Abstract
We report the identification of a virulent Streptococcus organism associated with fulminant endocarditis, using 16S rRNA gene amplification, sequencing and assembly from formalin-fixed, paraffin-embedded archival heart valve tissue, years after the autopsy of a patient.
Case Report
A 55-year-old white female had a congenitally abnormal aortic valve replaced with a porcine aortic valve to repair an associated left ventricular out-flow obstruction 8 years before her final illness. The same year the patient had a permanent pacemaker implanted for complete heart block. Several weeks before death the patient experienced increasing fatigue and abdominal complaints. An outpatient work-up, including an echocardiogram and blood culture, was negative at that time. Fever, cutaneous or eye findings suggestive of endocarditis were absent. The day before her death she presented to the Emergency Department with complaints of severe chest pain associated with nausea and diaphoresis. On admission, the patient was in mild respiratory distress and had bilateral rales. Her temperature was 99°F, pulse 85/minute, respirations 18/minute, and blood pressure 123/34 mm Hg. Physical examination of her heart and abdomen were normal. Her extremities showed no sign of deep vein thrombosis. Her chest X-ray showed no pulmonary edema and no evidence of a pneumothorax. An EKG showed no acute changes. Lab studies showed a white blood cell count of 13000/μl with a left shift, hemoglobin (Hgb) 10.9 g/dl, hematocrit (Ht) 35.8, platelets 227,000/μl, normal electrolytes and a blood urea nitrogen (BUN) and creatinine of 10 and 0.7. Creatin kinase (CK) level was 7 and the troponin was 0.06. The patient received multiple doses of nitroglycerin and metoprolol, aspirin, morphine, and furosemide. She was transferred to the Coronary Care Unit for further care. She was started on heparin protocol. A ventilation-perfusion (V/Q) scan ruled out pulmonary embolism. The following day the patient’s respiratory status worsened with increasing dyspnea, decreased oxygen saturation, and hypotension requiring additional supplemental oxygen via face mask. Subsequently the patient experienced respiratory and cardiac arrest and died despite aggressive resuscitative efforts.
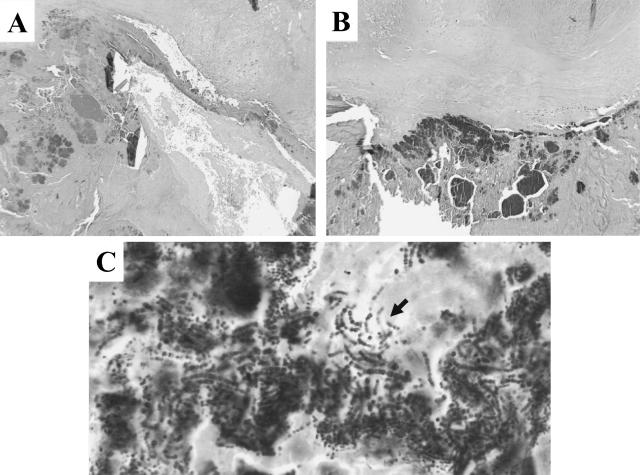
At autopsy, the prosthetic aortic valve showed almost complete dehiscence from its attachment to the aorta, involving the middle and posterior leaflets. The valve leaflets were thickened, partially nodular, and necrotic with focal perforation. Microscopically intense valvulitis characterized by necrosis and destruction of the aortic valve substance was seen (Figure 1, A and B). This process was contiguous with multiple vegetations containing fibrin, neutrophils and numerous bacterial colonies extending to the left ventricle endocardium. A gram stain showed numerous gram-positive cocci in aggregates and chains most consistent with streptococcal organisms (Figure 1C). Evidence of metastatic infection or thrombosis was not seen in other organs examined at autopsy. Neither blood nor valve culture was performed at the time of the autopsy.
Figure 1.
A and B: H&E-stained aortic valve slices obtained before and after three 20-μm thick slices cut for total genomic DNA purification (magnification, ×4). C: High-power magnification (magnification, ×60) of the gram-stained bacterial colonies. The arrow shows gram-positive cocci in chains.
Three years after the autopsy, a medico-legal issue raised questions about the identity of the apparent streptococcal organisms, and we attempted to identify the pathogenic bacterial strain from the paraffin-embedded valve tissue using 16S rDNA sequence determination.
Molecular Studies
We extracted total genomic DNA from three 20-μm thick slices of the formalin-fixed, paraffin-embedded, infected cardiac tissue (Figure 1) using a Puregene DNA purification kit (Gentra Systems, Minneapolis, MN) in the protocol as recommended by the manufacturer. The resulting total genomic DNA was dissolved in 40 μl PCR-grade distilled water. The same protocol was used to obtain genomic DNA from formalin-fixed, paraffin-embedded human tissue devoid of evidence of bacterial infection as a negative control for the subsequent PCR reactions. We also extracted genomic DNA from a colonial growth of a Moraxella species to serve as positive control. To amplify fragments of the bacterial 16S rRNA gene, we used primer pairs for the conserved regions of the 16S rRNA gene and PCR reaction parameters as described by Persing3 using a PTC 100 Programmable Thermal Controller (MJ Research, Inc., Waltham, MA) and Ampli Taq DNA polymerase (Applied Biosystems, Foster City, CA).
Our initial attempt to amplify a 800-bp fragment using primer pairs 8FPL-806R was unsuccessful, perhaps because the formalin-fixed, paraffin-embedded tissue suffered DNA degradation during the 3-year storage. We therefore targeted short DNA fragments using primer pair 515FPL-806R. We were able to amplify an approximately 300-bp fragment using the template purified from the paraffin block containing the colonized valve tissue (data not shown). This PCR fragment was isolated and then purified with a gel extraction kit and a PCR purification kit according to the manufacturer’s instructions (Qiagen, Germany). The nucleotide sequence of this 300-bp fragment was determined using the 515FPL forward and the 806R reverse primers for direct sequencing of the isolated and affinity-purified fragments by an ABI automatic sequencer at the Dartmouth College Molecular Biology Core Facility (Hanover, NH). The resulting sequence (515FPL-806R) was compared to the GenBank database using BLASTn.2 The result of the BLASTn search showed 100% homology of this 300-bp sequence to the corresponding 16S rDNA sequences of several well characterized (S. oralis, S. mitis, S. gordonii) and lesser known (S. sp. 3170A, S. sp. 3097C, S. sp. 2056B, S. sp. 4093B) oral Streptococcus species. To confirm the identity of the colonizing Streptococcus species we designed the following interim primers based on the conserved regions of the 16 rRNA genes of these streptococci:
315FP;CAGACTCCTACGGGAGGCAGCAGTAGGG AATC:
369RP;GACTTCCGTCCATTGCCGAAGATTCCCTACT GCTGC:
520RP;CTCGCTTTACGCCCAATAAATCCGGACAAC:
750FP;CTGAGGCTCGAAAGCGTGGGGAGCAAAC AGG:
1110FP;CATCATTCAGTTGGGCACTCTAGCGAGAC:
1150RP;GGTTTATTACCGGCAGTCTCGCTAGAGTGC,
(FP-forward primer; RP-reverse primer). These primers were then used along with the published conserved region primers (8FPL, 1492RPL)3 to amplify the entire 16S rDNA in the form of four additional overlapping fragments (8FPL-369R; 315F-520R; 750F-1150R; 1110F-1492RPL). There were two separate PCR reactions performed for each primer pair. The resulting PCR products were electrophoresed in and isolated from 1.1% agarose gels, affinity purified, and the nucleotide sequences were determined as described above. The assembly of the sequences of the five PCR fragments resulted in a 1464-bp near-complete 16S rDNA sequence (GenBank Accession Number AY256519).
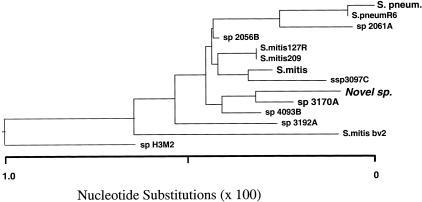
Comparison of this sequence to the GenBank nucleotide sequence database using BLASTn showed that this 16S rDNA sequence was closely related but not identical to a group of Streptococcus species. The results of the BLASTn search showed the highest homology to a recently described Streptococcus species, sp 3170A (99.795%) and less homology to S. mitis (99.658%) and S. pneumoniae (99.454%). Streptococcus sp. 3170A has recently been isolated from the human oral flora and has been found to be similar to S. mitis, S. pneumoniae, and S. oralis.4 Multiple alignment of these closely related 16S rDNA sequences along with our query sequence using the Jellyfish software (LabVelocity.com), showed the clustering of the nucleotide differences at mutational “hot spots” (not shown). The same sequences were used to create a phylogenetic tree using the LaserGene software (DNAStar, Inc., Madison, WI) (Figure 2).
Figure 2.
Phylogenetic tree of 12 closely related Streptococcus species generated by LaserGene software.
Discussion
Bacterial endocarditis has been classified historically as “acute” or “subacute” based on the rapidity of progression of the untreated disease. The acute form follows a fulminant course, with death occurring within a few days to a few weeks. Subacute endocarditis commonly occurs in the setting of prior valvular disease, is characterized by a slow, indolent course with vague systemic complaints and cutaneous findings and is frequently associated with viridans streptococci.1
Gram-positive organisms, mainly streptococci and staphylococci, continue to cause the majority of cases of infective endocarditis.5 A year-long population-based survey conducted in France, identified members of the genus Streptococcus as pathogens in 48% of infective endocarditis cases.6 S. pneumoniae has been associated with acute endocarditis, but the prevalence of this association has decreased in the antibiotic era from approximately 15% to less than 1%.7,8 In a recent prospective point prevalence study of oropharyngeal S. pneumoniae carriage rates among outpatients, only 30% of isolates presumed to be S. pneumoniae by standard identification criteria (optochin and bile solubility testing) could be confirmed by using a commercial DNA probe. More importantly, the 70% DNA probe-negative isolates showed a significantly reduced antibiotic susceptibility pattern compared to the DNA probe-positive isolates. Four streptococcus isolates, which were ambiguous by optochin and bile solubility testing, were identified by their 16S rRNA sequences.4 The 16S rRNA gene sequence of one of these isolates (sp 3170A) differed in only three nucleotides from the sequence of the 16S rDNA obtained from the infected cardiac tissue of our case report patient. Classification of infective endocarditis by etiological agent can have implications for the course of disease and the selection of antimicrobial agent(s). Several case reports have been published about pathogen identification using 16S rDNA sequence determination from resected colonized heart valves of infective endocarditis patients (eg, 9,10,11,12,13), and the method has recently been reported as a useful diagnostic adjunct to culture methods.14
The organism implicated in this case of fulminant endocarditis belongs to a group of streptococci that are very closely related and difficult to separate even by the genetic means we have used. No other accepted genetic or phenotypic differentiation method was available to us without recovery of a live organism. The fulminant nature of the disease, and the histological findings of an acute necrotizing process that led to valve dehiscence indicate that the organism likely to have phenotypic virulence characteristics similar to S. pneumoniae. Organisms now described in the literature as related to both S. mitis and S. pneumoniae may be capable of highly virulent infections.
In summary, we have identified a virulent streptococcus species by reconstructing its 16S rDNA sequence from a years-old archival paraffin block containing cardiac valve tissue from a patient with fulminant endocarditis.
Acknowledgments
We thank Beth Pingleton, Laure Tooke, and Arnold B. Hawk for their technical help and Dr. Robert H. Gross (Center for Biological and Biomedical Computing, Dartmouth College) and Jason Fladhammer (DNAStar, Inc) for their help with the LaserGene software.
References
- Mandell GD. New York: Churchill Livingstone, Inc.; Principles and Practice of Infectious Diseases. (ed 5.) 2000:857. [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persing DH, Smith TF, Tenover FC, White TJ, editors. Rochester, MN: Mayo Foundation; Diagnostic Medical MicrobiologyPrinciples and Applications. 1993:489–495. [Google Scholar]
- Wester CW, Ariga D, Nathan C, Rice TW, Pulvirenti J, Patel R, Kocka F, Ortiz J, Weinstein RA. Possible overestimation of penicillin-resistant Streptococcus pneumoniae colonization rates due to misidentification of oropharyngeal streptococci. Diagn Microbiol Infect Dis. 2002;42:263–268. doi: 10.1016/s0732-8893(01)00358-3. [DOI] [PubMed] [Google Scholar]
- Hoen B. Special issues in the management of infective endocarditis caused by gram-positive cocci. Infect Dis Clin North Am. 2002;16:437–452, xi. doi: 10.1016/s0891-5520(01)00004-6. [DOI] [PubMed] [Google Scholar]
- Hoen B, Alla F, Selton-Suty C, Beguinot I, Bouvet A, Briancon S, Casalta JP, Danchin N, Delahaye F, Etienne J, Le Moing V, Leport C, Mainardi JL, Ruimy R, Vandenesch F, Association pour l’Etude et la Prevention de l’Endocarditeinfectieuse (AEPEI) Study Group Changing profile of infective endocarditis: results of a 1-year survey in France. JAMA. 2002;288:75–81. doi: 10.1001/jama.288.1.75. [DOI] [PubMed] [Google Scholar]
- Whitby S, Pallera A, Schaberg DR, Bronze MS. Infective endocarditis caused by Streptococcus pneumoniae with high-level resistance to penicillin and cephalosporin. Clin Infect Dis. 1996;23:1176–1177. doi: 10.1093/clinids/23.5.1176. [DOI] [PubMed] [Google Scholar]
- Lefort A, Mainardi JL, Selton SC, Cassasus P, Guillevin L, Lortholary O. Streptococcus pneumoniae endocarditis in adults: a multi-center study in France in the era of penicillin resistance (1991–1998) Medicine (Baltimore) 2000;79:327–337. doi: 10.1097/00005792-200009000-00006. [DOI] [PubMed] [Google Scholar]
- Goldenberger D, Kunzli A, Vogt P, Zbinden R, Altwegg M. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J Clin Microbiol. 1997;35:2733–2739. doi: 10.1128/jcm.35.11.2733-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilck MB, Yanyun W, Howe JG, Crouch JY, Edberg SC. Endocarditis caused by culture-negative organisms visible by Brown and Brenn staining: utility of PCR and DNA sequencing for diagnosis. J Clin Microbiol. 2001:2025–2027. doi: 10.1128/JCM.39.5.2025-2027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Vermot I, Altwegg M, Zimmerli W, Fluckiger U. Duke criteria-negative endocarditis caused by Bartonella quintana. Infection. 1999;27:283–285. doi: 10.1007/s150100050032. [DOI] [PubMed] [Google Scholar]
- Qin X, Urdahl KB. PCR and sequencing of independent genetic targets for the diagnosis of culture-negative bacterial endocarditis. Diagn Microbiol Infect Dis. 2001;40:145–149. doi: 10.1016/s0732-8893(01)00263-2. [DOI] [PubMed] [Google Scholar]
- Tang Y-W, Hopkins MK, Kolbert CP, Hartley PA, Severance PJ, Persing DH. Bordetella holmesii-like organisms associated with septicemia, endocarditis, and respiratory failure. Clin Infect Dis. 1998;26:389–392. doi: 10.1086/516323. [DOI] [PubMed] [Google Scholar]
- Gauduchon V, Chalabreysse L, Etienne J, Celard M, Benito Y, Lepidi H, Thivolet-Bejui F, Vandenesch F. Molecular diagnosis of infective endocarditis by PCR amplification and direct sequencing of DNA from valve tissue. J Clin Microbiol. 2003;41:763–766. doi: 10.1128/JCM.41.2.763-766.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]