Abstract
The COBAS AmpliPrep instrument (Roche Diagnostics GmbH, D-68305 Mannheim, Germany) automates the entire sample preparation process of nucleic acid isolation from serum or plasma for polymerase chain reaction analysis. We report the analytical performance of the LightCycler Parvovirus B19 Quantification Kit (Roche Diagnostics) using nucleic acids isolated with the COBAS AmpliPrep instrument. Nucleic acids were extracted using the Total Nucleic Acid Isolation Kit (Roche Diagnostics) and amplified with the LightCycler Parvovirus B19 Quantification Kit. The kit combination processes 72 samples per 8-hour shift. The lower detection limit is 234 IU/ml at a 95% hit-rate, linear range approximately 104-1010 IU/ml, and overall precision 16 to 40%. Relative sensitivity and specificity in routine samples from pregnant women are 100% and 93%, respectively. Identification of a persistent parvovirus B19-infected individual by the polymerase chain reaction among 51 anti-parvovirus B19 IgM-negative samples underlines the importance of additional nucleic acid testing in pregnancy and its superiority to serology in identifying the risk of parvovirus B19 transmission via blood or blood products. Combination of the Total Nucleic Acid Isolation Kit on the COBAS AmpliPrep instrument with the LightCycler Parvovirus B19 Quantification Kit provides a reliable and time-saving tool for sensitive and accurate detection of parvovirus B19 DNA.
Beside driving seminal advances in uncovering and understanding gene function in all areas of life, polymerase chain reaction (PCR) analysis has improved health care by the remarkable sensitivity and specificity of its ability to detect viral pathogens in body fluids and tissues. Modern PCR thermal cyclers are heavily automated, but most assays have, up to now, required extensive hands-on time due to labor-intensive nucleic acid isolation from the sample.1
The COBAS AmpliPrep instrument (Roche Diagnostics GmbH, D-68305 Mannheim, Germany) was recently introduced to expand automation by isolating target sequences using biotinylated sequence-specific capture probes along with streptavidin-coated magnetic particles.1,2 It has lately been joined by an additional versatile tool, the Total Nucleic Acid Isolation (TNAI) Kit (Roche Diagnostics). This laboratory-use reagent allows the generic, not sequence-specific, isolation of all nucleic acids from plasma and serum on the COBAS AmpliPrep instrument based essentially on the method developed by Boom et al.3 We evaluated the analytical performance of this system using the LightCycler Parvovirus B19 Quantification Kit for PCR amplification (Roche Diagnostics).
Parvovirus B19 infection is a common childhood illness which usually runs a mild course in immunocompetent individuals, producing a characteristic rash known as erythema infectiosum or fifth disease.4 Infection may be complicated by severe arthralgia or a transient aplastic crisis in individuals suffering from chronic hemolytic disease.5 Congenital anemia and vasculitis have also been described.6 More recently the virus has been associated with hepatitis and myocarditis.7,8,9 Following maternal infection in pregnancy, the virus may be transmitted to the fetus, causing hydrops, spontaneous abortion, or intrauterine death.10 Besides transmission via the respiratory route, parvovirus B19 infection may also occur through contaminated blood and blood products.11 The latter has been recognized by the United States Food and Drug Administration, resulting in a proposal for parvovirus B19 nucleic acid testing (NAT) to be regarded as in-process testing rather than donor screening (www.fda.gov).
We present below data showing that the combination of the TNAI Kit, COBAS AmpliPrep instrument, and LightCycler Parvovirus B19 Quantification Kit provides a reliable and time-saving tool for sensitive and accurate parvovirus B19 DNA detection in the research laboratory.
Materials and Methods
Sample Material
Analytical performance data were generated using dilution series of either a parvovirus B19 DNA high-positive plasma donation (Transfusionszentrale, D-55131 Mainz, Germany) or the World Health Organization Standard (National Institute for Biological Standards and Control [NIBSC] first International Standard 2000 Parvovirus B19 DNA 500000 IU/ml; Code 99/800, South Mimms, UK). Dilutions were made in parvovirus B19 DNA-negative human EDTA plasma, citrate plasma, or serum. Research samples were derived mainly from pregnant women and were fully tested for the presence of anti-parvovirus B19 antibodies (Parvovirus B19 IgG EIA and Parvovirus B19 IgM EIA, Biotrin, Dublin, Ireland).
Nucleic Acid Testing
Fully automated preparation of viral nucleic acids was performed on the COBAS AmpliPrep instrument using the TNAI Kit according to the manufacturer’s instructions. In brief, samples were aliquoted into sample tubes (desired input volume plus 150 μl dead volume, chosen from the options between 50 and 850 μl) and placed in the instrument together with the TNAI Kit cassettes containing all necessary reagents. The amount of internal control (IC) was adjusted to 3.1 μl per 50 μl QS diluent. The resulting eluates were then analyzed directly or stored in output tubes at temperatures from −80°C through 37°C for eluate stability testing at different temperatures.
Eluates containing viral nucleic acids were analyzed by real-time PCR using the LightCycler Parvovirus B19 Quantification Kit on the LightCycler instrument following the manufacturer’s instructions. The reference was an experienced in-house PCR protocol with modifications in that sample extraction is now performed with the MagNA Pure LC Total Nucleic Acid Isolation Kit (Roche Diagnostics) and amplification is on the LightCycler.12,13
Experimental Design and Statistical Methods
Unprocessed samples were mostly stored at −80°C. The lower limit of detection was determined using a serial dilution of Parvovirus B19 DNA International Standard from 10 to 1000 IU/ml in the given matrix, 12 replicates each. Results were analyzed using the probit analysis algorithm.14 A dilution series of a parvovirus B19 DNA high-positive, single-blood donation processed in eight replicates/run and three independent runs was used to calculate precision using variance component analysis.15 Linearity was determined from the same data set using the equation f(x) = log(cn)-log[(cn + 1)/10], where cn was the concentration measured cycle n, with the specification Δf(x) ≤ 0.3.
Results and Discussion
Application of the LightCycler Parvovirus B19 Quantification Kit to the TNAI Kit on the COBAS AmpliPrep Instrument
The TNAI Kit is designed to be compatible with most downstream amplification systems. In the present study, it was tested in combination with the LightCycler Parvovirus B19 Quantification Kit. The IC delivered with the LightCycler Kit was generated by ligating the parvovirus B19 target amplicon into a pUC-based cloning vector suitable for amplification in E. coli. To allow detection of the IC distinct from the viral target, the nucleic acid sequence complementary to the LC640-labeled hybridization probe was exchanged for a sequence complementary to the LC705-labeled IC-specific hybridization probe (Figure 1). Thus, the IC is co-amplified with the target by the same set of primers and has to be positive with parvovirus B19 DNA-negative samples to validate a result but may be negative with parvovirus B19 DNA-positive samples due to competition for the primer pool. To allow 48 reactions to be set up per TNAI and LightCycler Parvovirus B19 Quantification Kit combination, the IC was entirely diluted in TNAI QS diluent resulting in a ratio of 3.1 μl IC per 50 μl diluent; the IC dropout rate was evaluated using 214 routine blood donations (Bavarian Blood Donation Service, Munich, Germany). The cp-value of the IC-corresponding signal was found in a range between 32 and 35 and was accompanied by a dropout rate of 0% (data not shown). It demonstrates that this amplification/detection system is not susceptible to putative plasma-borne inhibitors and that the results generated by the combination of the TNAI Kit on the COBAS AmpliPrep instrument with the LightCycler Parvovirus B19 Quantification Kit are reliable. An optimal workflow was then designed for the kit combination resulting in a throughput of 72 samples per 8-hour shift (Figure 2). To convert the LightCycler copy results to International Units, a correlation factor was calculated by extraction and quantification of the World Health Organization Standard diluted in parvovirus B19 DNA-negative dilution matrix (Table 1). The factor was found to be 2.77 IU/copy for EDTA plasma and 2.34 IU/copy for serum as dilution matrix, respectively, using a sample input volume of 200 μl. The stability of the parvovirus B19 DNA-positive eluate delivered by sample processing using the TNAI Kit was also determined at various temperatures over time (Table 2). The results showed that target recovery deviated by less than 0.3 log compared to the unstressed control, an observation allowing storage of the eluate for later re-testing. However, all data subsequently presented in this study were generated according to the workflow, which eliminated prolonged eluate storage.
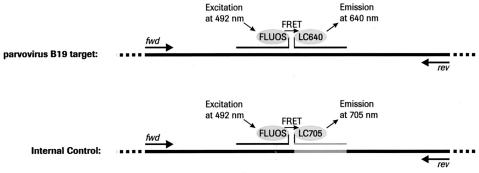
Figure 1.
Principle of simultaneous target and IC amplification and detection using hybridization probe technology. The target DNA as well as the IC DNA is co-amplified by means of PCR using the identical set of primers. During the annealing phase, the hybridization probes labeled with FLUOS at the 3′ end and with LC640 at the 5′ end, respectively, specifically bind to the single-stranded target sequence in direct proximity, thus allowing a fluorescence resonance energy transfer (FRET) resulting in a signal detectable at 640 nm. The IC is a plasmid that provides the original target amplicon with the exception for an exchange of the second hybridization probe binding site for a sequence complementary to the IC-specific hybridization probe being labeled with LC705 at the 5′ end. The signal generated in the presence of the IC is detectable at 705 nm and can thus be distinguished from the target signal in the LightCycler instrument. Since amplification of the target and IC sequence competes for the same pool of primer, the IC has to be positive with parvovirus B19 DNA-negative samples but may be negative with with parvovirus B19 DNA-positive samples.
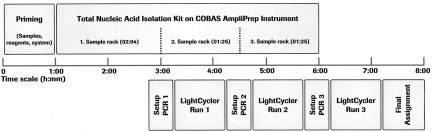
Figure 2.
Workflow scheme for the TNAI Kit on the COBAS AmpliPrep instrument with the LightCycler Parvovirus B19 Quantification Kit. After priming the COBAS AmpliPrep instrument, the first 24-sample rack is processed using the TNAI Kit, followed by setup of the first PCR. The first LightCycler run is started while the second sample rack is processed. This procedure is repeated with the second and third 24-sample racks, ending with complete data collection after a total of 8 hours.
Table 1.
Calculation of the Copy-IU Correlation Factor
| Measured copies* (copies) | Volume corrected* (copies/mL) | Correlation factor (IU/copy) | Converted to IU* (IU/mL) | |
|---|---|---|---|---|
| Mean (plasma/serum) | 48.4/57.0 | 3610/4270 | 2.77/2.34 | 10000/10000 |
| Min (plasma/serum) | 32.0/35.7 | 2400/2670 | 2.77/2.34 | 6648/6248 |
| Max (plasma/serum) | 66.8/74.1 | 5010/5560 | 2.77/2.34 | 13878/13010 |
Overall CV: 18%.
The WHO Parvovirus DNA international standard was diluted in EDTA plasma and serum to yield 1.00E+04 IU/mL and extracted in 24 replicates on the COBAS AmpliPrep instrument using the TNAI Kit. Five microliters per eluate were analyzed employing the LightCycler Parvovirus B19 Quantification Kit. The copy-IU correlation factor was calculated after volume correction from the mean of the replicates.
Table 2.
Determination of Eluate Stability
| Measured concentration (IU/mL)
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Temperature Time | Onboard
|
2–8°C
|
−20°C
|
Unstressed reference | ||||
| 17 hours | 1 day | 3 days | 7 days | 1 day | 7 days | 28 days | ||
| Low-positive sample | 5.45E+03 | 5.34E+03 | 5.69E+03 | 8.08E+03 | 6.63E+03 | 7.95E+03 | 5.38E+03 | 6.14E+03 |
| High-positive sample | 2.30E+06 | 1.85E+06 | 2.74E+06 | 1.48E+06 | 2.28E+06 | 3.54E+06 | 2.85E+06 | 2.87E+06 |
A high-positive and low-positive dilution of a single parvovirus B19 DNA-positive sample in K3-EDTA plasma was processed on the COBAS AmpliPrep instrument using the TNAI Kit. Resulting eluates were analyzed in triplicate employing the LightCycler Parvovirus B19 Quantification Kit either directly or after storage at the given temperature and time.
Analytical Performance
The criteria for evaluating the performance of the kit combination were sensitivity (lower limit of detection), dynamic range (lower and upper limit of quantification), and precision. Sensitivity was determined using serial dilutions of the World Health Organization standard for parvovirus B19 DNA in parvovirus B19 DNA-negative EDTA plasma, citrate plasma, and serum (Table 3). The correlation between sample input volume and sensitivity was optimal at 200 μl input volume with a detection limit of 234 IU/ml (95% hit-rate) using an EDTA plasma matrix. With a citrate plasma or serum matrix, sensitivity showed little increase with volume beyond 200 μl. This differential observation was not due to PCR inhibitors from EDTA plasma since the corresponding parvovirus B19 DNA-negative eluate did not influence signal recovery of spiked purified parvovirus B19 DNA (data not shown). However, all further analyses were performed using a 200 μl specimen input volume.
Table 3.
Dependence of Sensitivity on Specimen Type and Input Volume
| Specimen input volume (μl) | Lower limit of detection (IU/mL) at 95% hit-rate
|
||
|---|---|---|---|
| Dilution in K3-EDTA plasma | Dilution in citrate plasma | Dilution in serum | |
| 50 | 544 | ND | |
| 100 | 367 | ND | ND |
| 200 | 234 | 172 | 112 |
| 500 | 494 | ND | ND |
| 850 | >1000 | 87 | 110 |
ND, not determined.
The WHO Parvovirus DNA international standard was diluted in K3-EDTA plasma, citrate plasma, and serum to cover a range of 1000-10 IU/mL. Fifty to 850 microliters of the samples as taken from the options of the TNAI Kit were extrated using the TNAI Kit on the COBAS AmpliPrep instrument and the corresponding eluates analysed with the LightCycler Parvovirus B19 Quantification Kit. The lower limit of detection was calculated employing the PROBIT algorithm as described.14
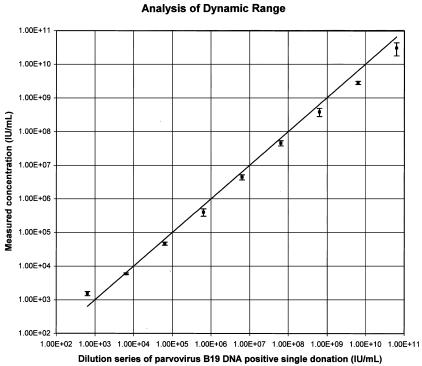
Dynamic range was evaluated using a dilution series of a single high-titer parvovirus B19 DNA blood donation in EDTA plasma calibrated to the World Health Organization standard. Linearity was good, with lower and upper quantification limits of approximately 104 IU/ml and 1010 IU/ml (Figure 3). Within this range, measured values exhibited intra-signal deviation of less than 0.3 log, although they were generally found slightly below the input concentration (Table 4).
Figure 3.
Dynamic range analysis with the combination of the TNAI Kit on the COBAS AmpliPrep with the LightCycler Parvovirus B19 Quantification Kit. A dilution series of a high titer parvovirus B19 DNA-positive single donation in K3-EDTA plasma calibrated to the World Health Organization standard was processed using the TNAI Kit on the COBAS AmpliPrep instrument in three independent runs with eight replicates each and then quantified using the LightCycler Parvovirus B19 Quantification Kit. Output results in copies were converted to IU using the correlation factor given in Table 1 and linearity determined as described in Materials and Methods. The line depicts 100% correlation.
Table 4.
Signal Recovery and Precision Testing
| Input concentration (IU/mL) | Measured concentration (mean IU/mL) | Deviation Cmeasured-Cinput (log) | Variance analysis
|
||
|---|---|---|---|---|---|
| CV 18 replicates (%) | Intra-assay CV 6 replicates/run (%) | Inter-assay CV 3 runs (%) | |||
| 6.36E+02 | 1.52E+03 | 0.29 | 40.42 | 41.62 | 0.00* |
| 6.36E+03 | 5.89E+03 | −0.13 | 22.97 | 23.95 | 0.00* |
| 6.36E+04 | 4.62E+04 | −0.23 | 22.88 | 22.61 | 4.21 |
| 6.36E+05 | 4.00E+05 | −0.29 | 26.00 | 16.40 | 24.02 |
| 6.36E+06 | 4.40E+06 | −0.25 | 21.95 | 17.42 | 15.89 |
| 6.36E+07 | 4.52E+07 | −0.24 | 16.45 | 7.14 | 17.64 |
| 6.36E+08 | 3.80E+08 | −0.32 | 25.65 | 12.21 | 26.85 |
| 6.36E+09 | 2.78E+09 | −0.45 | 31.11 | 31.75 | 0.00* |
| 6.36E+10 | 3.00E+10 | −0.42 | 38.34 | 16.64 | 41.11 |
Zero set by the software tool.
A dilution series of a parvovirus B19 DNA-positive single donation in K3-EDTA plasma was extracted using the TNAI Kit on the COBAS AmpliPrep instrument in six replicates per run and a total of three runs at 3 days and analyzed with the LightCycler Parvovirus B19 Quantification Kit. The results were taken to determine the deviation between measured and input concentration. The overall CV (overall precision), the intra-assay CV (precision within a single run), and the inter-assay CV (precision run-to-run) were calculated as described.13
Overall precision analysis showed a coefficient of variation (CV) of 16 to 40% (Table 4). The CV was lowest, 16 to 26%, at concentrations of approximately 104-109 IU/ml; it increased to around 40% at the upper limit of quantification and lower limit of detection. These analytical performance data meet the general requirements for NAT systems and are excellent in comparison with previous studies.16,17,18,19,20
Specificity and Sensitivity in Research Samples
Parvovirus B19 infection is a critical issue in pregnancy due to its potential transmission to the fetus, which can cause severe complications including fetal death. Since the infected fetus could benefit from timely intrauterine transfusion, accurate and sensitive detection of parvovirus B19 in pregnant women is of great importance.21 We compared the combination of the TNAI and LightCycler Parvovirus B19 Quantification kits with the reference NAT system set as gold standard in analyzing research samples largely obtained from routine blood tests in pregnant women (Table 5). The reference NAT system employs nucleic acid extraction with the MagNA Pure LC Total NA Kit with in-house real-time PCR targeting the VP1 gene region on the LightCycler instrument. The combination allowed semi-quantitative detection of parvovirus B19 DNA in serum and plasma down to a detection limit of approximately 600 copies/ml (approximately 1000 IU/ml according to the NIBSC). Quantitative correlation of the results was not possible due to the semi-quantitative character of the reference system.
Table 5.
Clinical Research Sample Test Results
| Samples (n) | Anti-parvovirus B19 serology
|
Parvovirus B19 DNA
|
||
|---|---|---|---|---|
| IgG | IgM | NAT system 1 | NAT system 2 | |
| 15 | −ve | −ve | −ve | −ve |
| 35 | +ve | −ve | −ve | −ve |
| 1 | +ve | −ve | +ve | −ve |
| 4 | +ve | +ve | −ve | −ve |
| 4 | +ve | +ve | −ve | +ve |
| 51 | +ve | +ve | +ve | +ve |
Clinical research samples were processed and analysed according to the workflow as described in the Material and Methods section. (NAT system 1: in-house reference method; NAT system 2: TNAI Kit on COBAS AmpliPrep instrument with LightCycler Parvovirus B19 Quantification Kit).
A first collective, consisting of 51 anti-parvovirus B19 IgM-negative samples, was analyzed in comparison to the reference system. One sample proved PCR-positive in both NAT systems, indicating persistent parvovirus B19 infection. As reported previously, parvoviral DNA may persist at low levels in immunocompetent individuals for up to 40 months post-infection.12,22,23 All remaining samples were parvovirus B19 DNA-negative in both NAT systems.
In a second collective, 51 of a total of 59 anti-parvovirus B19 IgM-positive samples were found parvovirus B19 DNA-positive by both NAT systems, whereas four anti-parvovirus B19 IgM low-positives did not react by PCR, suggesting clearance of viremia. Four samples were repeatedly parvovirus B19 DNA-positive using the Roche kit system but were found negative by the reference PCR and were therefore considered as false-positives.
In summary, the sensitivity of the combination of the TNAI Kit on the COBAS AmpliPrep instrument with the LightCycler Parvovirus B19 Quantification Kit in this limited number of samples is thus excellent at 100%. The specificity of the kit combination relative to the in-house NAT method was found to be 93%.
Acknowledgments
We thank S. Kunze and A. Schubert for excellent technical assistance, M. Wiedmann and S. Boehm for help on the sensitivity analysis, and D. Sizmann and J.C. Raymond for critical reading of the manuscript. COBAS, AmpliPrep, LightCycler, and MagNA Pure are trademarks of or members of the Roche Group.
References
- Jungkind D. Automation of laboratory testing for infectious diseases using the polymerase chain reaction: our past, our present, our future. J Clin Virol. 2001;20:1–6. doi: 10.1016/s1386-6532(00)00148-7. [DOI] [PubMed] [Google Scholar]
- Stelzl E, Kormann-Klement A, Haas J, Daghofer E, Santner BI, Marth E, Kessler HH. Evaluation of an automated sample preparation protocol for quantitative detection of hepatitis C virus RNA. J Clin Microbiol. 2002;40:1447–1450. doi: 10.1128/JCM.40.4.1447-1450.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ, Jones SE, Fischer-Hoch SP, Lewis E, Hall SM, Bartlett CLR. Human parvovirus, the cause of erythema infectiosum (fifth disease)? Lancet. 1983;1:1378. doi: 10.1016/s0140-6736(83)92152-9. [DOI] [PubMed] [Google Scholar]
- Pattison JR, Jones SE, Hodgson J, Davis LR, White JM, Stroud CE, Murtaza L. Parvovirus infections and hypoplastic crisis in sickle-cell anaemia. Lancet. 1989;1:664–665. doi: 10.1016/s0140-6736(81)91579-8. [DOI] [PubMed] [Google Scholar]
- Cohen B. Parvovirus B19: an expanding spectrum of disease. Br Med J. 1995;311:1649–1552. doi: 10.1136/bmj.311.7019.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoto Y, Kudoh T, Haseyama K, Suzuki N, Chiba S. Human parvovirus B19 infection associated with acute hepatitis. Lancet. 1996;347:868–901. doi: 10.1016/s0140-6736(96)91348-3. [DOI] [PubMed] [Google Scholar]
- Enders G, Dötsch J, Bauer J, Nützenadel W, Hengel H, Daffner D, Schalasta G, Searle K, Brown KE. Life-threatening parvovirus B19-associated myocarditis and cardiac transplantation as possible therapy: two case reports. Clin Infect Dis. 1998;26:355–358. doi: 10.1086/516295. [DOI] [PubMed] [Google Scholar]
- Dux S, Lentini S, Bock CT, Klingel K, Kandolf R, Bauriedel G. Parvovirus B19 myocarditis in a young man with previous non-bacterial meningitis. Dtsch Med Wochenschr. 2002;127:1584–1588. doi: 10.1055/s-2002-32941. [DOI] [PubMed] [Google Scholar]
- Enders E. Infections of the fetus and the neonate other than rubella. Collier L. London., editor. Edward Arnold; Topley & Wilson’s Microbiology and Microbial Infections. 1998:pp 873–915. [Google Scholar]
- Brown KE. Haematological consequences of parvovirus B19 infection. Baillieres Best Pract Res Clin Haematol. 2000;13:245–259. doi: 10.1053/beha.1999.0071. [DOI] [PubMed] [Google Scholar]
- Searle K, Schalasta G, Enders G. Development of antibodies to the nonstructural protein NS1 of parvovirus B19 during acute symptomatic and subclinical infection in pregnancy: implications for pathogenesis doubtful. J Med Virol. 1998;56:192–198. doi: 10.1002/(sici)1096-9071(199811)56:3<192::aid-jmv3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Searle K, Guilliard C, Wallat S, Schalasta G, Enders G. Acute parvovirus B19 infection in pregnant women: an analysis of serial samples by serological and semi-quantitative PCR techniques. Infection. 1998;26:139–143. doi: 10.1007/BF02771838. [DOI] [PubMed] [Google Scholar]
- Finney DJ. Cambridge: Cambridge University Press; Probit Analysis. (ed 3.) 1971 [Google Scholar]
- Neter J, Wassermann W, Kutner MH. IL, Irwin: Homewood; Applied Linear Statistical ModelsRegression, Analysis of Variance, and Experimental Designs. 1985 [Google Scholar]
- Allain JP, Thomas I, Sauleda S. Nucleic acid testing for emerging viral infections. Transfus Med. 2002;12:275–283. doi: 10.1046/j.1365-3148.2002.00386.x. [DOI] [PubMed] [Google Scholar]
- Knoell A, Louwen F, Kochanowski B, Plentz A, Stussel J, Beckenlehner K, Jilg W, Modrow S. Parvovirus B19 infection in pregnancy: quantitative viral DNA analysis using a kinetic fluorescence detection system (TaqMan PCR). J Med Virol. 2002;67:259–266. doi: 10.1002/jmv.2216. [DOI] [PubMed] [Google Scholar]
- Heegaard ED, Brown KE. Human parvovirus B19. Clin Microbiol Rev. 2002;15:485–505. doi: 10.1128/CMR.15.3.485-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder TC, Hufnagel M, Zahn K, Beutel K, Schmitt HJ, Ullmann U, Rautenberg P. New LightCycler PCR for rapid and sensitive quantification of parvovirus B19 DNA guides therapeutic decision-making in relapsing infections. J Clin Microbiol. 2001;39:4413–4419. doi: 10.1128/JCM.39.12.4413-4419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fini F, Gallinella G, Girotti S, Zerbini M, Musiani M. Development of a chemiluminescence competitive PCR for the detection and quantification of parvovirus B19 DNA using a microplate luminometer. Clin Chem. 1999;45:1391–1396. [PubMed] [Google Scholar]
- Fairley CK, Smoleniec JS, Caul OE, Miller E. Observational study of effect of intrauterine transfusions on outcome of fetal hydrops after parvovirus B19 infection. Lancet. 1995;346:1335–1337. doi: 10.1016/s0140-6736(95)92346-2. [DOI] [PubMed] [Google Scholar]
- Cassinotti P, Siegl G. Quantitative evidence for persistence of human parvovirus B19 DNA in an immunocompetent individual. Eur J Clin Microbiol Infect Dis. 2000;19:886–887. doi: 10.1007/s100960000384. [DOI] [PubMed] [Google Scholar]
- Musiani M, Zerbini M, Gentilomi G, Plazzi M, Gallinella G, Venturoli S. Parvovirus B19 clearance from peripheral blood after acute infection. J Infect Dis. 1995;172:1360–1363. doi: 10.1093/infdis/172.5.1360. [DOI] [PubMed] [Google Scholar]