessed by determining the coefficient of variation of all triplicate measurements within an assay averaged 1.4% for samples, 1.8% for a positive control, and 1.1% for a negative control created by pooling DNA from all healthy controls. Interassay variation (based on the coefficient of variation of log10 values; n = 15 assays) was 4% for the positive control and 25% for the negative control.
Samples of DNA from paired granulocyte (n = 17) and lymphocyte (n = 17) preparations were sequenced for semiquantitative detection of the mutation. All lymphocyte DNA failed to demonstrate the mutation by sequencing. The assay showed 100% concordance with sequence-based typing for detecting the presence of the mutation in granulocyte DNA samples with at least 0.08 copies per cell (n = 14). Three samples of granulocyte DNA with low mutational load showed no mutation by sequence-based typing, presumably because of limitations in assay sensitivity.
Quantitative determination of JAK2 V617F in monocytes and granulocyte preparations were similar for three individuals with high levels of the mutation. Measurements in lymphocyte fractions were just above those observed in healthy controls, presumably because of contamination by the myeloid lineage (data not shown). Amplifications in the granulocyte fraction occurred 11 to 12 cycles before detectable amplification in the lymphocyte fraction, indicating good discrimination between amplification of mutant wild-type alleles.
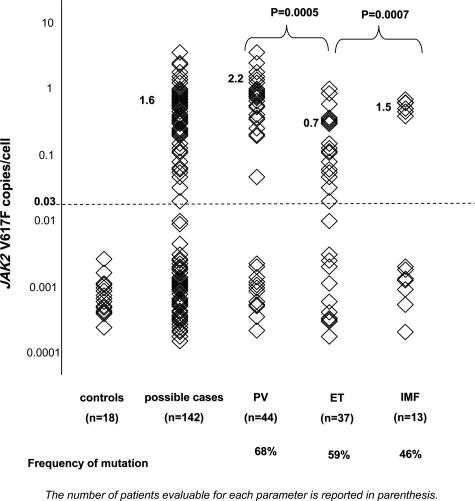
Variation in JAK2 V617F copies per cell among individuals referred for mutational typing and among 18 controls is given in Figure 2. We observed a strongly bimodal distribution of the data for cases that grouped lower levels of mutation in the healthy controls and a number of the cases separately from logarithmically higher levels of mutation in a subset of the cases. Restricting to values that fell in the higher half of this bimodal distribution, mutational load was significantly increased among cases of PV compared with ET (P < 0.001). A small number of cases of IMF (n = 7) showed increased mutation load compared with cases of ET (P < 0.001). The frequency of the mutation, based on assigning the occurrence of the mutation to cases with mutation load above the threshold defining the bimodal distribution is given in Figure 2 and was within previously reported ranges.
Figure 2.
Distribution of JAK2 V617F copies per cell among cases and controls.
For 13 cases with PV and one case with ET, we measured more than two copies of JAK2 V617F per cell. Of these 14 individuals, only two had cytogenetic data that did not identify chromosomal abnormalities. These patients tended to have grossly hypercellular bone marrows, and most were being treated with hydroxy urea.
Clinical associations with occurrence of JAK2 V617F (based on assigning occurrence of mutation to levels of mutation load in the higher bimodal distribution) among myeloproliferative disease (MPD) cases (n = 93) of PV, ET, or IMF that we detected were restricted to increases in white cell and neutrophil counts at diagnosis and on treatment (Table 1).
Table 1.
Clinical Associations with Occurrence of JAK2 V617F Among MPD Cases (n = 93) of PV, ET, or IMF
| Laboratory and clinical features | V617F present (n = 56) | V617F absent (n = 37) | P value (Mann-Whitney) |
|---|---|---|---|
| PRV-1 overexpression | 0.96 (25) | 0.92 (32) | 0.272 |
| Hb at diagnosis (g/L) | 162 (49) | 151 (30) | 0.185 |
| WCC at diagnosis (×109/L) | 11.4 (14) | 8.1 (16) | <0.0005 |
| Neutrophil at diagnosis (×109/L) | 8.4 (44) | 5.5 (25) | 0.005 |
| MCV at diagnosis | 89 (41) | 92 (29) | 0.20 |
| RCC at diagnosis (×109/L) | 5.7 (43) | 5.3 (25) | 0.67 |
| Platelets at diagnosis (×109/L) | 549 (47) | 517 (30) | 0.22 |
| Log mtDNA/ cell on treatment | 1.8 (34) | 1.8 (27) | 0.99 |
| WCC on treatment (×109/L) | 17 (9) | 7 (6) | 0.026 |
The number of data points evaluated for each parameter is reported in parentheses. Hb, hemoglobin; WCC, white cell count; MCV, mean corpuscular volume; RCC, red cell count.
Multiple regression analyses identified jointly significant (P = 0.015) variables at diagnosis as PRV-1, hemoglobin, white cell count, neutrophil count, and red cell count. These variables were also significant in univariate analyses with the exception of PRV-1 overexpression, which required adjustment of other variables before reaching significance. Although platelet count was significantly negatively associated with mutational load in univariate analyses (P = 0.001), this significance was lost after accounting for other variables (Table 2).
Table 2.
Associations with Levels of JAK2 V617F
| Laboratory features at diagnosis | Spearman rank correlations | P value (univariate) |
|---|---|---|
| Hemoglobin (g/L) | 0.397 | 0.005 |
| Hematocrit | 0.385 | 0.02 |
| Neutrophil count (×109/L) | 0.439 | 0.003 |
| Platelet count (×109/L) | −0.488 | 0.001 |
| White cell count (×109/L) | 0.274 | 0.06 |
n = 56 cases MPD+ mutation+.
Discussion
The aim of the current study was to develop and validate a method for measuring the number of JAK2 V617F mutations in samples from patients presenting with possible myeloproliferative diseases. Although there are already a wide range of molecular-based approaches for achieving this,8,9,10,11,12,13,14,15 we wished to develop a system that would provide information of copy number per cell in absolute terms. In this respect, our assay represents an advance in providing absolute rather than relative quantitation and in enabling detection of more than two copies per cell.
The precision and efficiency of the PCR is similar to that attained for real-time PCR quantitation of the Philadelphia chromosome (BCR-ABL) transcript,16 and the method lends itself to high throughput and cheap and readily available set-up. The sensitivity is favorably comparable with pyrosequence and amplification refractory mutation system-based methods.17,18,19 The performance characteristics of the assay presented here seem very similar to those reported recently15 in which locked nucleic acids are used with molecular beacons to quantify the mutant allele in a real-time PCR assay, with the notable exception that in our study, we have shown low-level detection of the mutation in control samples to be an artifact of the system.
Mutational load among healthy controls was extremely low, and as previously stated, we assumed that this represented nonspecific amplification of the wild-type allele because of mispriming or the presence of 3′-shortened mutation-specific primers (because it is only the very 3′-end base that confers specificity and 3′-shortened primers will amplify from both wild-type and mutant DNA). Groups using real-time PCR assays have previously reported that nonspecific amplification of a negative allele occurs after a certain number of cycles,20 and our assay also does not maintain specificity at these low levels of detection. For the purpose of diagnosing myeloproliferative disease, we set a threshold for reporting the presence of the mutation based on the ability to discriminate healthy controls from individuals with disease and based on the ability of the assay to show a linear relationship with dilutions of the mutation. Using this approach, we report a mutational frequency that is consistent with previous publications,8 although lower than might be expected for cases of PV, given the high sensitivity of the assay. One possible explanation may relate to differences in the ascertainment of cases of PV for different studies.
We chose to measure JAK2 V617F levels in granulocytes in an effort to maximize sensitivity of detection. However, our data indicate that the assay would easily detect the presence of the mutation in whole buffy coats, bone marrow aspirates, or mononuclear populations.
Our study detected an average of more than two copies per cell in a number of individuals who were diagnosed MPD (Figure 2). The veracity of these high mutational loads requires confirmation by an alternative approach. However, currently, our method is uniquely designed to detect changes in copy number greater than two, essentially because it is not based on ratios of wild-type to mutant alleles. Although probably unlikely, a copy number of greater than two copies per cell could also be due to loss of an HGH allele, and confirmation using an unrelated marker at a different location should be pursued. Despite these limitations, the study provides supporting data for a hypothesis that disease evolution involves further gene duplication associated with genetic instability. Homozygosity in the JAK2 mutation has been reported to result from uniparental disomy. However, we require an alternative model to explain more than two copies per cell. One possibility is that in-frame internal tandem duplication occurs, similar to that reported for the mutation in FLT-3.21
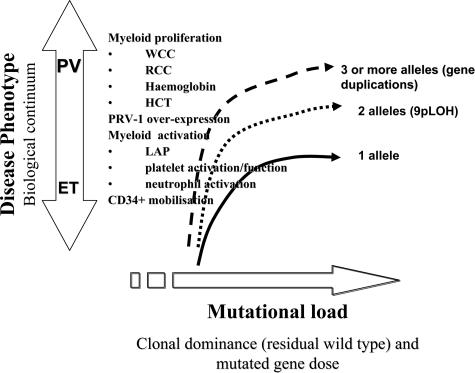
Interpreting the clinical relevance of mutational load to disease evolution and phenotype variation within the spectrum of myeloproliferative disorders represents the next significant challenge. A continuum model22 would predict significant correlations between clinical phenotype and mutational load that extends across current classifications of essential thrombocythemia and polycythemia vera (Figure 3). Our investigation certainly shows that cross-sectional measures of mutational load are significantly different according to disease phenotypes observed in MPD. We also observe a significant positive relationship between mutational load with PRV-1 overexpression and with myeloproliferative markers, consistent with the findings of Vannucchi et al18 and Lippert et al.23 The Lippert et al group also reported significant correlations between JAK2 V617F levels and leukocyte counts. However, unlike our study, they did not find that mutational levels at diagnosis were linked with hemoglobin level, hematocrit level, or platelet counts. Further investigation that accounts for these differences across studies is therefore required. If, as our data indicate, mutational load does reflect myeloproliferative aspects of PV and ET phenotype, then its measurement would provide a useful tool for monitoring disease progression and for providing insights into the molecular process of disease. Potentially, this type of assay could be similar to the way that real-time PCR quantitation of BCR-ABL mRNA in the blood of patients with chronic myeloid leukemia has become the predominant molecular monitoring technique. Other studies have indicated that the mutation load is also correlated with increases in a number of activation markers of myeloid cells, although exactly how these cellular activations relate to thrombo-hemorrhagic and fibrotic complications is still unclear.18,24
Figure 3.
Relationship between mutational load and disease phenotype. In terms of a pathogenic model, our data support a significant relationship between mutational load and the different disease phenotypes observed in MPD. Our data have also demonstrated a significant positive relationship between mutational load and myeloproliferative markers and PRV-1 overexpression. Other studies have indicated that the mutation load is also correlated with increases in a number of activation markers of myeloid cells, although exactly how these cellular activations relate to thrombo-hemorrhagic and fibrotic complications is still unclear.
A next step toward an enhanced understanding of the pathogenesis of MPD and improvements in MPD management may involve the development of specific JAK2 inhibitors (as for imatinib for chronic myeloid leukemia). In this setting, a standardized approach to molecular monitoring with the potential for translation into routine clinical practice will become increasingly important for evaluating new therapies.
In summary, the data indicate that the TaqMan quantitative JAK2 V617F test provides an attractive approach for monitoring disease progression, the efficacy of curative interventions, or early indications of emerging drug resistance. Further investigation is now needed to determine whether variation in this measurement can elucidate the pathogenic mechanism and contribute to clinical management decisions.
Footnotes
Supported by the Ray and Bill Dobney Trust.
References
- James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, Garcon L, Raslova H, Berger R, Bennaceur-Griscelli A, Villeval JL, Constantinescu SN, Casadevall N, Vainchenker W. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- Schafer AI. Molecular basis of the diagnosis and treatment of polycythemia vera and essential thrombocythemia. Blood. 2006;107:4214–4222. doi: 10.1182/blood-2005-08-3526. [DOI] [PubMed] [Google Scholar]
- Moliterno AR, Williams DM, Rogers O, Spivak JL. Molecular mimicry in the chronic myeloproliferative disorders: reciprocity between quantitative JAK2 V617F and Mpl expression. Blood. 2006;108:3913–3915. doi: 10.1182/blood-2006-03-008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100:2292–2302. doi: 10.1182/blood-2002-04-1199. [DOI] [PubMed] [Google Scholar]
- Hammond E, Sayer D, Nolan D, Walker U, deRonde A, Montaner J, Cote H, Gahan M, Cherry CL, Wesselingh S, Reiss P, Mallal S. Assessment of precision and concordance of quantitative mitochondrial DNA assays: a collaborative international quality assurance study. J Clin Virol. 2003;27:97–110. doi: 10.1016/s1386-6532(02)00134-8. [DOI] [PubMed] [Google Scholar]
- Nolan D, Hammond E, James I, McKinnon E, Mallal S. Contribution of nucleoside-analogue reverse transcriptase inhibitor therapy to lipoatrophy from the population to the cellular level. Antivir Ther. 2003;8:617–626. [PubMed] [Google Scholar]
- Klippel S, Strunck E, Temerinac S, Bench AJ, Meinhardt G, Mohr U, Leichtle R, Green AR, Griesshammer M, Heimpel H, Pahl HL. Quantification of PRV-1 mRNA distinguishes polycythemia vera from secondary erythrocytosis. Blood. 2003;102:3569–3574. doi: 10.1182/blood-2003-03-0919. [DOI] [PubMed] [Google Scholar]
- Steensma DP. JAK2 V617F in myeloid disorders: molecular diagnostic techniques and their clinical utility. A paper from the 2005 William Beaumont Hospital Symposium on Molecular Pathology. J Mol Diagn. 2006;8:397–411; quiz 526. doi: 10.2353/jmoldx.2006.060007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khwaja A. The role of Janus kinases in haemopoiesis and haematological malignancy. Br J Haematol. 2006;134:366–384. doi: 10.1111/j.1365-2141.2006.06206.x. [DOI] [PubMed] [Google Scholar]
- Murugesan G, Aboudola S, Szpurka H, Verbic MA, Maciejewski JP, Tubbs RR, Hsi ED. Identification of the JAK2 V617F mutation in chronic myeloproliferative disorders using FRET probes and melting curve analysis. Am J Clin Pathol. 2006;125:625–633. doi: 10.1309/TK0X-L917-XK2V-LRPQ. [DOI] [PubMed] [Google Scholar]
- Olsen RJ, Tang Z, Farkas DH, Bernard DW, Zu Y, Chang CC. Detection of the JAK2(V617F) mutation in myeloproliferative disorders by melting curve analysis using the LightCycler system. Arch Pathol Lab Med. 2006;130:997–1003. doi: 10.5858/2006-130-997-DOTJMI. [DOI] [PubMed] [Google Scholar]
- Lay M, Mariappan R, Gotlib J, Dietz L, Sebastian S, Schrijver I, Zehnder JL. Detection of the JAK2 V617F mutation by LightCycler PCR and probe dissociation analysis. J Mol Diagn. 2006;8:330–334. doi: 10.2353/jmoldx.2006.050130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure R, Mai M, Lasho T. Validation of two clinically useful assays for evaluation of JAK2 V617F mutation in chronic myeloproliferative disorders. Leukemia. 2006;20:168–171. doi: 10.1038/sj.leu.2404007. [DOI] [PubMed] [Google Scholar]
- Stevenson WS, Hoyt R, Bell A, Guipponi M, Juneja S, Grigg AP, Curtis DJ, Scott HS, Szer J, Alexander WS, Tuckfield A, Roberts AW. Genetic heterogeneity of granulocytes for the JAK2 V617F mutation in essential thrombocythaemia: implications for mutation detection in peripheral blood. Pathology. 2006;38:336–342. doi: 10.1080/00313020600820906. [DOI] [PubMed] [Google Scholar]
- Sidon P, Heimann P, Lambert F, Dessarsi B, Robin Y, Housni HE. Combined locked nucleic acid and molecular beacon technologies for sensitive detection of the JAK2V617F somatic single-base sequence variant. Clin Chem. 2006;52:1436–1438. doi: 10.1373/clinchem.2006.066886. [DOI] [PubMed] [Google Scholar]
- Branford S, Hughes T. Diagnosis and monitoring of chronic myeloid leukemia by qualitative and quantitative RT-PCR. Methods Mol Med. 2006;125:69–92. doi: 10.1385/1-59745-017-0:69. [DOI] [PubMed] [Google Scholar]
- Jones AV, Kreil S, Zoi K, Waghorn K, Curtis C, Zhang L, Score J, Seear R, Chase AJ, Grand FH, White H, Zoi C, Loukopoulos D, Terpos E, Vervessou EC, Schultheis B, Emig M, Ernst T, Lengfelder E, Hehlman R, Hochhaus A, Oscier D, Silver RT, Reiter A, Cross NCP. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood. 2005;106:2162–2168. doi: 10.1182/blood-2005-03-1320. [DOI] [PubMed] [Google Scholar]
- Vannucchi AM, Pancrazzi A, Bogani C, Antonioli E, Guglielmelli P. A quantitative assay for JAK2V617F mutation in myeloproliferative disorders by ARMS-PCR and capillary electrophoresis. Leukemia. 2006;20:1055–1060. doi: 10.1038/sj.leu.2404209. [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Scott LM, Buck G, Wheatley K, East CL, Marsden JT, Duffy A, Boyd EM, Bench AJ, Scott MA, Vassiliou GS, Milligan DW, Smith SR, Erber WN, Bareford D, Wilkins BS, Reilly JT, Harrison CN, Green AR, United Kingdom Myeloproliferative Disorders Study Group, Medical Research Council Adult Leukaemia Working Party, Australasian Leukaemia and Lymphoma Group Definition of subtypes of essential thrombocythaemia and relation to polycythaemia vera based on JAK2 V617F mutation status: a prospective study. Lancet. 2005;366:1945–1953. doi: 10.1016/S0140-6736(05)67785-9. [DOI] [PubMed] [Google Scholar]
- Maas F, Schaap N, Kolen S, Zoetbrood A, Bun I, Dolstra H, de Witte T, Schattenberg A, van de Wiel-van Kemenade E. Quantification of donor and recipient hemopoietic cells by real-time PCR of single nucleotide polymorphisms. Leukemia. 2003;17:630–633. doi: 10.1038/sj.leu.2402857. [DOI] [PubMed] [Google Scholar]
- Medeiros BC, Zhang T, Lipton JH, Kamel-Reid S: Absence of FTL3 mutations in patients with JAK2(V617F) mutation negative essential thrombocythemia. Am J Hematol 2006, [Epub ahead of Print] [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Green AR. Management of polycythemia vera and essential thrombocythemia. Hematology Am Soc Hematol Educ Program. 2005:201–208. doi: 10.1182/asheducation-2005.1.201. [DOI] [PubMed] [Google Scholar]
- Lippert E, Boissinot M, Kralovics R, Girodon F, Dobo I, Praloran V, Boiret-Dupre N, Skoda RC, Hermouet S. The JAK2–V617F mutation is frequently present at diagnosis in patients with essential thrombocythemia and polycythemia vera. Blood. 2006;108:1865–1867. doi: 10.1182/blood-2006-01-013540. [DOI] [PubMed] [Google Scholar]
- Bellucci S, Michiels JJ. The role of JAK2 V617F mutation, spontaneous erythropoiesis and megakaryocytopoiesis, hypersensitive platelets, activated leukocytes, and endothelial cells in the etiology of thrombotic manifestations in polycythemia vera and essential thrombocythemia. Semin Thromb Hemost. 2006;32:381–398. doi: 10.1055/s-2006-942759. [DOI] [PubMed] [Google Scholar]