Abstract
Infection with mucosotropic human papillomavirus (HPV) is the necessary cause of cervical intraepithelial neoplasia. Several epidemiological studies suggest that HPV viral load can be a risk factor of cervical dysplasia. The purpose of the present study was to evaluate a methodology to determine HPV viral load of eight oncogenic HPV types (16, 18, 31, 39, 45, 51, 52, and 58). The quantitation assay is based on a high-throughput real-time PCR. The E6-E7 region of HPV types 16, 18, 45, and 51 were used to amplify specific DNA sequences and cloned into a plasmid vector. The constructs for HPV types 16, 18, 45, and 51, and the whole genome for HPV types 31, 39, 52, and 58 were quantitated using a limiting dilution analysis and used to create standard curves. Type-specific HPV clones were used to determine specificity, linearity, and intra- and inter-assay variation. The sensitivity of our assay covered a dynamic range of up to seven orders of magnitude with a coefficient of intra-assay variation less than 6% and the inter-assay variation less than 20%. No cross-reactivity was observed on any of the type-specific standard curves when phylogenetically close HPV types were used as templates. Our real-time PCR methodologies are highly reproducible, sensitive, and specific over a sevenfold magnitude dynamic range.
Among all types of human papillomavirus (∼100 types) the ones classified as oncogenic types are of clinical significance due to their association with cervical neoplastic development and carcinoma. Although most HPV infections are transient, a number of host and viral factors may lead to a persistent infection thus increasing the risk of developing neoplasia.1 Understanding the natural history of oncogenic HPV infection and the predictive value of one or several HPV test, is, therefore, essential for the development of appropriate cervical cancer prevention and control programs. The limitations of cytological analysis have pressured the development of alternative screening methods in population studies, with molecular diagnosis emerging as a promising diagnostic tool in cervical cancer prevention.2 Quality assurance of these emerging technologies is paramount for large-scale clinical investigations.3 In population-based studies, questions of reliability, sensitivity, specificity, reproducibility, and scalable capacity for automation are central in selecting assays to determine factors associated with cervical carcinogenesis.
Prevalence studies conducted worldwide indicate that approximately 30% of women are infected with HPV. Therefore, the use of HPV testing as a primarily screening tool is not sufficient.4 Alternative measures that may have greater predicted value, such as HPV viral load, are currently under investigation. Several studies have correlated HPV viral load and its prognostic significance in the evolution of cervical intraepithelial neoplasia.5,6 The methodologies used to quantitate viral burden have varying levels of specificity and sensitivity and performance evaluation is frequently not comprehensive. Hybrid capture has been a common technique used to determine HPV viral load, but does not have the capacity to differentiate specific types of HPV.7,8 PCR-based methods using densitometric analysis have frequently been used to determine HPV viral burden and its association with cervical lesion development.9,10,11 However, the estimation of viral load using semi-quantitative PCR may be inaccurate due to the inherent efficiency of the detection system as well as the presence of inhibitors in the sample.12
Real-time PCR is a more recent addition in the molecular diagnosis of HPV infection.13 Real-time PCR has the advantage of being highly specific, reproducible, and capable of detecting HPV viral load up to eight orders of magnitude in a linear range.14 These qualities make real-time PCR attractive for use in epidemiological studies and as a potential diagnostic test. Several studies reported the predictive value of HPV viral burden determined by real-time PCR and the persistence of HPV infection.13,15,16,17 These studies, however, only focused on the quantitation of viral load for HPV type 16.
To our knowledge, there has been no systematic evaluation of real-time PCR to determine viral load for oncogenic HPV types based on standardized criteria of quality control assurance. Here, we present a quality control evaluation to assess the sensitivity, specificity, and reproducibility (intra- and inter-assay variability) of viral load quantitation using the ABI Prism 7900 Sequence Detection System for eight oncogenic HPV types (16, 18, 31, 39, 45, 51, 52 and 58). The analytical performance was based on the National Committee for Clinical Laboratory Standards (NCCLS) guideline EP10-T, using a 10-μl volume reaction in a high-throughput format (384-well system for the processing of large number of samples).
Materials and Methods
Primer and Probe Design
The E6/E7 regions of HPV types 16 (nt 65–857), 18 (nt 87–907), 31 (nt 39–856), 39 (nt 44–921), 45 (nt 75–906), 51 (nt 88–865), 52 (nt 93–852), and 58 (nt 77–869) were obtained from Los Alamos HPV database (http://hpv-web.lanl.gov/stdgen/virus/hpv/). The nucleotide sequence for each type was aligned to identify highly heterologous regions and used to design type-specific probes and primers by the software Primer Express (Applied Biosystems). Primers and probes spanning a targeted region <200bp were selected according to the specifications in the software manual. Type specificity was determined by no-predicted cross-hybridization with other HPV types using the NCBI database BLAST search. Primers and probes for HPV types 16 and 18 were selected as previously reported.14 The sequences and nucleotide location of the selected primers and probes are shown in Table 1. Primers and probes were synthesized by Integrated DNA Technologies (Coralville, IA). Probes were labeled to the 5′ end with the reporter dye 6-carboxy-fluorescein (FAM) or 6-carboxy-4,7,2′7′-tetrachlorofluorescein (TET) and the 3′ end was blocked with the non-fluorescent quencher Black Hole-1 (Integrated DNA Technologies). Probes and primers were resuspended in sterile HPLC-grade water to form a 200 nmol/L and 100 nmol/L solution, respectively, stored in 20-μl volume aliquots at −20°C until used.
Table 1.
Primers and Dual-Labeled Fluorogenic Probes Used in the TaqMan Assay
| Name | Oligonucleotide sequence* | Labels† |
|---|---|---|
| HPV-16:520U25-primer‡ | TTGCAGATCATCAAGAACACGTAGA | |
| HPV-16:671L24-primer‡ | CTTGTCCAGCTGGACCATCTATTT | |
| HPV-16:558U33-probe‡ | AATCATGCATGGAGATACACCTACATTGCATGA | FAM-BH1 |
| HPV-18:530U19-primer‡ | CAACCGAGCACGACAGGAA | |
| HPV-18:729L21-primer‡ | CTCGTCGGGCTGGTAAATGTT | |
| HPV-18:580U37-probe‡ | AATATTAAGTATGCATGGACCTAAGGCAACATTGCAA | TET-BH1 |
| HPV-31:449F-primer | ATTCCACAACATAGGAGGAAGGTG | |
| HPV-31:524R-primer | CACTTGGGTTTCAGTACGAGGTCT | |
| HPV-31:474T-probe | ACAGGACGTTGCATAGCATGTTGGA | FAM-BH1 |
| HPV-39:470F-primer | CAGGACAGTGTCGACGGTGCT | |
| HPV-39:570R-primer | TGGGCTTTGGTCCACGCATAT | |
| HPV-39:501T-probe | ACGGGAGGACCGCAGACTAACACG | FAM-BH1 |
| HPV-45:425F-primer | GGACAGTACCGAGGGCAGTGTAA | |
| HPV-45:495R-primer | TCCCTACGTCTGCGAAGTCTTTC | |
| HPV-45:450T-probe | CATGTTGTGACCAGGCACGGCA | FAM-BH1 |
| HPV-51:358F-primer | AAAGCAAAAATTGGTGGACGA | |
| HPV-51:438R-primer | TGCCAGCAATTAGCGCATT | |
| HPV-51:392-probe | CATGAAATAGCGGGACGTTGGACG | FAM-BH1 |
| HPV-52:78F-primer | GTGCATGAAATAAGGCTGCAGT | |
| HPV-52:213R-primer | GTAGGCACATAATACACACGCCA | |
| HPV-52:101T-probe | TGTGCAGTGCAAAAAAGAGCTACAACG | FAM-BH1 |
| HPV-58:64F-primer | CCACGGACATTGCATGATTTG | |
| HPV-58:144R-primer | CTTTTTGCATTCAACGCATTTCA | |
| HPV-58:95T-probe | TGGAGACATCTGTGCATGAAATCGAA | FAM-BH1 |
Sequences are given from 5′ → 3′.
Fluorochrome is at 5′, non-fluorescent quencher Black Hole-1 is at 3′.
Sequences taken from (14).
Construction of External Standards
Clones for HPV types 16, 18, 45, and 51 were constructed by cloning a fragment containing the targeted region for real-time PCR described above. Cloning regions for HPV-16 consisted of nucleotides 351–855, nucleotides 295-1064 for HPV-18, nucleotides 80–922 for HPV-45, and nucleotides 49–895 for HPV-51. Cloning primers were designed using Oligo software version 5.0 (Molecular Biology Insights, Inc., Cascade, CO). HPV-16 (F-primer 5′-TGTATGGAACAACATTAGAAC-3′; R-primer 5′-TCAGCCATGGTAGATTAT-3′), HPV-18 (F-primer 5′-CATGCCATAAATGTATAGATT-3′; R-primer 5′-AAATGTTCCTTGTGTATCAAT-3′), HPV-45 (F-primer 5′-GATCCAAAGCAACGACCCTA-3′; R-primer 5′-TCCTCTGCCGAGCTCTCTAC-3′) and HPV-51 cloning primers (F-primer 5′-GAAAACGGTGCATATAAAAGTGC-3“; R-primer 5′-CCTCTGTACCTTCACAGTCCATC-3′) were used to generate HPV type-specific amplicons using DNA from CaSki and HeLa cell lines for HPV-16 and -18, respectively, and clinical samples for HPV-45 and HPV-51. The specificity of the amplification was evaluated by agarose-gel electrophoresis and by sequence analysis. Amplified fragments from HPV-16 were cloned into the pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA) and the amplified fragments from HPV-18, HPV-45, and HPV-51 were cloned into the pGEM-T easy vector (Promega, Madison, WI) according to the specifications of the manufacturers. To verify cloning effectiveness, isolated plasmids from 5-ml bacterial cultures were used as template in PCR reactions using both external (cloning) and internal primers, and by digestion with the restriction enzyme EcoRI (Boehringer, Petersburg, VA) and conventional agarose gels. Clones for HPV types 31, 39, 52, and 58 were kindly provided by Dr. Attila Lorincz, Digene Corp., Professor Gerard Orth, Institute Pasteur, Dr. Wayne D. Lancaster, Wayne State University, and Dr. Toshihiko Matsukura, National Institute of Health in Japan, respectively. All HPV clones were transformed into One Shot Top-10 chemically competent cells (Invitrogen) and stored as glycerol stocks at −20°C until used. HPV clones were amplified by culturing 5-ml amp-LB media overnight at 37°C and re-inoculated in 1 Lt LB media containing 5 mmol/L ampicillin and grown overnight at 37°C. HPV plasmids were purified using the QIAGEN plasmid Maxi purification kit (QIAGEN, Valencia, CA) according to the manufacturer’s instructions. The relative plasmid DNA concentration was determined by fluorescence using the PicoGreen dsDNA quantitation reagent (Molecular Probes, Eugene, OR).
Limiting Dilution
The approximate plasmid DNA concentration for each HPV type was determined by a modification of the PCR-based limiting dilution assay described elsewhere18,19 and used as reference material. Briefly, using a preliminary calculation of plasmid copy number based on PicoGreen quantitation, ten replicates of a 10-fold dilution series for the plasmid DNA were created (from 1:10 to 1:1 × 108). The last dilution of the 10-fold dilution series was used to create a twofold dilution series (from 1:2 to 1:4096) covering a range from 2 × 108 to 4.096 × 1011 copies/μl. One μl of each dilution from the twofold dilution series was added to a 10-μl PCR reaction and subjected to 50 PCR cycles using the ABI PRISM 7900HT (Applied Biosystems). Annealing temperatures for each HPV type were used as determined by the primer titration study, and non-template controls were used in each case to establish the threshold. The proportion of negative end points (fluorescence values not significantly different at 5% level above background) served to estimate the concentration of the undiluted reference material by Poisson probability distribution using the computer program QUALITY V1.1.4 described by Rodrigo et al.18
PCR Optimization
To assess the optimal primer ratio and annealing temperature for each HPV type, a series of six primer ratios were tested covering a range of 50 to 900 mmol/L. Each primer ratio was run in triplicate in a 10-μl PCR reaction using the TaqMan universal PCR master mix (Applied Biosystems). Each PCR was run for 45 cycles covering a gradient of annealing temperature from 52°C to 65°C. The PCR reaction was transferred to a 384-well plate and the fluorescence signal was read using the ABI PRISM 7900HT (Applied Biosystems). The primer ratio and annealing temperature were chosen according to the highest fluorescence signal normalized with a passive internal reference dye in the PCR reaction.
Standard Curves and Controls
External standard curves were generated by serial dilutions of known input concentrations of HPV DNA covering a range from 1 copy to 1 × 107 copies/10 μl reaction. Each dilution was aliquoted in 20-μl volume and stored at −20°C until used. Human blood DNA (known to be HPV negative) was used as background DNA for an equivalent of 100 cells per PCR reaction. DNA from human blood, exfoliated cervical cells, and cell lines were extracted using the QIAamp purification kit (QIAGEN) according to the instructions of the manufacturer. DNA concentration was determined using a real-time PCR quantification assay for the housekeeping gene RNase P (Applied Biosystems). Eight points for each standard curve covering a dynamic range from 100 to 107 were run in triplicate along with non-template controls to set the threshold value. DNA extracted from CaSki cells or HeLa cells was used as positive control for HPV-16 or HPV-18, respectively. Specific HPV clones were used as homologous templates for HPV-31, -39, -45, -51, -52, and -58.
Real-Time PCR
All PCR reactions were performed in a 10-μl volume using the ABI PRISM 7900HT (Applied Biosystems). Each individual reaction contained 5 μl 2X TaqMan universal PCR master mix with uracil-N-glycosylase (Applied Biosystems), 1 μl 200 nmol/L fluorogenic probe, 1 μl primer ratio, 0.65 ng human blood DNA, and up to 3 μl HPV DNA or sterile HPLC-grade water in non-template controls. The amplification profile was initiated by a 2-minute incubation at 50°C, followed by a 10-minute incubation at 95°C, and a two-step amplification of 15 seconds at 95°C and 60 seconds at 59.3°C or 54.1°C for 45 cycles. Data were collected at the end of the amplification step. All experiments were performed in triplicate including the positive controls and non-template controls.
Validation Assay
The accuracy, efficacy, and reliability of HPV DNA quantification by real-time PCR were evaluated using the National Committee for Clinical Laboratory Standards EP10-T guidance: “Preliminary Evaluation of Chemistry Clinical Methods.”20 In sum, four concentrations of HPV DNA (101 copies/μl, 102 copies/μl, 104 copies/μl, and 106 copies/μl) for HPV types 16, 18, 31, 39, 45, 51, 52, and 58 were created by diluting the reference solution in TE buffer and analyzed in triplicate in eight independent assays to evaluate intra- and inter-assay variation. Specificity was evaluated using human blood DNA (∼5 ng) spiked with clones for HPV-16, HPV-18, HPV-31, HPV-39, HPV-45, HPV-51, HPV-52, or HPV-58 as heterologous DNA templates at an input copy number of 1 × 106 copies/PCR reaction. Each sample was used as unknown in type-specific HPV quantitation assays. Four clinical samples obtained from exfoliated cervical cells with known high and low HPV-16 copy numbers were run to verify accuracy of the assay and their DNA concentration was determined by the RNase-P assay. DNA extracted from CaSki and HeLa cell lines was used as positive controls for HPV-16 and HPV-18, respectively.
Statistical Analysis
Repeatability or intra-assay variation (defined as the degree of agreement among individual tests) was calculated by computing the relative SD of three replicates per assay. Reproducibility was assessed by computing the coefficient of variation (%CV) among the mean values in eight independent assays. The efficiency of the standard curves was calculated based on the slope from eight independent experiments.
Results
Experimental Design of a TaqMan Assay for HPV Load Quantitation
The probe and primer sequences generated by the software Primer Express were chosen when no predicted cross-reactivity with homologous sequences were obtained from a search on the EMBL and GenBank databases (Table 1). Primers and probes were optimized for primer concentration and annealing-extension temperature and the conditions were chosen according to the maximum fluorescent signal generated after 45 PCR cycles. For subsequent experiments, the forward-reverse primer ratio used to generate standard curves for each HPV type corresponded to the annealing-extension temperature of 59.3°C for HPV types 16, 18, 31, 39, 52, and 58; and the annealing temperature for HPV-45 and HPV-51 was 54.1°C. For HPV-16, the optimal forward/reverse primer ratio was 900/600 mmol/L; for HPV-18, 900/600 mmol/L; for HPV-31, 600/900 mmol/L; for HPV-39, 600/900 mmol/L; for HPV-45, 900/600 mmol/L; for HPV-51, 900/600 mmol/L; for HPV-52, 600/900 mmol/L; and for HPV-58, 300/300 mmol/L.
Validation of Reference Curves for HPV Quantitation
Linearity
The reference curves of all HPV types covered a dynamic range being able to discriminate from one up to 107 copies in a linear fashion as indicated in Table 2. The limit of detection for HPV-16 was determined to be somewhere between 1 to 10 copies based on the percentage of positive reactions at 100 (54% of replicates positive). It was not possible to detect fluorescence at one copy level for HPV-52 in any of the 24 replicates suggesting the reference solution was diluted beyond this point. At 10 copies per tube, however, all 24 replicates for both HPV-16 and HPV-52 were positive and we determined this value to be the lowest detection limit for these HPV-reference curves.
Table 2.
Reproducibility of HPV-16, -18, -31, -39, -45, -51, -52, and -58 Standard Curves
|
|
HPV-16
|
HPV-18
|
HPV-31
|
HPV-39
|
||||
|---|---|---|---|---|---|---|---|---|
| Linearity range (# copies) | 10 − 1 × 107 | 1 − 1 × 106 | 1 − 1 × 107 | 1 − 1 × 107 | ||||
| Mean ± SD | %CV | Mean ± SD | %CV | Mean ± SD | %CV | Mean ± SD | %CV | |
| Slope (n = 8) | −3.76 ± 0.112 | 2.98 | −3.69 ± 0.065 | 1.76 | −3.39 ± 0.057 | 1.67 | −3.45 ± 0.067 | 1.94 |
| Efficiency (n = 8) | 1.84 ± 0.035 | 1.91 | 1.87 ± 0.020 | 1.09 | 1.98 ± 0.018 | 0.93 | 1.95 ± 0.026 | 1.31 |
| Y-intercept (n = 8) | 43.59 ± 0.724 | 1.66 | 40.46 ± 0.782 | 1.93 | 38.09 ± 0.376 | 0.99 | 39.03 ± 0.632 | 1.62 |
| Value of fit (R2) (n = 8) | 0.993 ± 0.004 | 0.42 | 0.994 ± 0.003 | 0.28 | 0.995 ± 0.005 | 0.53 | 0.995 ± 0.002 | 0.23 |
| Standard curve (n = 24) | Ct ± SD | %CV | Ct ± SD | %CV | Ct ± SD | %CV | Ct ± SD | %CV |
| 100 | Nd | 38.97 ± 0.857 | 2.20 | 36.37 ± 0.313 | 0.86 | 38.96 ± 0.653 | 1.68 | |
| 101 | 39.43 ± 0.841 | 2.13 | 35.30 ± 0.875 | 2.47 | 34.85 ± 0.580 | 1.66 | 35.41 ± 0.677 | 1.91 |
| 102 | 36.28 ± 0.953 | 2.64 | 31.73 ± 0.849 | 2.68 | 31.59 ± 0.408 | 1.29 | 31.84 ± 0.450 | 1.41 |
| 103 | 31.75 ± 1.066 | 3.36 | 27.52 ± 0.820 | 2.98 | 28.00 ± 0.313 | 1.12 | 28.22 ± 0.408 | 1.45 |
| 104 | 27.63 ± 1.053 | 3.81 | 23.59 ± 0.694 | 2.94 | 24.71 ± 0.405 | 1.64 | 25.09 ± 0.366 | 1.46 |
| 105 | 23.66 ± 0.949 | 4.01 | 19.69 ± 0.481 | 2.44 | 21.27 ± 0.316 | 1.49 | 21.38 ± 0.427 | 1.99 |
| 106 | 20.65 ± 0.846 | 4.09 | 17.41 ± 0.460 | 2.64 | 17.64 ± 0.374 | 2.12 | 17.67 ± 0.351 | 1.98 |
| 107 | 17.27 ± 0.657 | 3.81 | 14.91 ± 0.335 | 2.25 | 14.73 ± 0.330 | 2.24 | ||
Standard curves for eight high-risk HPV types were constructed with control plasmids that either contained the E6–E7 region cloned in a vector (HPV-16, -18, -45, and -51) or harbored the whole HPV genome (HPV-31, -39, -52, and -58). The control plasmids were diluted in TE buffer containing human blood DNA equivalent of a 100 cells (0.65ng) to cover a linear range of seven orders of magnitude (from 100 to 107 per sample). Each curve was run in triplicate in eight independent assays.
Table 2.
Continued
| HPV-45
|
HPV-51
|
HPV-52
|
HPV-58
|
|||||
|---|---|---|---|---|---|---|---|---|
| 1 − 1 × 107 | 1 − 1 × 107 | 10 − 1 × 106 | 1 − 1 × 107 | |||||
| Mean ± SD | %CV | Mean ± SD | %CV | Mean ± SD | %CV | Mean ± SD | %CV | |
| −3.16 ± 0.308 | 0.98 | −3.27 ± 0.069 | 2.10 | −3.72 ± 0.057 | 1.53 | −3.46 ± 0.082 | 2.38 | |
| 1.98 ± 0.018 | 0.93 | 2.03 ± 0.031 | 1.52 | 1.86 ± 0.018 | 0.97 | 1.98 ± 0.018 | 0.93 | |
| 39.38 ± 0.24 | 0.62 | 37.36 ± 0.28 | 0.76 | 40.27 ± 0.694 | 1.72 | 40.75 ± 0.665 | 1.63 | |
| 0.995 ± 0.001 | 0.08 | 0.993 ± 0.002 | 0.10 | 0.995 ± 0.003 | 0.30 | 0.994 ± 0.003 | 0.29 | |
|
|
||||||||
| Ct ± SD | %CV | Ct ± SD | %CV | Ct ± SD | %CV | Ct ± SD | %CV | |
| 37.64 ± 0.366 | 0.97 | 35.89 ± 0.486 | 1.36 | Nd | 38.92 ± 0.418 | 1.07 | ||
| 32.51 ± 0.525 | 1.62 | 32.05 ± 0.346 | 1.08 | 35.40 ± 0.735 | 2.07 | 35.80 ± 0.806 | 2.25 | |
| 29.45 ± 0.288 | 0.98 | 28.39 ± 0.414 | 1.46 | 31.89 ± 0.672 | 2.11 | 32.42 ± 0.692 | 2.13 | |
| 25.86 ± 0.271 | 1.05 | 25.26 ± 0.372 | 1.48 | 28.12 ± 0.517 | 1.83 | 28.62 ± 0.496 | 1.73 | |
| 22.55 ± 0.156 | 0.69 | 22.04 ± 0.361 | 1.63 | 23.89 ± 0.666 | 2.79 | 24.95 ± 0.602 | 2.41 | |
| 19.57 ± 0.265 | 1.35 | 19.96 ± 0.451 | 2.26 | 20.23 ± 0.563 | 2.78 | 21.43 ± 0.482 | 2.25 | |
| 17.43 ± 0.203 | 1.17 | 17.18 ± 0.232 | 1.35 | 16.94 ± 0.445 | 2.63 | 17.91 ± 0.377 | 2.11 | |
| 13.76 ± 0.139 | 1.01 | 14.94 ± 0.276 | 1.85 | 15.23 ± 0.342 | 2.25 | |||
To evaluate the performance of the standard curves we compared the cycle threshold (Ct) for each point of the curve since this value should theoretically be invariable in replicate samples run under the same conditions. As shown in Table 2, the reproducibility of the standard curves was high as the variation coefficient for each point was less than 5% for all HPV standard curves. Measurements of linearity such as slope, Y-intercept, and linearity correlation coefficient (R2) for each type-specific reference curve did not significantly vary among the eight assays (inter-assay %CV <2%).
Intra- and Inter-Assay Variability
Different dilutions of the reference solutions were used as controls to assess the precision and reproducibility of the standard curves to quantitate HPV viral load. Three replicates of the control samples containing either high (106), medium (104 and 102), or low (101) copy number for each HPV type were run as unknowns in eight independent assays. The coefficient of variation was determined based on the values obtained from the replicates (intra-assay variation) and between the experiments (inter-assay variation). The performance results are summarized in Table 3. As observed in this table, the intra-assay coefficient of variation for all HPV types ranged from 0.45% to 5.77%. The inter-assay coefficient of variation, however, increased proportionally with the dilution of the samples where the smallest variation was observed in the 106 controls and the highest variation was observed in the 101 controls (%CV = 3.91 and 19.71, respectively).
Table 3.
Intra- and Inter-Assay Variation of HPV-16, -18, -31, -39, -45, -51, -52, and -58 Quantification by Real-Time PCR
| Sample | Intra-assay variation (assays 1–8)
|
Inter-assay variation
|
||
|---|---|---|---|---|
| Quantity range* | %CV range | Quantity mean† | %CV | |
| HPV-16 106 | (5027903–6214029) | (0.85 –3.08) | 5288299 | 10.27 |
| HPV-16 104 | (18454 –26481) | (0.75–3.76) | 22276 | 11.81 |
| HPV-16 102 | (256–415) | (0.91 –4.61) | 324 | 16.43 |
| HPV-16 101 | (30 –50) | (1.96 –4.40) | 37 | 19.71 |
| Clinical sample 1 | (2.25 –3.35) | (0.72–4.29) | 2.64 | 14.63 |
| Clinical sample 2 | (2640 –4213) | (0.45 –3.82) | 3489 | 13.02 |
| Clinical sample 3 | (1.07–1.96) | (1.09–4.43) | 1.57 | 16.84 |
| Clinical sample 4 | (0.073–0.119) | (1.89–4.91) | 0.094 | 18.92 |
| CaSki | (2320–3025) | (1.56–4.33) | 2729 | 8.68 |
| HPV-18 105 | (809525–1014517) | (0.85–2.22) | 947017 | 6.97 |
| HPV-18 104 | (32831–47795) | (1.01 –2.99) | 41788 | 11.15 |
| HPV-18 102 | (200–326) | (0.66–2.22) | 270 | 16.83 |
| HPV-18 101 | (32 –54) | (1.05 –3.95) | 42 | 17.39 |
| HeLa | (158 –230) | (0.97 –3.92) | 175 | 14.30 |
| HPV-31 106 | (2231407–2599950) | (1.37–3.59) | 2463187 | 4.58 |
| HPV-31 104 | (20104–24975) | (1.22–3.14) | 21355 | 7.42 |
| HPV-31 102 | (126 –178) | (0.58 –3.82) | 153 | 12.86 |
| HPV-31 101 | (12 –19) | (1.45 –4.48) | 14 | 14.53 |
| HPV-39 106 | (7996214–9057376) | (0.74–2.74) | 8499714 | 3.91 |
| HPV-39 104 | (66156–82400) | (1.10–2.08) | 73513 | 7.43 |
| HPV-39 102 | (250 –346) | (1.08 –3.28) | 304 | 13.40 |
| HPV-39 101 | (22 –39) | (2.05 –4.67) | 32 | 16.86 |
| HPV-45 106 | (4252977–5118239) | (0.86–3.51) | 4660059 | 6.92 |
| HPV-45 104 | (57995–77422) | (2.12–4.83) | 67129 | 10.49 |
| HPV-45 102 | (562 –853) | (2.75 –4.98) | 665 | 14.33 |
| HPV-45 101 | (52 –82) | (3.40 –5.44) | 66 | 16.32 |
| HPV-51 106 | (623008–738661) | (0.47–3.96) | 686336 | 6.77 |
| HPV-51 104 | (11540–15910) | (2.29 –4.58) | 13447 | 11.56 |
| HPV-51 102 | (855 –1235) | (1.43 –5.09) | 984 | 13.43 |
| HPV-51 101 | (54 –82) | (2.78 –5.77) | 63 | 14.27 |
| HPV-52 106 | (3262783–4160203) | (1.83–3.86) | 3735372 | 7.21 |
| HPV-52 104 | (19151–25694) | (1.42–4.72) | 22695 | 10.52 |
| HPV-52 102 | (160 –266) | (0.87 –3.54) | 208 | 14.81 |
| HPV-52 101 | (24 –41) | (1.59 –4.71) | 30 | 17.28 |
| HPV-58 106 | (4131939 –4829752) | (1.09–3.60) | 4437483 | 5.17 |
| HPV-58 104 | (22696–30833) | (1.45 –4.22) | 26394 | 11.22 |
| HPV-58 102 | (214 –296) | (1.05 –4.62) | 250 | 15.00 |
| HPV-58 101 | (23 –37) | (2.25–5.28) | 30 | 16.38 |
Average viral load values and the coefficient of variation range (intra-assay variation) were calculated based on the mean and standard deviation of three replicate run on the same assay.
Average viral load mean values for eight independent assays. The correspondent coefficient of variation (inter-assay variation) was calculated based on the individual mean and standard deviation of eight independent assays (n = 3 × 8).
The DNA reference solution for HPV types 16, 18, 31, 39, 45, 51, 52, and 58 were diluted in TE buffer to obtain four concentrations (101 copies/μl, 102 copies/μl, 104 copies/μl, and 106 copies/μl). Each dilution was used as control and run in triplicate in eight independent assays. Clinical samples 1 to 4 are the biological samples obtained from exfoliated cervical cells. The viral load values of the biological samples are given in copy number per cell equivalent as calculated by RNase-P.
To estimate the variability of measuring viral burden in biological samples we analyzed four DNA samples obtained from exfoliated cervical cells, as well as DNA from CaSki and HeLa cells known to contain HPV-16 and HPV-18, respectively. DNA concentration was determined by real-time PCR using a commercially available kit to quantitate RNase P. Approximately 3 ng of each DNA sample was subjected to real-time PCR for HPV-16 and HPV-18. This analysis was conducted in triplicate and conducted in eight independent experiments. The results of the overall variability for the viral load measurement are shown in Table 3. As indicated in this table, the intra- and inter-assay coefficients of variation were less than 5% and 18.92%, respectively.
Accuracy
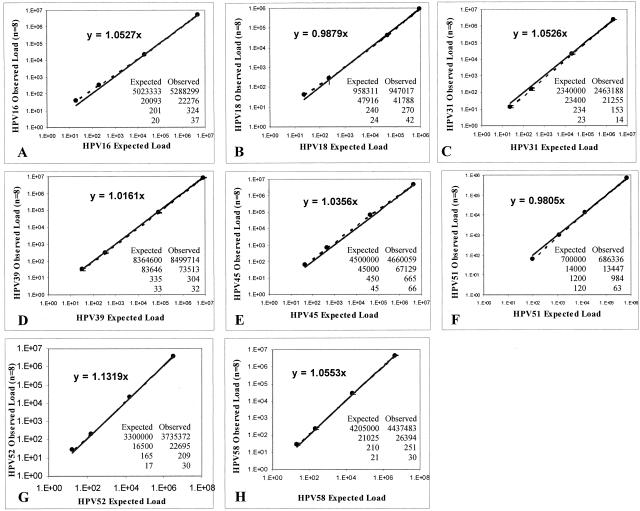
To assess assay accuracy we plotted, in a log-log scale, the theoretical input copy number of HPV controls at four levels of analyte concentration against the mean copy values of three replicates obtained from eight independent experiments. The slopes created by plotting the observed and expected values for each specific HPV type are shown in Figure 1. The deviation from the line of identity (slope = 1) was within the 90% CI for all type-specific HPV plots and ranged between 0.0121 and 0.0553.
Figure 1.
Accuracy of HPV quantitation. Experimentally determined viral load values (y axis) for HPV-16 (A), HPV-18 (B), HPV-31 (C), HPV-39 (D), HPV-45 (E), HPV-51 (F), HPV-52 (G), and HPV-58 (H) were plotted against the expected input copy number (x axis) in a log-log scale. Each point represents the mean value with the SE for triplicate determinations of eight independent assays (n = 3 × 8). Accuracy was determined using the slope value of the tendency line (allowed error window, m = 1.0 ± 0.05).
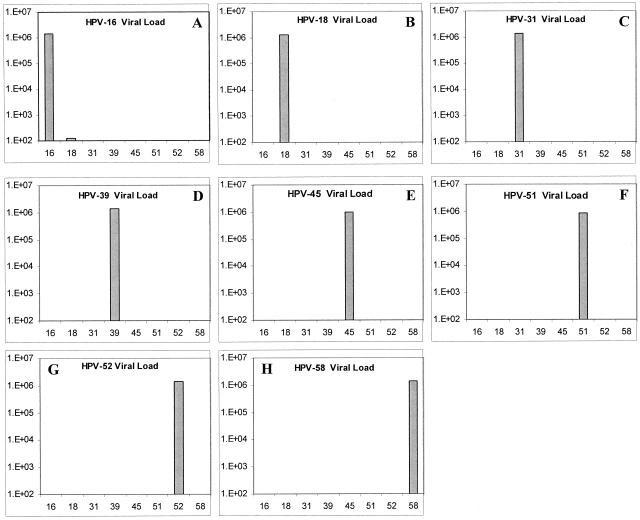
Specificity of the Quantitation Assay
Assay specificity (the ability to discriminate among phylogenetically related HPV types) was evaluated using different HPV clones as heterologous templates in type-specific reactions that were quantitated against the correspondent HPV standard curve. As observed in Figure 2, all HPV reactions showed a high degree of type specificity with the exception of HPV-16, which showed cross-reactivity with HPV-18 when an input copy number of 1 × 106 was used (100 copies detected after 45 cycles). No variation in specificity was observed in a duplicate assay performed at a different time (data not shown).
Figure 2.
Specificity assay for HPV-16 (A), HPV-18 (B), HPV-31 (C), HPV-39 (D), HPV-45 (E), HPV-51 (F), HPV-52 (G), and HPV-58 (H) using heterologous templates and type-specific fluorogenic oligo-probes. The bar (▪) in each panel corresponds to the mean viral load value for three replicates on the same assay ± SD.
Discussion
Our objective was to design real-time quantitative assays to determine viral load in a high-throughput format using a 10-μl volume reaction in the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). The performance evaluation of these assays was conducted following the protocol EP10-P of the NCCLS. For probe and primer design, we chose the E6-E7 region of the HPV genome since this region is highly conserved among the oncogenic HPV types and is intact in both the episomal and integrated forms of infectious HPV.
The selection of an appropriate material to be used as external control presents a particular challenge in developing a quantitative method for HPV since the viral genome can be present in either episomal or integrated forms at different stages during the natural history of HPV infection. Tucker et al14 reported the design of a real-time fluorogenic assay targeting the E6-E7 region to quantitate HPV-16 and HPV-18 viral load. However, this assay used a 50-μl volume reaction and the standard curve was created using PCR amplicons from the targeted regions diluted in buffer and using tRNA as carrier potentially leading to an overestimation of the sensitivity of the assay due to a less complex environment of the standard in comparison with DNA extracted from cells. A recent study using real-time PCR to determine HPV-16 viral load in cervical scrapes used a plasmid clone that contained the full-length HPV-16 DNA to create standard curves.17 However, the standard curves contained only diluted plasmids and the authors did not mention the usage of carrier DNA, and therefore may introduce systematic error in the sensitivity of the assay. In our quantification assay, human blood DNA was spiked with plasmids harboring the targeted HPV region or the whole HPV clone to create standard curves. The addition of human cell equivalents was conducted to account for the natural complexity of the DNA extracted from exfoliated cervical cells since the ultimate goal for our HPV quantitation assay is to use it as a tool in population-screening studies. We tested a range of input DNA and we observed that the dynamic range of the standard curve decreased with increasing concentrations of human DNA (data not shown). Although a large quantity of DNA can be obtained from cervical swabs, the samples have to be diluted to the optimal range of input DNA concentration to have a more accurate reading of viral load.
Perhaps the single most important factor in quantitative assays is the accuracy of the reference solution used as an external standard. Several studies using real-time PCR for the determination of viral load reported different methods to quantitate their reference solutions such as UV absorption,21 fluorescence,14,15,22 or they simply failed to describe the method used. Without a doubt, assessing the exact amount of DNA to be used as reference presents a major challenge for absolute quantification particularly while generating the low-end region of the standard curve. Several reports have used limiting dilution to quantitate targeted analytes with high accuracy and precision.23,24,25 Consequently, we used a modification of a novel and highly accurate determination of DNA concentration based on the quantitation of low copy number of the analyte using the PCR-based limiting dilution assay described by Rodrigo et al.18 Based on the concentration of the reference materials determined by limiting dilution, the dynamic range for all HPV standard curves cover a ranged of up to seven orders of magnitude (Table 2). It is worth noting, however, that the quantitation of reference materials using limiting dilution can introduce bias within the context of sensitivity since it assumes that one copy of the targeted analyte can be amplified by PCR and that enough signal is actually detected. This can create ambiguity in defining the lower limit of detection as observed in the HPV-16 and HPV-52 standard curves where less than 100% of replicates showed no positive amplification during the PCR reaction at one copy per reaction (Table 2). Another possibility explaining the reduction of sensitivity of the HPV-16 and HPV-52 standard curves may be related to the probe sequence itself and the efficiency to anneal when the targets are present in a diluted environment. In the case of the HPV-16 standard curve, not all of the 100 standards showed a positive signal in the PCR amplification reaction reflecting a lack of sensitivity at this concentration. Also, the efficiency of the HPV-16, HPV-18, and HPV-52 standard curves were ∼1.8 which reflects the need for optimization of the PCR conditions, either Mg+2 concentration, type of Taq polymerase, annealing temperature, or designing alternative sets of probes. The low efficiency observed of these standard curves is reflected in the high variability in the low-end of the standard curve (Figure 1). For HPV-16 and -18, these results reflect the difficulties in translating a previously described method to a different format.
The performance evaluation of the eight HPV quantitation assays showed an intra-assay variation of less than 6% and an inter-assay coefficient of variation less than 20% illustrating the high reproducibility of the method (Table 3). The inter-assay variability in similar validation protocols for real-time PCR assays has been reported to be up to 50%.21,25,26 The high inter-assay variability observed in clinical samples compared to controls is a reflection of the complexity and heterogeneity of DNA extracted from exfoliated cervical cells. The accuracy of the eight HPV quantitation assays was high as determined by the small deviation from the line of identity as depicted in Figure 1. The increment in variation of the observed versus expected relation is a common observation in PCR reactions since the HPV controls were obtained from bacteria and thus some variations in the PCR reaction may be due to the presence of inhibitors.12 Another possibility includes the variation during PCR preparation where bias can be introduced either by mechanical failure of measuring the correct volume in dilutions or by the stochastic process of targeting analytes in a more diluted environment where the homogeneity of sample plays a key role. In addition, the inherent variability in the PCR reaction per se is an important factor for the PCR performance (ie, enzymatic efficiency, primer-probe annealing, and buffer variation). The robustness of the quantitation assay (the capacity of the method to remain unaffected by small variations in the main parameters) was tested by a multiplex assay to simultaneously quantitate HPV-16 and HPV-18 in the same reaction. The results, however, showed a marked decrease in sensitivity thus suggesting that increasing the complexity of the reaction can lead to competition for PCR components (data not shown). The limitation of the present quantitation method is the need to determine a housekeeping gene as a measure of cell equivalent in a separate reaction, thus introducing variability with respect to viral load quantitation from the same sample.
One of the advantages of using in-house standards is the ability to mimic the complexity of the biological sample being tested. In our assay, the intra- and inter-assay variation showed good reproducibility when DNA from exfoliated cells was used. It was important to evaluate this parameter since the intent is to use the quantitative protocol in population-based screening studies. Although the variability in our quantitative assay was low, the estimation of inter-laboratory reliability is hard to assess since each research laboratory creates its own reference material making the results from independent labs difficult to compare (Gravitt PE, personal communication). In addition, the stability of the in-house reference material can vary overtime even if kept at frozen temperatures, probably due to bacterial remnants during the purification process and the liability of DNA to degrade. It is, therefore, of great importance to develop a reference material with enough robustness in both precision and accuracy that introduces minor systematic error or bias due to batch-to-batch variation for inter-laboratory evaluation.
Our assay showed high degree of specificity for seven of the eight type-specific reactions. The cross-reactivity with HPV-18 observed in the HPV-16 reaction is in agreement with the results obtained by Tucker et al.14 This cross-reactivity could be the result of non-specific primer and probe hybridization under the PCR conditions used. However, this represents only a small variation in the specificity of the HPV-16 reaction since only 100 copies are quantitated after 45 PCR cycles with an input DNA of approximately 1 × 106 copies per reaction. The output/input ratio of 0.001 for HPV-16 indicates that the HPV-16 primers anneal with low efficiency to a similar yet unspecific region. Strategies to improve specificity include changing parameters in the PCR reaction such as the addition of DNA-stabilizing co-solvent to increase the melting temperature and use of conditions that are more stringent in the PCR reaction.27
To our knowledge, this is the first time that an absolute quantitation assay to determine HPV viral load has been evaluated using quality assessment protocols for its potential to be transferred from the research lab to the analytical lab. However, the clinical significance of HPV viral load and the robustness of real-time PCR measure as primary screening tools in cervical cancer prevention and control remains to be determined. The real-time PCR assay developed in our laboratory is highly specific for eight oncogenic HPV types, highly reproducible in a wide dynamic range, performed in minimal reaction volume, and suitable for high-throughput format.
Acknowledgments
We thank Dr. Luisa L. Villa, Dr. Patty E. Gravitt, Dr. David C. Swan, Andrea Trevisan, and Linda Vaught for helpful discussions and comments on the experiments.
Footnotes
Supported in part by a grant from the National Cancer Institute Grant RO1-CA81310.
References
- Ho GYF, Bierman R, Beardsley L, Chang CJ, Burk RD. Natural history of cervicovaginal papillomavirus infection in young women. N Engl J Med. 1998;338:423–428. doi: 10.1056/NEJM199802123380703. [DOI] [PubMed] [Google Scholar]
- Davies P, Kornegay J, Iftner T. Current methods of testing for human papillomavirus. Best Practice Res Clin Obstet Gynecol. 2001;15:677–700. doi: 10.1053/beog.2001.0214. [DOI] [PubMed] [Google Scholar]
- Schiffman MH, Schatzin A. Test reliability is critically important to molecular epidemiology: an example from studies of human papillomavirus infection and cervical neoplasia. Cancer Res. 1994;54:1944–1947. [PubMed] [Google Scholar]
- Solomon D, Schiffman M, Tarone R. Comparison of three management strategies for patients with atypical squamous cells of undetermined significance: baseline results from a randomized trial. J Natl Cancer Inst. 2001;93:293–299. doi: 10.1093/jnci/93.4.293. [DOI] [PubMed] [Google Scholar]
- Ylitalo N, Sorensen P, Josefsson AM, Magnusson PK, Andersen PK, Ponten J, Adami HO, Gyllensten UB, Melbye M. Consistent high viral load of human papillomavirus 16 and risk of cervical carcinoma in situ: a nested case-control study. Lancet. 2000;355:2194–2198. doi: 10.1016/S0140-6736(00)02402-8. [DOI] [PubMed] [Google Scholar]
- Joseffson AM, Magnusson PK, Ylitalo N, Sorensen P, Qwarforth-Tubbin P, Andersen PK, Melbye M, Adami HO, Gyllensten UB. Viral load of human papilloma virus 16 as a determinant for development of cervical carcinoma in situ: a nested case-control study. Lancet. 2000;355:2189–2193. doi: 10.1016/S0140-6736(00)02401-6. [DOI] [PubMed] [Google Scholar]
- Peyton CL, Schiffman M, Lorincz AT. Comparison of PCR- and hybrid capture-based human papillomavirus detection systems using multiple cervical specimen collection strategies. J Clin Microbiol. 1998;36:3248–3254. doi: 10.1128/jcm.36.11.3248-3254.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzato F, Singer A, Mould T, Santos LC, Maia A, Cariri L. Cervical cancer detection by hybrid capture and evaluation of local risk factors. Int J Gynecol Obstet. 2001;73:41–46. doi: 10.1016/s0020-7292(00)00390-8. [DOI] [PubMed] [Google Scholar]
- Ho GYF, Burk RD, Klein S, Kadish AS, Chang CJ, Palan P, Basu J, Tachezy R, Lewis R, Romney S. Persistent genital human papillomavirus infection as a risk factor for persistent cervical dysplasia. J Natl Cancer Inst. 1995;87:1365–1371. doi: 10.1093/jnci/87.18.1365. [DOI] [PubMed] [Google Scholar]
- Swan DC, Tucker RA, Tortolero-Luna G, Mitchel MF, Wideroff L, Unger ER, Nisenbaum RA, Reeves WC, Icenogle JP. Human papillomavirus (HPV) DNA copy number is dependent on grade of cervical disease and HPV type. J Clin Microbiol. 1999;37:1030–1034. doi: 10.1128/jcm.37.4.1030-1034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlecht NF, Trevisan A, Duarte-Franco E, Rohan TE, Ferenczy A, Villa LL. Viral load as a predictor of the risk of cervical intraepithelial neoplasia. Int J Cancer. 2003;103:519–524. doi: 10.1002/ijc.10846. [DOI] [PubMed] [Google Scholar]
- Bickley J, Hopkins D. Inhibitors and enhancers of PCR. Saunders G, Parkes H, editors. London, England: LGC, UK. Royal Society of Chemistry; Analytical Molecular Biology. (ed 1.) 1999:81–102. [Google Scholar]
- Joseffson A, Livak K, Gyllensten U. Detection and quantitation of human papillomavirus by using the fluorescent 5′ exonuclease assay. J Clin Microbiol. 1999;37:490–496. doi: 10.1128/jcm.37.3.490-496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RA, Unger E, Holloway BP, Swan DC. Real-time PCR-based fluorescent assay for quantitation of human papillomavirus types 6, 11, 16, and 18. Mol Diagn. 2001;6:39–47. doi: 10.1054/modi.2001.21899. [DOI] [PubMed] [Google Scholar]
- Klussmann JP, Weissenborn SJ, Wieland U, Dries V, Kolligs J, Jungehuelsing M, Eckel HE, Dienes HP, Pfister HJ, Fuchs PG. Prevalence, distribution, and viral load of human papillomavirus 16 DNA on tonsillar carcinomas. Cancer. 2001;92:2875–2884. doi: 10.1002/1097-0142(20011201)92:11<2875::aid-cncr10130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Beskow AH, Gyllensten UB. Host genetic control of HPV 16 titer in carcinoma in situ of the uterine cervix. Int J Cancer. 2002;101:526–531. doi: 10.1002/ijc.90010. [DOI] [PubMed] [Google Scholar]
- Van Duin M, Snijders PJF, Schrijnrmakers HJF, Voorhorst FJ, Rozendaal L, Nobbenhuis MAE, Van den Brule AJC, Verheijen RHM, Helmerhost TJ, Meijer C. Human papillomavirus 16 load in normal and abnormal cervical scrapes: an indicator of CIN II/III and viral clearance. Int J Cancer. 2002;98:590–595. doi: 10.1002/ijc.10232. [DOI] [PubMed] [Google Scholar]
- Rodrigo AG, Goracke PC, Rowhanian K, Mullins JI. Quantitation of target molecules from PCR-based limiting dilution assays. AIDS Res Hum Retroviruses. 1997;13:737–742. doi: 10.1089/aid.1997.13.737. [DOI] [PubMed] [Google Scholar]
- Taswell C. Limiting dilution assays for the determination of immunocompetent cell frequencies. J Immunol. 1981;126:1614–1619. [PubMed] [Google Scholar]
- Passey RB, Maluf KC. Foundations for validation of quantitative analytical methods in the clinical laboratory. Arch Pathol Lab Med. 1992;116:732–738. [PubMed] [Google Scholar]
- Locatelli G, Santoro F, Veglia F, Gobbi A, Lusso P, Malnati M. Real-time quantitative PCR for human herpesvirus 6 DNA. J Clin Microbiol. 2000;38:4042–4048. doi: 10.1128/jcm.38.11.4042-4048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan DC, Tucker RA, Holloway BP, Icenogle JP. A sensitive, type-specific, fluorogenic probe assay for detection of human papillomavirus DNA. J Clin Microbiol. 1997;35:886–891. doi: 10.1128/jcm.35.4.886-891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. Quantitation of targets for PCR by use of limiting dilution. Biotechniques. 1992;13:444–448. [PubMed] [Google Scholar]
- Morrison TB, Ma Y, Weis JH, Weis JJ. Rapid and sensitive quantification of Borrelia burgdorferi-infected mouse tissues by continuous fluorescent monitoring of PCR. J Clin Microbiol. 1999;37:987–992. doi: 10.1128/jcm.37.4.987-992.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha BK, Tian B, Bucy P. Quantitation of HIV-1 by real-time PCR with a unique fluorogenic probe. J Virol Methods. 2001;93:33–42. doi: 10.1016/s0166-0934(00)00288-3. [DOI] [PubMed] [Google Scholar]
- Broccolo F, Locatelli G, Sarmati L, Pergiovanni S, Veglia F, Andreoni M, Butto S, Ensoli B, Lusso P, Malnati MS. Calibrated real-time PCR assay for quantitation of human herpesvirus 8 DNA in biological fluids. J Clin Microbiol. 2002;40:4652–4658. doi: 10.1128/JCM.40.12.4652-4658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevet E, Lemaitre G, Katinka MD. Low concentrations of tetramethylammonium chloride increase yield and specificity of PCR. Nucleic Acid Res. 1995;23:3343–3344. doi: 10.1093/nar/23.16.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]