Primary immunodeficiencies are a heterogeneous group of disorders, which affect cellular and humoral immunity or non-specific host defense mechanisms mediated by complement proteins, and cells such as phagocytes and natural killer (NK) cells.1,2,3 These disorders of the immune system cause increased susceptibility to infection, autoimmune disease, and malignancy. There are over 80 primary immunodeficiencies, many of which are very rare, and in most cases associated with inherited genetic defects. One in 500 individuals, in the United States, is born with a defect in some component of the immune system.4 In the majority of cases, the primary immunodeficiencies manifest in the first year of life.4 They can, however, present at any age, including adulthood.5 The advent of molecular genetic analyses now allows for the detection and confirmation of immunodeficiencies that were otherwise not severe enough during childhood to have led to a specific diagnosis. In addition, effective treatment for many disorders has led to increased survival of many children with primary immunodeficiency into adult life.
Recent advances in molecular techniques have led to the identification and characterization of more than 25 newly recognized immunological disease genes since 1997.6,7 The identification of many genes responsible for primary immunodeficiencies has provided insights regarding the spectrum of clinical severity seen in a particular disorder and the phenotypic overlap resulting from mutations of different genes.
This review focuses on the molecular genetic features of primary immunodeficiencies with emphasis on the molecular pathophysiology of the diseases. In large part, the molecular detection of gene mutations leading to these diseases is carried out in research laboratories. Thus, information regarding comparison of analytical methods and prioritization of specific targets for study is largely lacking. Diagnostic perspectives for disease entities for which this information exists are included in their respective sections. We have organized the review into categories of B cell, T cell, severe combined immunodeficiencies, and defects of phagocytes and other miscellaneous immunodeficiencies. The clinico-pathological and immunological aspects are beyond the scope of this review and are available elsewhere. Various websites containing pertinent databases including those organized by the Pan-American Group for Immunodeficiencies (PAGID) http://www.clinimmsoc.org/pagid/, mutation registries, and the large European Society for Immunodeficiencies (ESID) registry which contains clinical data for over 7000 patients from 24 countries are listed in Table 1.8 In addition, the website for GeneTests-GeneClinics which provide information regarding specific laboratories and the nature of the molecular assays is also shown in Table 1.
Table 1.
Immunodeficiency Mutation Databases and Other Related Websites
| Database
|
Immunodeficiency disorder
|
Internet address of database
|
|---|---|---|
| ATbase | Ataxia-telangiectasia | http://www.ent.ki.se/ATbase/ |
| ATM | Ataxia-telangiectasia | http://www.vmresearch.org/atm.htm |
| BLMbase | Bloom syndrome | http://www.uta.fi/imt/bioinfo/BLMbase/ |
| BTKbase | X-linked agammaglobulinemia (XLA) | http://www.uta.fi/imt/bioinfo/BTKbase/ |
| CD3Ebase | CD3epsilon deficiency | http://www.uta.fi/imt/bioinfo/CD3Gbase/ |
| CD40Lbase | X-linked hyper-IgM syndrome (XHIM) | http://www.uta.fi/imt/bioinfo/CD40Lbase/ |
| CYBAbase | Autosomal recessive CGD p22 phox deficiency | http://www.uta.fi/imt/bioinfo/CYBAbase/ |
| CYBBbase | X-linked chronic granulomatous disease (XCGD) | http://www.uta.fi/imt/bioinfo/CYBBbase/ |
| FAA and FAC | Fanconi anemia | http://www.rockefeller.edu/fanconi/mutate/ |
| IL2RGbase | X-linked severe combined immunodeficiency (XSCID) | http://www.nhgri.nih.gov/DIR/LGT/SCID/ |
| JAK3base | Jak3 deficiency | http://www.uta.fi/imt/bioinfo/JAK3base/ |
| NCF1base | Autosomal recessive CGD p47 phox deficiency | http://www.uta.fi/imt/bioinfo/NCF1base/ |
| NCF2base | Autosomal recessive CGD p67 phox deficiency | http://www.uta.fi/imt/bioinfo/NCF2base/ |
| RAG1base | RAG1 deficiency | http://www.uta.fi/imt/bioinfo/RAG1base/ |
| RAG2base | RAG2 deficiency | http://www.uta.fi/imt/bioinfo/RAG2base/ |
| SH2D1Abase | X-linked lymphoproliferative syndrome (XLP) | http://www.uta.fi/imt/bioinfo/SH2D1Abase/ |
| ZAP70base | ZAP70 deficiency | http://www.uta.fi/imt/bioinfo/ZAP70base/ |
| Immune Deficiency Foundation | http://www.primaryimmune.org | |
| National Organization for Rare Disorders | http://www.raredisease.org | |
| Online Mendelian Inheritance in Man | http://www3.ncbi.nim.nih.gov | |
| GeneTests | http://www.genetests.org |
Classification
The International Union of Immunological Society’s Scientific Committee on Primary Immunodeficiency Diseases/World Health Organization4 define several major categories of primary immunodeficiencies including: defects in non-specific host defense (phagocytes, natural killer cells, complement); defects of specific humoral immunity (B lymphocytes, antibodies); combined deficiency of cellular (T cell mediated) and humoral immune defense; immune defects associated with other major defects; and immunodeficiencies associated with or secondary to other diseases. Selected primary immune disorders, their presumed pathogenesis, inheritance pattern, and diagnostic tests are summarized in Table 2.
Table 2.
Gene Defects, Inheritance, and Diagnostic Tests for Primary Immunodeficiencies
| Predominantly antibody deficiencies
| |||
|---|---|---|---|
| Designation | Pathogenesis/defect | Inheritance | Diagnostic tests |
| 1. X-linked agammaglobulinaemia | Mutations in btk | XL | Btk by immunoblotting or FACS analysis, mutation analysis |
| 2. Autosomal recessive agammaglobulinaemia | Mutations in μ or λ5 genes; others | AR | — |
| 3. Ig heavy-chain gene deletions | Chromosomal deletion at 14q32 | AR | — |
| 4. κ chain deficiency mutations at AR | Point mutations at chromosome 2p11 in some patients | AR | — |
| (a) IgG subclass deficiency | Defects of isotype differentiation | Unknown | — |
| (b) IgA deficiency | Failure of terminal differentiation in IgA = B cells | Variable | Autoimmune and allergic disorders |
| 5. Common variable immunodeficiency | Variable; undetermined | Variable | See text |
| T cell deficiencies | ||||
|---|---|---|---|---|
| 1. Purine nucleoside phosphorylase (PNP) deficiency | Deficiency of PNP | AR | Red cell PNP levels and metabolite | |
| Mutation analysis | ||||
| 2. CD3 γ or CD3ε deficiency | Defect in TCR signaling | AR | CD3 fluorescence intensity | |
| Mutation analysis | ||||
| 3. ZAP-70 deficiency | ZAP70 | AR | ZAP70 expression and activation | |
| Mutation analysis | ||||
| 4. X-linked lymphoproliferative syndrome | Uncontrolled B cell stimulation SH2DIA(Xq25) | XL | Mutation analysis, SAP expression under development | |
| Combined immunodeficiencies | ||||
|---|---|---|---|---|
| 1. T-B SCID | ||||
| (a) X-linked (γc deficiency) | Mutations in γ chain of IL-2, 4, 7, 9, 15 receptors | XL | CD154 expression on activated T cells by FACS analysis | |
| Mutation analysis | ||||
| (b) Autosomal recessive (Jak3 deficiency) | Mutation in Jak3 | AR | JAK3 expression/activation | |
| Mutation analysis | ||||
| 2. T-B-SCID | ||||
| (a) RAG ½ deficiency | Mutation in RAG 1/2 genes | AR | RAG1 and RAG2 mutation analysis | |
| (b) Adenosine deaminase (ADA) deficiency | T-cell and B-cell defects from toxic metabolites (eg dATP, S-adenosyl homocysteine) | AR | Red cell ADA levels and metabolites | |
| Mutation analysis | ||||
| (c) Other | Defective VDJ recombination | AR | ||
| 3. T + B-SCID | ||||
| (a) Omenn Syndrome | Missense mutations in RAG 1 and 2 genes | AR | RAG 1 and RAG2 | |
| Mutation analysis | ||||
| (b) IL-2Rα deficiency | Mutations in IL-2Rα gene | AR | IL-2R FACS expression | |
| 4. X-linked hyper IgM | Mutations in CD40 ligand gene | XL | CD145 expression on activated T cells by FACS analysis | |
| Mutation analysis | ||||
| 5. MHC class II deficiency | Mutation in transcription factors (CIITA bare lymphocyte syndrome, or RFX5, RFXAP, RFXANK genes) for MHC class II molecules | AR | HLA-DR expression | |
| Mutation analysis | ||||
| 6. TAP-2 deficiency | Mutations in TAP-2 gene | AR | HLA class I expression | |
| 7. CD45 deficiency | PTPRC | Mutation analysis | ||
| Congenital defects of phagocytic number and/or function
| |||
|---|---|---|---|
| Pathogenesis/gene defect | Inheritance | Diagnostic tests | |
| 1. Severe congenital neutropenia | Mutation in ELA2 | AR | Mutation analysis |
| 2. Cyclic neutropenia | Mutation in ELA2 | AR | Mutation analysis |
| 3. Leucocyte adhesion defect 1 [deficiency of beta chain (CD18) of LFA-1, Mac 1, p150, 95] | Chemotaxis, adherence, endocytosis, Integrin B2 | AR | Flow cytometry |
| 4. Leukocyte adhesion defect 2 (failure to convert GDP mannose to fucose) | Chemotaxis, rolling | AR | ND |
| 5. Chediak-Higashi syndrome | Defective cytotoxic T/NK cells, LYST gene | AR | Mutation analysis |
| 6. Specific granule deficiency | Chemotaxis/C/EBP gene | AR | Mutation analysis |
| 7. Shwachman syndrome | Chemotaxis, mutations in SBDS gene | AR | Mutation analysis |
(Table continues)
Epidemiology
Many immunodeficiencies are rare, however, the incidence of primary immunodeficiencies has increased by 10-fold since 1969 to 1 in 10,000.4 This is partly due to increased identification of affected patients, reduced morbidity and mortality from the introduction of antibiotics, enhanced methods of detection of immunological abnormalities, and the identification of gene mutations responsible for the disorders. Approximately 400 new cases of primary immunodeficiency are diagnosed per year in the U.S. See Table 3 for prevalence of primary immunodeficiencies.
Table 3.
Prevalence of Primary Immunodeficiencies
| Disease | Prevalence |
|---|---|
| IgA deficiency | 1/600 |
| X-linked agammaglobulinemia (XLA) | 1/200,000 |
| X-linked lymphoproliferative syndrome | <1/1,000,000 live births in males |
| X-linked immunodeficiency with hyper-IgM | <1/1,000,000 live births in males |
| X-linked severe combined immunodeficiency | 1/50,000 to 1/100,000 |
| JAK3-deficient severe combined immunodeficiency (autosomal recessive T-B+ SCID) | <1/500,000 live births |
| Adenosine deaminase deficiency | ½-1/100 births |
| B-cell negative SCID, Omenn syndrome | ∼1/100,000 |
| Ataxia-telangiectasia | ∼1/100,000 |
| Leukocyte adhesion deficiency type I | 1/100,000 |
| X-linked chronic granulomatous disease (X-CGD) | 1/250,000 |
| Autosomal CGD − p22phox deficiency | <1/2,000,000 |
| Autosomal CGD − p47phox deficiency | ∼1/500,000 |
| Autosomal CGD − p67phox deficiency | <1/2,000,000 |
Approach to Diagnosis
The primary immunodeficiencies characteristically present in childhood with infections that persist for long duration with multiple recurrences that are resistant to antibiotics. Failure to thrive and developmental delay are significant clues to the seriousness of their infections. Many immunodeficient children develop other symptoms such as skin rashes, and many have associated developmental anomalies of the face, skeletal system, heart, and pigmentation.
The nature of the pathogens and sites of infections can provide insight as to the underlying immunodeficiency. Defects involving B cell function result in recurrent sinopulmonary infections, often with bacterial septicemia. The lack of antibody production may also increase susceptibility to invasive disease with enteroviruses, resulting in chronic viral meningitis, and giardiasis. T cells are essential for the control of viral and fungal disease, however they also provide helper function to B cells for effective antibody responses. Thus, T cell disorders present as combined T and B cell immunodeficiency with susceptibility to both bacterial and chronic, invasive viral, and fungal pathogens. Patients with disorders of granulocytes are susceptible to staphylococcal diseases and gram-negative infections.
The primary immunodeficiencies are commonly inherited disorders, thus a family history is one of the best diagnostic clues. Unfortunately, because these diseases are rare with low carrier frequencies, a negative family history does not rule out a primary immunodeficiency. Furthermore, occurrence of new mutations, especially for X-linked disorders, is so high that the majority of patients with proven X-linked immunodeficiency mutations have no history of affected male relatives.
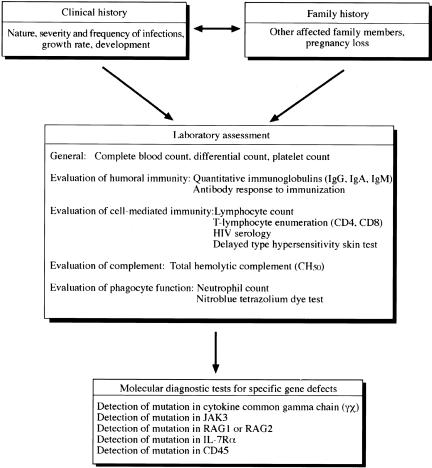
Laboratory Assessment
Initial laboratory evaluation for immunodeficiency should include a minimum of tests that can be performed reliably by any laboratory.9 The initial screening should include a complete blood count and quantitation of serum IgG, IgM, and IgA levels. Laboratories should provide age-matched normal values for cell counts, immunoglobulin measurements and proper controls for functional studies. Other readily available tests are: 1) Quantification of blood mononuclear cell populations: T cells (CD3, CD4, CD8, TCR αβ, TCR γδ); B cells (CD19, CD20, CD21, Ig); NK cells (CD16/CD56); monocytes (CD15); activation markers (HLA-DR, CD25, CD80 for B cells), CD154 for T cells. 2) T cell functional evaluation: delayed hypersensitivity skin tests (PPD, Candida, histoplasmin, and tetanus toxoid); proliferative response to mitogens (anti-CD3 antibody, phytohemagglutinin, concanavalin A) and allogeneic cells (mixed lymphocyte response); cytokine production. 3) B cell functional evaluation: natural or commonly acquired antibodies; response to immunization proteins and carbohydrate antigens; and quantitative IgG subclass determination. 4). Phagocyte function: reduction of nitroblue tetrazolium; chemotaxis assays, and bactericidal activity.
Diagnostic Criteria
The PAGID and ESID10 have established diagnostic criteria for immunodeficiencies which are divided into three categories: definitive, probable, and possible. These criteria provide objective guidelines that ensure that the same definitions are being used universally for diagnosis and clinical research studies. Patients with a definitive diagnosis are assumed to have a greater than 98% probability that in 20 years they would still be given the same diagnosis. Detection of a gene mutation is the most reliable method of making a diagnosis. In some disorders, the absence of the specific transcript or protein is diagnostic, although in others this may be hampered by transient or low levels of expression. The clinical and laboratory findings in several of the X-linked immunodeficiencies are sufficiently distinctive that if coupled with a family history that is specific to X-linked inheritance, a definitive diagnosis can frequently be made. In families with a known mutation in a particular gene, pre- or peri-natal testing can be used to establish a definitive diagnosis in a newborn or fetus. Table 4 highlights the major genetic abnormalities required for definitive diagnosis of primary immunodeficiency disorders.
Table 4.
Genetic Abnormalities Required for Definitive Diagnosis of Primary Immunodeficiency Disorders
| Disease | Gene defect |
|---|---|
| SCID | |
| Mutation in the cytokine gamma chain (γ) | |
| Mutation in JAK3 | |
| Mutation in RAG1 or RAG2 | |
| Mutation in IL-7Rα | |
| Mutations in both alleles of adenosine deaminase | |
| DiGeorge syndrome | |
| Deletion of chromosome 22q11.2 | |
| MHC-class II deficiency | |
| Mutation in one of the following genes: CIITA, RFX-B, RFX-5, RFX-AP | |
| Leukocyte adhesion defect | |
| Mutation in the β2 integrin gene | |
| Absence of the β2 integrin mRNA in leukocytes | |
| Chronic granulomatous disease | |
| Mutation in gp91, p22, p47 or p67 phox | |
| Absent mRNA for one of the above genes by Northern blot analysis | |
| X-linked SCID | |
| Mutation in the cytokine common gamma chain (γ) | |
| Absent γc mRNa on Northern blot analysis of lymphocytes | |
| X-linked Agammaglobulinemia | |
| Mutation in Btk | |
| Absent Btk mRNa on Northern blot analysis of neutrophils or monocytes | |
| Absent Btk protein in monocytes or platelets | |
| X-linked hyper IgM | |
| Mutation in the CD40L gene | |
| Ataxia telangiectasia | |
| Mutations in both alleles of ATM | |
| Wiskott-Aldrich syndrome | |
| Mutation in WASP | |
| Absent WASP mRNA on Northern blot analysis of lymphocytes | |
| Absent WASP protein in lymphocytes | |
| X-linked lymphoproliferative syndrome | |
| Mutation SH2D1 A/SAP/DSHP |
Modified from10.
Patients with a probable diagnosis are those with all of the clinical and laboratory characteristics of a particular disorder but do not have a documented abnormality in the gene, mRNA, or protein that is known to be abnormal in the given disorder. They are assumed to have a greater than 85% probability that in 20 years they will be given the same diagnosis. Patients with a possible diagnosis are those that have some but not all of the characteristic clinical or laboratory findings of a particular disorder. Since early diagnosis can prevent serious consequences, timely diagnosis of an index case can provide opportunity for genetic counseling, carrier detection, and prenatal diagnosis. Figure 1 outlines the algorithm for evaluation and diagnosis of patients with suspected severe combined immunodeficiency. As defined above, the identification of a gene mutation involving any of the cytokine common gamma chain (γχ), JAK3, RAG1, RAG2, IL-7Rα, or CD45 would lead to a definitive diagnosis of SCIDs. Those without evidence of gene mutation but the clinical and laboratory features of SCIDs will have a “probable” diagnosis of the disease.
Figure 1.
Algorithm for evaluation and diagnosis of a patient with suspected severe combined immunodeficiency.
B Cell Immunodeficiencies-Predominantly Antibody Defects
X-Linked Agammaglobulinemia
X-linked agammaglobulinemia (XLA) is a typical antibody deficiency in which production of antibodies is prevented due to a block in B cell maturation. Serum concentrations of IgG, IgA, and IgM are markedly reduced. Levels of circulating B cells are significantly decreased and plasma cells are absent from lymph nodes and bone marrow, although the number of T cells is normal or even increased. The clinical phenotype may be variable and even members of the same family can have different symptoms. The majority of affected boys present with recurrent bacterial infections from the age of 4 to 12 months following the disappearance of maternal immunoglobulin.11 Infections caused by pyogenic bacteria are the most common clinical manifestations. Rare cases of XLA have been described in adults.12
Molecular and Cellular Defects
X-linked agammaglobulinemia was the first immunodeficiency to be characterized at the genetic level.13 XLA is caused by a block in B cell differentiation due to mutations involving the Bruton kinase gene, Btk, which encodes a tyrosine kinase that regulates the activity of signaling pathways by phosphorylation.14,15 BTK is activated by immunoglobulins and other membrane receptors that in turn activate phospholipase Cγ and calcium influx. Although the specific defect in the signaling pathway that impairs B-cell development is unknown, it is proposed that BTK is important in mediation of survival signals.16 The phenotypic heterogeneity seen in patients does not reflect the consequences of different mutations as most of the mutations identified thus far lead to complete absence of BTK protein. Modifier genes and environmental factors have been postulated to play a role in influencing the expression pattern of the B cell deficiency. Flow cytometric methods to detect expression of BTK protein expression17 and molecular diagnostic tests allow the detection of the carrier state and enable prenatal diagnosis of the disorder.18 However, since approximately one third to one half of XLA cases are sporadic, alternative mechanisms of diagnosis are necessary. Furthermore, deleterious mutations of the μ heavy chain gene,19 lambda 5/14–1 surrogate light chain gene,20 Blnk gene which encodes an adaptor protein that has a critical role in pro-B to pre-B cell progression,21 and the CD79a gene22 have been identified in agammaglobulinemia patients.
Hyper-IgM Syndrome
Hyper-IgM syndrome (HIM) represents a group of distinct entities characterized by defective normal or elevated IgM in the presence of diminished IgG and IgA levels.23 Seventy per cent of the cases are X-linked in inheritance,24 and others are autosomal recessive.25 Male patients with X-linked hyper-IgM have a history of recurrent pyogenic infections, and are particularly susceptible to Pneumocystis carinii. They are also prone to profound neutropenia, autoimmune hemolytic anemia, and thrombocytopenic purpura. Liver disease including sclerosing cholangitis, viral hepatitis as well as hepatic lymphoma are common and their frequencies increase with age.26 The long-term survival rate for patients with XHIM is poor despite regular use of intravenous immunoglobulins. Less than 30% of the patients are alive at 25 years of age. Major causes of death include Pneumocystis carinii pneumonia early in life, liver disease, and malignancies in later life.27 Allogeneic bone marrow transplantation28 or non-myeloablative bone marrow transplantation from matched, unrelated donors29 have been successful in the treatment of hyper-IgM syndrome.
Molecular and Cellular Defects
The genetic anomaly in X-linked hyper-IgM syndrome has been mapped to Xq26, and resides in mutations of the CD40 ligand gene now known as CD154.30 CD40L is a member of the tumor necrosis factor family that binds to its receptor (CD40) expressed in B cells. Interactions between the CD40 ligand present on activated T cells and CD40 on B cells is required for productive isotype switching in B cells.31 The defect in X-linked hyper-IgM syndrome is a failure of isotype switch. Failure of this switch results in defective formation of germinal centers and immunoglobulin switching. Studies of CD40L (CD154) knockout mice that are also susuceptible to Pneumocystis carinii infections show that the defect of CD40L expression prevents CD40-mediated up-regulation of CD80/CD86 expression in B cells and other antigen presenting cells, ultimately resulting in poor T cell priming and defective type 1 immune response.32
Another form of X-linked hyper-IgM is associated with ectodermal dysplasia (XHM-ED). Interestingly, the expression of CD40 and CD40L are normal but a specific missense mutation in the putative zinc-finger domain of the gene that encodes nuclear factor κB (NF-κB) essential modulator (NEMO, also known as IKKγ) have been described in these patients.33 The mutations of NEMO prevent CD40L-mediated degradation of inhibitor of NF-κB (IκB-α) resulting in defects in immunoglobulin class-switching, inability of antigen-presenting cells to synthesize the NF-κB-regulated cytokines such as IL-12 and TNF-α when stimulated with CD40L.33
A minority of patients show an autosomal recessive inheritance pattern named hyper IgM-2. A mutation of the activation-induced cytidine deaminase (AID) gene, the product of which is selectively expressed in B cells from germinal centers34 has been reported. There is a similar defect in immunoglobulin switching, however significant differences exist in that they have lymph nodes with hyperplastic germinal centers and patients do not suffer from opportunistic infections. The B cells characteristically exhibit few somatic mutations of the variable part of the rearranged immunoglobulin genes. The exact mechanism of how a member of a RNA editing enzyme family leads to the defect is unknown.
More recently, homozygous mutations of the CD40 gene leading to lack of surface expression of CD40 have been reported in another form of autosomal recessive hyper IgM. The clinical and immunological features are indistinguishable from those of the X-linked form.35
Common Variable Immunodeficiency
Common variable immunodeficiency (CVID) refers to a heterogeneous group of disorders which are characterized by defective antibody formation.4 CVID is the most frequent of the primary immunodeficiency diseases among populations of European descent and affects both sexes equally.36 The feature common to all patients is hypogammaglobulinemia, generally affecting all antibody classes but sometimes only IgG. Several modes of inheritance (autosomal recessive, autosomal dominant, and X-linked) have been reported, however sporadic cases are most common. The usual age at presentation is in the second or third decade of life. CVID is the most common clinically significant primary immunodeficiency disease that can present initially in adult life.5
The clinical manifestations of CVID include manifestations of antibody deficiency, ie, recurrent pyogenic sinopulmonary infections. Recurrent attacks of herpes simplex are common, and herpes zoster develops in about 20% of patients.3 Some patients may have unusual enteroviral infections with a chronic meningoencephalitis and a dermatomyositis-like illness. In addition, CVID patients are also prone to persistent episodes of diarrhea caused by Giardia lamblia. A high frequency of autoimmune diseases including rheumatoid arthritis, pernicious anemia, hemolytic anemia, thrombocytopenia, and neutropenia, is also seen.3 A syndrome resembling sarcoidosis can also affect some patients with CVID. The granulomas tend to involve the lung, liver, spleen, and conjunctivae. They are also at risk for inflammatory bowel diseases such as Crohn’s disease, celiac disease, and nodular lymphoid hyperplasia. A few patients present with opportunistic infections such as Pneumocystis carinii, mycobacteria, or fungal infections.
Molecular and Cellular Defects
The primary cause of CVID is not known. In part because CVID comprises a heterogeneous group of disorders, non-random recurrent cytogenetic abnormalities unique to CVID have not been identified. Current evidence suggests that the humoral defect in CVID patients is as a result of insufficient in vivo stimulus for B cell activation rather than an intrinsic inability of the B lymphocytes to undergo terminal differentiation into plasma cells.37 Furthermore, circulating B cells from CVID patients failed to undergo somatic hypermutation in immunoglobulin-variable region genes, similar to cord blood B cells. They were also unable to produce IgA on engagement of the Ig receptor suggesting the presence of severe deficiency of switched memory B cells (CD27+ IgM−IgD−) in CVID patients.38,39
Abnormalities in T cell signaling and thus defective T cell to B cell interactions, are thought to underlie the diminished in vivo stimulation of B cell activation and differentiation into immunoglobulin secreting plasma cells.3,7 Experimental evidence indicates that the molecular basis of abnormal B cell differentiation is varied. Some CVID patients have mutations interfering with the regulation of the expression of immunoglobulin genes.40,41 Others have functional abnormalities of CD4+ (helper) cells or CD8+ (suppressor) T cells, as well as defective B cells42 and apoptosis.43 An aberrantly spliced lck transcript, a key molecule in T cell receptor-mediated signaling, lacking the entire exon 7 associated with decreased expression of LCK protein has been described in a patient with CVID.44 Low levels of serum interleukin 2 (IL-2) and interferon gamma (IFN-γ)45 due to defects in these cells can contribute to hypogammaglobulinemia.
Recent studies have also implicated genes within the HLA complex predisposing to CVID; many patients have deletions of the C4A gene or possess rare alleles of the C2 gene. Both CVID and isolated IgA deficiency may affect different individuals of the same family, suggesting that they may be related disorders with a common genetic defect.41 A subset of male patients with CVID have mutations in the X-linked lymphoproliferative disease gene SH2D1A/SAP46 suggesting a link between defects in the SAP gene to abnormal B cell development. The diagnosis is based on the exclusion of other known causes of humoral immune defects.
IgA Deficiency
Selective IgA deficiency is the most common form of immunodeficiency in the Western world, affecting approximately 1 in 600 individuals.47 Only about one third of the patients are particularly prone to infections. Most patients have IgA levels below 5 mg/dl. The serum concentrations of the other immunoglobulins are usually normal, but patients have a high incidence of autoantibodies, many with allergies including food reactions, allergic conjunctivitis, rhinitis, urticaria, atopic eczema, and bronchial asthma.48 In about two thirds of the cases, the deficiency does not lead to an increased occurrence of infections, whereas the remaining patients suffer from bacterial infections in both the upper and lower respiratory tract.
Molecular and Cellular Defects
The major role of IgA is to facilitate presentation of antigen to mucosal T cells. The pathogenesis of IgA deficiency involves a block in B cell differentiation that occurs due to defective interaction between T and B cells. This is demonstrated by the observation that IL-12 treatment can overcome IgA deficiency by providing adequate T cell priming in mice.49 The pathogenesis of IgA deficiency is associated with genes within the major histocompatability complex such as HLA-B8, SC01, and DR3.50 Other studies have implicated genomic polymorphisms in the tumor necrosis factor gene51 as a protective factor in IgA deficiency. The defect is manifested at the stem cell level as transfer of bone marrow from an IgA-deficient donor to a normal recipient results in IgA deficiency in the recipient.52
Selective IgG Subclass Deficiencies
Selective deficiencies of IgG subclasses, with or without IgA deficiency, are caused by defects in several genes. IgG2 deficiency is most common in children, whereas adults more often have low levels of IgG3.4 Hyper-IgE syndrome is a rare immunodeficiency state characterized by recurrent skin and pulmonary abscesses and extremely elevated serum IgE levels. It is a single-locus disease with an autosomal dominant pattern of transmission with variable expressivity.53 Genotypic analysis of 19 kindreds with multiple cases of hyper-IgE syndrome with polymorphic markers in a candidate region on human chromosome 4 suggests the proximal 4q region as a candidate locus for the disease.54 Single-strand conformation polymorphism analysis followed by DNA sequencing revealed mutations in the α subunit of the interleukin-4 receptor α55 in 3 of 3 patients with hyper-IgE. The R576 is a novel IL-4 receptor α allele causing a change from glutamine to arginine at position 576 (R576) in the cytoplasmic domain of the IL-4 receptor α protein. However, this allele is also commonly found in those patients with atopic dermatitis suggesting that the mutation may predispose persons to allergic disease rather than a primary genetic cause of the disease.
T Cell Immunodeficiencies
Purine Nucleoside Phosphorylase Deficiency
The ubiquitous purine nucleoside phosphorylase (PNP) plays a key role in the purine salvage pathway.56 PNP deficiency is a lethal, autosomal recessive disease that causes profound T cell deficiency with variable deficiencies in the humoral system. Patients with PNP deficiency experience recurrent bacterial, viral, and fungal infections usually beginning early in life. Immunodeficiency is accompanied by a neurological disorders and developmental retardation.57
Molecular and Cellular Defects
The PNP protein reversibly catalyzes the degradation of the purine nucleosides inosine and deoxyinosine to hypoxanthine and that of guanosine and deoxyguanosine to guanine. Deficiency of nucleoside phosphorylase leads to a dysfunction of the purine salvage pathway and accumulation of deoxyguanosine triphosphate (dGTP) which is preferentially toxic to T cells compared to B cells.58 Mutations result in truncated proteins leading to variable levels of enzymatic activity.59 Recent mouse models of purine nucleoside phosphorylase deficiency suggest that accumulation of dGTP in the mitochondria result in impaired mitochondrial DNA repair with enhanced sensitivity of T cells to spontaneous DNA damage leading to T cell apoptosis.60
Zap-70 Deficiency
ZAP-70 deficiency is inherited in an autosomal recessive manner. Recurrent and opportunistic infections occur within the first year of life.4 There is lymphopenia involving CD8+ T cells, with normal numbers of non-functional CD4+ T cells and severe combined immunodeficiency.61 Zap-70 (ζ-associated polypeptide of 70 kd) is a tyrosine kinase that binds to the TCR’s phosphorylated immunoreceptor tyrosine-based activation motif (ITAM) sequences. Recruitment of ZAP-70 to the TCR and its subsequent phosphorylation and activation, largely by Lck, is essential for downstream signaling events.62 In Zap-70 deficiency, a mutation within the kinase domain of ZAP-70 results in abolished protein expression. Signaling through TCR is defective, influencing T cell development with a selective block in positive selection of CD8+ cells. There is failure of peripheral CD4+ T cell proliferative response to mitogens or anti-CD3 antibody. By contrast, the activity of natural killer cells, B cells, and serum immunoglobulin levels are normal. The severe combined immunodeficiency associated with ZAP-70 deficiency is fatal unless treated by allogeneic bone marrow transplantation.
CD3ε and CD3γ Deficiencies
Rare congenital immunodeficiencies are caused by mutations in the γ63 and ε64 subunits of CD3. They are inherited in an autosomal recessive (11q23) manner and result in moderate to severe immunodeficiency due to decreased circulating CD3+ T cells, and poor responses to T cell mitogens. They show decreased lymphocyte membrane expression of TCR/CD3.
Severe Combined Immunodeficiencies (SCIDs)
The severe combined immunodeficiencies (SCIDs) syndrome is characterized by gross impairment of both the humoral and cell-mediated immunity and by susceptibility to overwhelming fungal, bacterial, and viral infections. The syndrome comprises a heterogeneous group of primary immunodeficiencies associated with various defects of the immune system involving T, B, and sometimes natural killer cells.
T-B-SCID
RAG1 Deficiency, RAG2 Deficiency, and Omenn Syndrome
Two related deficiencies of recombination activating genes, RAG1 and RAG2 result in a spectrum of severe combined immunodeficiencies called RAG1/RAG2 deficiency and Omenn syndrome. Both conditions are inherited as autosomal recessive diseases. RAGs are crucial proteins that play a role in activating V(D)J recombination in the B cell and T cell receptor genes required for generation of the diversity of the recognition sites. Absence or defective V(D)J recombination results in the arrest of B and T cell development such that most of the circulating lymphocytes in affected patients are natural killer cells. Mutations that lead to total absence of RAG1 or RAG2 gene product (null mutations) are known to lead to severe combined immune deficiency without mature lymphoid cells,65 whereas mutations that result in partial V(D)J recombinase activity due to missense mutation on at least one allele lead to Omenn syndrome.66 A clinical phenotype similar to Omenn syndrome has been also seen as a result of engraftment of maternal T cells as a complication of a transplacental transfusion.67 Thus, analysis of the RAG genes by direct sequencing may be an effective way to provide accurate diagnosis of RAG-deficient as opposed to RAG-independent V(D)J recombination defects.
Omenn syndrome is an immunodeficiency disease with autoimmune features resembling graft-versus-host disease.68 It is characterized by the absence of circulating B cells and infiltration of many organs by activated oligoclonal T cells. Most present with infantile diffuse erythrodermia, alopecia, protracted diarrhea, lymphadenopathy, hepatosplenomegaly, fever, hypereosinophilia, and elevated serum IgE levels leading to failure to thrive and ultimately death. Protein loss due to diarrhea and exudative erythrodermia often leads to generalized edema. The condition is fatal unless it is corrected by bone marrow transplantation.
Molecular and Cellular Defects
Molecular studies of patients with Omenn syndrome66 have revealed missense mutations of the RAG1 and RAG2 genes which result in impaired but not absent rearrangement of the B cell receptor and T cell receptor genes. These findings indicated that the immunodeficiency manifested in patients with Omenn syndrome arises from mutations of RAG1 and RAG2 genes that decrease the efficiency of V(D)J recombination. More recent studies have shown that some cases of Omenn syndrome are characterized by mutations identical to those seen in T-B-SCID patients suggesting the role of additional factors that may play a part in the development of Omenn syndrome.69 An analysis of TCR repertoire demonstrates exquisite restriction of TCR clonotypes indicating antigen-driven proliferation of T cells. The TCR from some patients lacked N- or P-nucleotide insertions and used proximal variable and joining gene segments, suggesting abnormal intrathymic T cell development. Abnormal assembly of gene segments and truncated rearrangements within non-productive alleles also suggest abnormalities in the TCR rearrangement mechanism.70
Adenosine Deaminase Deficiency
Adenosine deaminase (ADA) deficiency accounts for about half of the autosomal recessive forms of SCIDs. It is one of the most severe immunodeficiencies and is associated with severe depletion of B cells, T cells, and NK cells. It is the second-most prevalent form of SCID accounting for approximately 20% of the group. Affected individuals die from overwhelming opportunistic infections within the first few months of life if untreated. ADA follows purine nucleoside phosphorylase in purine nucleoside catabolism, but deficiency in this enzyme causes even more severe symptoms than PNP deficiency that is largely limited to T cells. In addition to immunological defect, most patients with ADA deficiency also have skeletal abnormalities.
Molecular and Cellular Defects
ADA, an enzyme of the purine salvage pathway, catalyzes the conversion of adenosine and 2′-deoxyadenosine to inosine and 2′-deoxyinosine, respectively. ADA deficiency results in accumulation of the toxic metabolites, adenosine and deoxyadenosine, which accumulate in the cells of affected patients. Deficiency of adenosine deaminase results in a profound decrease in the maturation of lymphocyte precursors.71 Defective enzymatic activity in lymphocyte precursors results in selective accumulation of dGTP that inhibits cellular division. Typical patients are diagnosed by age 6 months and rarely survive beyond 1 to 2 years unless immune function is restored by stem cell transplation or enzyme replacement therapy. Partial ADA deficiency has been found in less severely affected patients with delayed or late/adult onset of immune deficiency72,73 and autoimmunity.71 About 70 known mutations, the majority missense, span the 32-kb, 12 exon ADA gene on chromosome 20q.74 Expression studies demonstrate that missense mutations appear more deleterious yielding 0.005% to 0.6% of the ADA activity from wild-type cDNA whereas splicing mutations are associated with mild severity.74 Studies of ADA-deficient mice show multiple defects of T cells including increased thymic apoptosis as well as defective T cell receptor signaling.75 Treatment of ADA deficiency by gene transfer has also been attempted.76 Flow cytometric analysis of ADA expression can be used to assess the expression of ADA in follow-up of patients treated in clinical gene transfer protocols.77
T-B+ SCID
SCIDs with lack of circulating T cells but a normal number of B cells accounts for 30 to 50% of all cases of human SCIDs. The X-linked form is the most common form that provides rationale for the prevalence of SCID being three times as common in boys as in girls. The second most common variant is autosomal recessive and due to mutations of the JAK3 gene.
X-Linked SCID
This group of immunodeficiency diseases appears to be as a result of several genetic causes. Affected infants are susceptible to recurrent severe infections caused by a wide range of pathogens including, Candida albicans, Pneumocystis carinii, Pseudomonas, cytomegalovirus, and varicella. Death from varicella, herpes, adenovirus, or cytomegalovirus may occur very soon after infection. Infants with severe combined imunodeficiency invariably have profound lymphopenia.3 The number of natural killer cells may be normal or high. In contrast to the autosomal recessive forms of SCID in which both T and B cells are profoundly deficient, the X-linked form of SCID is characterized by the presence of normal number of peripheral blood B-cells.
Molecular and Cellular Defects
Approximately 50% of patients with SCID have an X-linked recessive pattern of inheritance. It has been mapped to Xq13.78 These patients have a mutation in the common cytokine receptor gamma chain gene that encodes a shared, essential component of the receptors for IL-2, IL-4, IL-7, IL-9, and IL-15.79 The mutation results in the expression of the gamma chain which exhibits reduced binding to JAK3.80 Thus, the early lymphoid progenitor cells lacking intact interleukin receptors, fail to be stimulated by these growth factors that are vital to the normal development of T and B cells. Affected newborns are profoundly deficient in thymic size and there is progressive T cell deficiency resulting from ineffective rescue from apoptosis or replication senescence.81 Prenatal mutational screening by single-strand conformational polymorphism, heteroduplex analysis and dideoxy fingerprinting (ddF) followed by direct sequencing for mutations in the gamma chain reveal that ddF is the most sensitive method for detection of heterozygous mutations.82 Allogeneic stem cell transplantation even in utero, as well as ex vivo gene therapy can correct the immunodeficiency in these patients.83
JAK3 Deficiency
Mutations of the JAK3 gene lead to a form of non-X-linked autosomal recessive form of SCID. Clinically, they resemble infants with X-linked SCID with elevated levels of B cells and very low levels of T cells and natural killer cells in the blood.79 The JAK3 gene maps to chromosome 19p12–13.1 and encodes the JAK3 protein, an intracellular tyrosine kinase, which is crucial for signal-transmission of cytokine receptors to signal transducers and activators of transcription (STATs). Once recruited into the cytokine receptor complex, STATs are phosphorylated and then translocated into the nucleus to regulate transcription at multiple sites. As a result, there is almost complete absence of JAK3 kinase activity with impairment of IL-2 and IL-4 signaling.84,85 Due to the multiple cytokines using this signaling pathway, an early and severe block in T and natural killer cell development combined with impaired B cell function is observed. The identification of the genomic organization of the human JAK3 gene into 23 exons86 has made rapid mutation detection and prenatal diagnosis feasible.87 Twenty-seven unique mutations have been identified that affect all seven structural Jak homology (JH) functional domains. The ability to test the function of a specific and expressed mutant in a phosphorylation assay using physiological substrates (JAK3 and STAT5) has enabled the verification of the consequences of the observed mutations. While some mutations result in absence of protein activity, others such as the C759R substitution results in constitutive phosphorylation of JAK3 which cannot be up-regulated by cytokine stimulation and thus block signal transduction.88 Thus, demonstration of JAK3 protein expression by Western blot does not rule out functional JAK3 deficiency until cytokine-induced phosphorylation of JAK3 itself and/or STAT5 is excluded. JAK3 deficiency is a candidate for gene therapy. In vitro biochemical correction of JAK3-deficient human B-cell derived cell lines has been accomplished.89
Other SCIDs
Bare Lymphocyte Syndrome Type I (TAP1 and TAP2 Deficiency)
Bare lymphocyte syndrome is characterized by a severe decrease of HLA class I and/or class II molecules. Patients show reduced numbers of CD8+ T cells and lack natural killer cell activity. HLA class I expression depends on the formation of a peptide-loading complex composed of class I heavy chain, β2-microglobulin, the transporter associated with antigen processing (TAP) and tapasin which links TAP to the heavy chain. Type I bare lymphocyte syndrome is caused by a deficiency in the TAP proteins encoded by genes within the major histocompatibility complex and plays a role in presentation of antigenic peptides to T cells. TAP transports peptides from the cytoplasm into the inner lumen of the endoplasmic reticulum and thus defects in TAP induce poor peptide loading on class I heavy chains. The TAP complex is a composed of TAP1 and TAP2 which, via the ATP-binding cassette transporter, translocates peptides from the cytosol to the waiting MHC class I molecules in the endoplasmic reticulum. TAP1 and TAP2 deficiency have identical clinical presentations that manifest within the first 6 years of life with recurrent bacterial infections of the upper respiratory tract.
Molecular and Cellular Defects
Mutations of both TAP1 and TAP2 genes result in deficient expression of class I HLA proteins on the cell surface with defects in natural killer cell cytotoxicity.90,91 A novel genetic cause of type I bare lymphocyte syndrome has been identified within the Tapasin gene resulting from a deletion of 4 exons by Alu-mediated recombination.92
Bare Lymphocyte Syndrome Type II (MHC Class II Deficiency)
The cell surface glycoproteins of the MHC class II are crucial players in the immune response. Defective expression of major histocompatibility complex class II molecules account for 5% of SCID.93 Children with autosomal recessive form of hereditary MHC class II deficiency, or bare lymphocyte syndrome type II are extremely susceptible to bacterial, viral, and fungal infections, beginning in the first year of life. Mortality rate is high, with most children dying from overwhelming infections by the age of 4 years.94
Molecular and Cellular Defects
The genetic lesions responsible for this syndrome do not lie within the MHC-II locus itself, but reside instead in genes encoding transcription factors controlling MHC-II expression. Different subtypes of human MHC II molecules are transcribed from TATA-less promoters that contain conserved S, X, and Y boxes. Protein complexes that bind to these proximal promoter elements attract the class II transactivator (CIITA) by an unknown mechanism. S and X boxes bind a tripartite regulatory factor X (RFX) complex, while the Y box binds the nuclear factor Y (NFY) complex. While the genes for MHC-II determinants remain intact, different mutations have been found in four trans-acting factors, RFX5, RFXAP, RFXANK(B), and CIITA.95 MHC II molecules expressed on the surface of B cells present processed peptide fragments to the TCR of CD4 + T helper cells, triggering the antigen-specific T cell response. The regulatory factor (RF) X, a complex binding to the X-box of MHC-II promoters in the nucleus, is mutated in one type.
CD4 +T cells are decreased in all forms, although circulating lymphocyte numbers are normal and immunoglobulin numbers can also be decreased. Mutant forms of CIITA and all of the known subunits of RFX have been found among patients with the bare lymphocyte syndrome because all mutations cause absolute deficiency of MHC class II proteins.96,97
CD45 Deficiency
A molecular defect of CD45 is a rare cause of severe combined immunodeficiency with an autosomal recessive inheritance.98,99 CD45 is an abundant transmembrane tyrosine phosphatase expressed on all leukocytes and is required for efficient lymphocyte signaling, integrin-mediated adhesion, and migration of immune cells. Mutations leading to loss of a component of the extracellular domain of CD45 result in very low circulating T cells but a normal number of B cells. The T cells are unresponsive to mitogens and serum immunoglobulin levels usually decrease with age. A homozygous 6-bp deletion in the CD45 gene resulting in a loss of glutamic acid 339 and tyrosine 340 in the first fibronectin type II module of the extracellular domain of CD45 has been reported.98 A male patient with a deficiency in CD45 due to a large deletion at one allele and a point mutation at the other resulting in aberrant splice site99 has also been reported.
Treatment of SCIDs
SCIDs have been among the first diseases to be cured by bone marrow stem cell transplantation, and there is no need for pretransplant immunosuppression. Preliminary studies show that they are also good candidates for gene therapy. Retroviral reconstitution of the gene defect into autologous marrow hematopoeitic cells have led to full correction of X-linked SCIDs.100
Defects of Phagocytic Cells
Most congenital phagocytic disorders are diagnosed in the first year of life, but leukocyte adhesion deficiencies and chronic granulomatous disease may not be diagnosed until adulthood. Chronic granulomatous disease tends to exhibit increased susceptibility to catalase-positive organisms, whereas defects of the interferon-γ-interleukin 12-pathway are characteristically associated with mycobacteria and other intracellular pathogens. Myeloperoxidase deficient individuals will generally not be susceptible to infections except for patients with diabetes who develop severe candidiasis.
Cyclic Neutropenia
Cyclic neutropenia occurs sporadically and by an autosomal-dominant inheritance pattern. The typical presentation is that of recurrent, severe neutropenia (an absolute neutrophil count of less than 200 cells per cubic millimeter) lasting 3 to 6 days of every 21-day period. Diagnosis depends on serial measurements of absolute neutrophil counts over a period of several weeks.101 Patients are usually asymptomatic, however, during periods of severe neutropenia, they develop recurring episodes of fever, aphthous ulcers, gingivitis, and cellulitis. Deep tissue infections and bacteremia from Clostridium species are the most serious complications.
Molecular and Cellular Defects
Although neutropenia in these disorders has been attributed to impaired or ineffective neutrophil production, the molecular and cellular basis for these diseases has remained largely unknown. Positional cloning studies have led to the mapping of candidate genes to chromosome 19p13.3, a region containing the genes for three neutrophil proteases: azurcidin, proteinase 3, and neutrophil elastase. Sequence analysis of the PCR-amplified genomic DNA revealed that all affected members in 13 families and one sporadic case of cyclic neutropenia harbored a mutation within the neurophil elastase gene (ELA2), with most of the mutations occurring at the junction of exons 4 and 5.102 Neutrophil elastase is a serine protease chiefly synthesized by promyelocytes. The mutations affect the catalytic site of the enzyme, resulting in inactive elastase.103 The link between inactive elastase and the cyclic changes in the level of neutrophils remains to be determined. However, defects in the granulocyte colony-stimulating factor signaling pathway is thought to destabilize normal steady-state conditions and increase the number of circulating lymphocytes, reticulocytes, and platelets during neutropenia.104
Subsequent studies also showed that 90% of patients with classic congenital neutropenia also had mutations in ELA2 gene, however a greater diversity of mutations was found.105 Those mutations associated with cyclic neutropenia occur in proximity to the active site and the binding pocket for the enzyme’s substrate whereas the mutations responsible for congenital neutropenia would be predicted to change the molecular folding of the protein possibly affecting the storage of the enzyme in the primary granules.106
Severe Congenital Neutropenia
Severe congenital neutropenia is a heterogeneous disorder related to cyclic neutropenia. There is severe neutropenia with an absolute neutrophil count of less than 500 cells per cubic milliter, recurrent bacterial infections, and absence of myeloid maturation with arrest at the promyelocyte stage.107 The disease manifests in the first months of life with infectious complications from Staphylococcus aureus and Burkholderia aeruginosa resulting in cellulitis, perirectal abscess, meningitis, and stomatitis.108 The disease is more severe than cyclic neutropenia and is three to four times more common.109 The disease was initially described as an autosomal recessive disease, however it can occur sporadically and as an autosomal dominant disorder.
Molecular and Cellular Defects
The underlying genetic abnormality in severe congenital neutropenia is largely unknown. In about 10% of the patients, a heterozygous mutation of the granulocyte colony-stimulating factor receptor is identified. Germline mutations of the gene encoding neutrophil elastase (ELA2) have been observed in a large percentage of (22 of 25) patients studied according to a recent report.105 This gene mutation has been found to play a role in other neutropenic disorders including cyclic neutropenia, but not in Shwachman-Diamond syndrome patients. Accelerated apoptosis of neutrophil precursors is a common feature of both cyclic and severe congenital neutropenia. Nevertheless, the mechanism by which mutations of the ELA2 gene cause the premature death of these cells is unclear.110 The enzyme neutrophil elastase is synthesized in neutrophil precursors early in the process of primary granule formation. It is currently presumed that the mutant elastase functions aberrantly within the cells to accelerate apoptosis of the precursors resulting in ineffective and oscillatory production. Although mutations in ELA2 may be necessary for the phenotype of congenital neutropenia, it may not be sufficient.111 In most patients, treatment with G-CSF diminishes the number of infections.107,112 ELA2 mutations may provide a background for the G-CSF receptor mutations that occur in the transformation to acute myeloid leukemia. More recently, a novel mutation affecting the the GTPase binding domain of the Wiskott-Aldrich syndrome protein (WASP) has been described in an X-linked form of severe congenital neutropenia.113
Shwachman-Diamond Syndrome
Shwachman-Diamond syndrome (SDS) is a rare autosomal recessive disorder characterized by exocrine pancreatic insufficiency, skeletal abnormalities, bone marrow dysfunction, and recurrent infections.108 Neutropenia, either cyclic or intermittent, with or without pancytopenia can occur.114 Recurrent infections involving sinuses, lungs, bones, skin, and urinary tract begin during the first year of life.108 Patients have increased risk of bone marrow aplasia, myelodysplasia, and myeloid leukemia.115
A variety of immune defects have been observed including decreased in vitro B lymphocyte proliferation, lack of specific antibody production, low numbers of CD4+ T cells and decreased numbers of natural killer cells.116 Additionally, there is hyperactivation of the Fas-mediated apoptotic pathway resulting in higher tendency of bone marrow mononuclear cells to undergo apoptosis.117 The significant immune dysfunction seen in these patients suggests that Shwachman-Diamond syndrome may involve a marrow defect accounting for aberrant function of hematopoietic and lymphopoietic lineages. The mapping of the SDS locus to the centromeric region of chomosome 7 (7p10–7q11)118 is interesting as both myelodysplastic syndrome and AML frequently exhibit monosomy 7, isochromosome 7, and deletion of 7q.119
More recently, mutations involving a previously uncharacterized gene, SBDS, have been identified in patients with Shwachman-Diamond syndrome.120 The mutations are located in the interval of 1.9 cM at 7q11 resulting from gene conversion involving exon 2 in 89% of unrelated individuals with Shwachman-Diamond syndrome (141 of 158 families) with 60% (95 of 158 families) carrying two converted alleles. SDBS encodes a predicted protein of 250 amino acids and thought to be involved in RNA processing. The complex and pleiotropic phenotype associated with this syndrome suggests however, the presence of several mutations responsible for the disease.
Leukocyte Adhesion Defect
Leukocyte adhesion deficiency is a rare, autosomal recessive genetic disorder in which neutrophils fail to mobilize and migrate to sites of injury. There is delayed separation of the umbilical cord in infancy, followed by severe, scarring skin infections, gingivitis, and systemic bacterial infections. Because of abnormal aggregation and migration, even when there is no infection, the patients have twice the normal number of neutrophils in the peripheral blood.121
Molecular and Cellular Defects
The gene defects associated with this disorder involve CD18 and CD11c, both of which are components of surface integrin complexes that are essential for neutrophil aggregation and attachment to endothelial surfaces.122 Leukocyte adhesion deficiency type 1 is an autosomal recessive disorder resulting from a lack of β2 integrin adhesion molecules on neutrophils.123
The second type of leukocyte adhesion deficiency is a defect of carbohydrate fucosylation and is associated with growth retardation, dysmorphic features, and neurological deficits.124 These patients lack CD15s, sialyl-LewisX, a ligand for the selectin family. In these patients, there is no fucosylation of other glycoconjugates that are required for interactions with P-selectins and E-selectins on endothelial cells.125 The genetic defect has not been determined, however treatment with oral fucose has reduced the frequency of infections and fevers.125
Rac2 Deficiency
Dominant-negative mutations resulting in deficiency of ras-related C3 botulinum toxin substrate (Rac2), the predominant hematopoeitic-specific Rho GTPase in neutrophils also leads to leukocyte adhesion deficiency.126 The Rho family of GTPases plays key roles in regulating cytoskeletal reorganization, integrin complex formation, and cell adhesion. The neutrophils from this disorder exhibit abnormal chemotaxis and secretion of primary granules and defective generation of superoxide.127 Rac2 is integral to the function of the actin cytoskeleton and its deficiency results in the inability of neutrophils to migrate normally in response to bacterial peptides.128 Furthermore, biochemical and biological characterization of the human Rac2 GTPase dominant-negative mutant suggests that neutrophils with the mutant Rac2 GTPase exhibit increased apoptosis as well as defective neutrophil function.129
Defects of Interferon-γ-Interleukin-12 Axis
The interferon-γ-interleukin-12 axis is critical for defense against intracellular microbes such as mycobacteria, salmonella, and listeria. Defects in the ligand binding and signaling chain of the interferon-γ receptor, the interleukin-12 receptor or interleukin-12 itself result in profound deficiency of T cells and susceptibility to mycobacterial infection and severe combined immunodeficiency.130 Interferon-γ is important for activation of macrophages and neutrophils with subsequent secretion of tumor necrosis factor α and activation of NADPH oxidase. IL-12 is stimulated when pathogens activate dendritic cells and macrophages which in turn leads to secretion of interferon-γ.131 Mutations have been identified in the genes for both chains of the interferon-γ receptor with autosomal recessive and autosomal dominant inheritance.132 Children with a mutation that cause a complete loss of the ligand-binding chain have severe disease that begins in early infancy. The main features are disseminated atypical mycobacterial disease or fatal Bacilli Calmette-Guerin infection after vaccination and inability to form granulomas.132
Specific Granule Deficiency
Neutrophil-specific granule deficiency (SGD) is a rare disorder characterized by a lack of specific or secondary granules containing lactoferrin, transcobalamin, gelatinase B, and collagenase in developing and mature neutrophils.133 Due to the absence of proteins necessary for the normal respiratory burst, the neutrophils show defects in bactericidal activity as well as abnormal migration and disaggregation.134 The expression of the primary granules, myeloperoxidase and lysozyme, is unaffected. Patients present with frequent bacterial infections with S. aureus, S. epidermidis, and enteric bacteria primarily of the skin and lungs. The neutrophils display atypical bilobed nuclei, and lack expression of primary, secondary and tertiary granule proteins. Defects in chemotaxis, disaggregation, receptor up-regulation, and bactericidal activity are also characteristic features.
Molecular and Cellular Defects
The functional loss of the myeloid-specific transcription factor CCAAT/enhancer binding protein (C/EBPvarepsilon) due to frame-shift mutations leads to deficiencies in mulple myeloid lineages and the development of specific granule deficiency.135,136
Chronic Granulomatous Disease (CGD)
CGD is a rare inherited immunodeficiency that is caused by a functional defect of the various components of NADPH oxidase of phagocytes. Patients with CGD present with typical responses to infection such as fever, leukocytosis, and localized inflammation. As a result, many present with lymphadenopathy, hepatosplenomegaly, and recurrent infections. Due to defective microbial killing however, there is incomplete clearing of the infections, with subsequent formation of gramulomatous lesions. The disease is inherited in either an X-linked form, which accounts for two-thirds of cases and an autosomal recessive form.137
Molecular and Cellular Defects
The basic cellular defect of CGD is a deficiency of the respiratory burst.138 Structural components of the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase system is required for the generation of superoxide and other reactive species that are important in the killing of ingested microorganisms. The NADPH oxidase is composed of four polypeptide subunits and mutations in the corresponding genes are responsible for the four different genetic subgroups of CGD. Approximately 2 of 3 of CGD cases arise from mutations involving the X-linked gene encoding the gp91phox subunit. The CYBA gene encoding p22phox is localized on chromosome 16q24.139 Mutations in CYBA, a subunit of the cytochrome b558, the redox element of NADPH oxidase, result in a rare autosomal recessive form of the disorder. Twenty-six different mutations in the CYBA gene have been described, including deletions, insertions, and substitutions leading to missense, nonsense, frame-shift, and splice-site mutations with no “hot-spots.”139,140,141,142 About one quarter of the CGD patients have mutations in the NCF1 gene encoding p47phox. It involves a GT deletion at the beginning of exon 2 of NCF1, most likely caused by recombination events between the NCF1 and one of its pseudogenes.143 Gene-scan methods have been used to detect one NCF1 gene (carriers) from controls and from NCF1-deficient patients.144 Prenatal diagnosis of p47phox-mutant CGD has been reported.145 It is the most common autosomal form of CGD. The remaining cases of autosomal recessive CGD involve defects in p47phox or cx139,146,147 with those involving p67phox being the rarest. As a group, those with X-linked form, p22phox, p67phox CGD tend to have a worse clinical course compared to those with p47phox CGD.
Myeloperoxidase Deficiency
Hereditary myeloperoxidase (MPO) deficiency is the most common biochemical defect of neutrophils. An estimated prevalence of 1 of 2000 individuals has been reported in the United States.4 The clinical outcome of hereditary MPO deficiency is much less severe than other genetic diseases of neutrophils such as chronic granulomatous disease (CGD) or Chediak-Higashi syndrome. Approximately half of those affected have a complete deficiency of myeloperoxidase while the rest have structural or functional defects in the enzyme.148 Myeloperoxidase is a principal component of the azurophilic (primary) granules and an increased susceptibility to infections (particularly Staphylococcus aureus and Candida albicans) has been reported for some MPO-deficient patients.
Molecular and Cellular Defects
The mutations in the gene encoding myeloperoxidase are heterogeneous and result in transcriptional or post-transcriptional defects.149 More recent studies demonstrate that hereditary MPO deficiency is not a simple autosomal recessive trait but a result of compound heterozygosity with respect to the R569W missense mutations of the MPO gene.148
Other Immunodeficiencies
Wiskott-Aldrich Syndrome
Wiskott-Aldrich syndrome (WAS) is inherited as an X-linked recessive disease and characterized by immune dysregulation and microthrombocytopenia. The estimated incidence in the U.S. is 4.0 per million male live births, accounting for six new cases per year.4 Affected males have eczema, recurrent bacterial infections, and profound thrombocytopenia with platelets of reduced size and function.150 Serum IgM concentrations are reduced, whereas IgA and IgE concentrations are elevated, and IgG is normal. Antibody formation, especially to bacteria with a polysaccharide capsule, is defective. In addition, affected patients have a poor response to protein antigens. The number of B cells progressively increases with time, while the number of T cells progressively decreases.151 Further evidence of T cell dysfunction is demonstrated by poor in vitro T cell response to the mitogenic effects of anti-CD3.152 Immune dysregulation may also manifest as food allergy, autoimmune disease including hemolytic anemia, vasculitides, and inflammatory bowel disease. In its less severe form known as X-linked thrombocytopenia, mutations in the same gene produce characteristic platelet abnormalities, but minimal immunological disturbances. The immune defects appear later and are variable and progressive.153,154 In the past, WAS patients generally died within the first decade of life from infection, hemorrhage, or malignancy. However, the recent improved management modalities including intravenous immune globulin therapy and splenectomy, have improved the life expectancy of WAS patients with median survival of 15 years.
Molecular and Cellular Defects
Several nonsense and missense mutations as well as deletions and insertions of the WAS gene (Xp11.22)155 have been identified in WAS patients.156 The normal WAS gene encodes the Wiskott-Aldrich syndrome protein (WASP),155 that belongs to a member of a unique family of proteins which are responsible for transduction of signals from the cell membrane157 to the actin cytoskeleton. The interaction between WAS protein, the Rho family GTPase CDC42 and the cytoskeletal organizing complex, Arp2/3 is disturbed and leads to defects in cell signaling, polarization, motility, and phagocytosis.158 The cell-surface sialoglycoproteins, most notably CD43 (sialophorin/leukosialin), are unstable with decreased expression in the cell membranes of lymphocytes.155 Although the neutrophils and macrophages in WAS patients have been shown to exhibit impaired chemotactic responses,159 other functional properties of these cells appear to be unimpaired.157 Deficiency of WAS protein can be reliably detected in peripheral blood mononuclear cells of affected patients by flow cytometry.160,161 Prenatal molecular diagnosis of WAS patients can be carried out by SSCP and heteroduplex assays followed by automatic DNA seqencing from chorionic villus samples.162
Most patients with X-linked thrombocytopenia have missense mutations within exons 1 and 2 of the WAS gene leading to decreased but detectable protein expression whereas a wide spectrum of mutations, most often leading to complete absence of protein have been detected in classical WAS.163 The observation of missense mutations of the WAS gene in two families with an intermittent form of X-linked thrombocytopenia164 broadens the spectrum of clinical phenotypes associated with defects of the WAS gene and indicates the need for molecular analysis in males with reduced platelet volume, regardless of platelet number. The only definitive therapy for the disease at present is allogeneic bone marrow transplatation, however retrovirus-mediated WAS gene transfer has been shown to correct the T cell abnormalities.165
DiGeorge/Velocardiofacial Syndrome
DiGeorge/velocardiofacial syndrome (DGS) is a congenital immune disorder characterized by lack of embryonic development or underdevelopment of the thymus and surrounding organs. The syndrome is associated with several defects including cardiac outflow tract anomalies, abnormal facies, thymic hypoplasia, cleft palate, and hypocalcinemia.166 The immune deficit is caused by hypoplasia or aplasia of the thymus. The degree of thymic abnormality varies and only about 20% of the patients have decreased T cell number and function. Autoimmune diseases and recurrent infections are other common features.167 Although mostly diagnosed in infancy, in some cases hypocalcemic tetany during adulthood have resulted in the diagnosis of DiGeorge syndrome with underlying cardiovascular malformations.168
Molecular and Cellular Defects
The DiGeorge anomaly was originally considered as a clinical paradigm for thymus deficiency but has been redefined as a member of a group of disorders that have in common a chromosome deletion involving 22q11.2 (the DiGeorge syndrome chromosome region, or DGCR) that commonly include 24 continguous genes.166 The embryologic defect is due to inadequate development of facial neural-crest tissues resulting in defective organogenesis of the pharyngeal pouch derivatives. No single gene or combination of genes has been demonstrated to contribute to the complex phenotypic spectrum of DGS. Knockout mice studies have shown that mice heterogyzous for a transcription factor of the T-box gene, Tbx1, dsplayed a wide range of developmental anomalies encompassing almost all of the common DGS features169,170 Other candidate genes that have been studied are RanBP1, a GTPase important for nucleocytoplasmic transport,171 and CRKL which encodes an SH2-SH3-SH3 adaptor protein. Mice defective in these candidate genes result in phenotypes with similarities with human DiGeorge syndrome.172 With the rapid progress in molecular cytogenetics, the investigation of choice is now a standard karyotype to exclude major rearrangements. Polymerase chain reaction and fluorescence in situ hybridization using probes from within the deletion segment are also available for prenatal diagnosis of the 22q11.2 deletion.173
Ataxia Telangiectasia
Ataxia telangiectasia (AT) is an autosomal recessive disorder characterized by various neurological deficits including, cerebellar degeneration with progressive ataxia, oculocutaneous telangiectasia, and immunodeficiency.4 Most patients develop recurrent bacterial sinopulmonary infections and increased susceptibility to hematological cancer.174,175 The immunological defects are of variable severity and may affect both T and B cells. Other important features include thymic hypoplasia, growth retardation, hypogonadism, and consistently elevated levels of α-fetoprotein, which may be used to establish the diagnosis.174 In addition to these findings, 80% of patients are IgA deficient and serum levels of IgG4 and IgG2 may also be decreased. AT patients also exhibit increased susceptibility to the effects of ionizing radiation, due to defective DNA repair and chromosomal instability.176
Molecular and Cellular Defects
The ATM gene has been mapped to chromosome 11q22–23.177 The gene designated ATM has been found to encode a polypeptide with a phosphatidyl inositol-3′ (PI-3) kinase domain.178 It has similarities to the catalytic subunit of DNA-dependent protein kinase179 and can directly phosphorylate and activate many genes involved in cell cycle checkpoint responses including p53,180 CHK2,181 Nbs1,182 and BRCA1.183 ATM is involved in mitogenic signal transduction, meiotic recombination, response to DNA damage and control of the cell cycle, and apoptosis.184 Biallelic loss of the ATM gene or its substrates results in a defect in the radiation-activated S phase checkpoint. The ATM gene is mutated in AT patients from all four complementation groups, indicating that it is probably the sole gene responsible for the disorder.178 In addition, in AT patients of any age, approximately 10% of all T lymphocytes show the presence of translocations and inversions, mainly involving chromosomes 7 and 14 at specific breakpoints; the chromosomal location of the T cell antigen receptor genes, as well as the immunoglobulin gene loci.174 Whereas truncating mutations are the most common type of mutation observed in individuals affected with AT,185 those that occur in sporadic breast cancer are missense mutations. Furthermore analysis of ATM mutations in sporadic lymphomas and leukemias show that there is a mixture of both missense and truncating alterations distributed across the whole of the ATM coding sequence. Interestingly, mutations in sporadic T-prolymphocytic leukemia were predominantly missense, clustering in the region encoding the PI-3 kinase catalytic domain of the protein, while those seen in mature B cell lymphomas such as chronic lymphocytic leukemias and mantle cell lymphomas were distributed across the whole of the ATM coding sequence.186 The highest percentage of ATM mutations is found within mantle cell lymphoma.187 ATM heterozygous mice harboring an in-frame deletion corresponding to the human 7636del9 mutation show an increased susceptibility to malignancies. Expression of ATM cDNA containing the 7636del9 mutation had a dominant-negative effect inhibiting radiation-induced ATM kinase activity.188 These findings indicate that different ATM mutations might differ in the degree of associated cancer risk. There is emerging data that show that many chromosomal instability disorders share components of converging signaling pathways. For example, phosphorylation of the Fanconi anemia protein FANCD2 by ATM kinase following ionizing radiation is required for activation of the S phase checkpoint.189
Immunodeficiency with Albinism
Chediak-Higashi Syndrome
Chediak-Higashi syndrome (CHS) is a rare autosomal recessive disorder characterized by partial skin and ocular albinism, increased susceptibility to infections, and progressive neuropathy.190 About 85 to 90% of the patients present in early childhood with life-threatening bacterial infections due to neutropenia and lack of natural killer cell function. Most patients with CHS develop lethal complications, called the “accelerated phase” during which T cells and macrophages undergo overwhelming activation involving multiple organs.191
Molecular and Cellular Defects
Mutations of the gene encoding a protein named LYST in humans (lysosomal trafficking regulator/CHS) or Beige in mice, that is a cytoplasmic protein that controls lysosome traffic is thought to cause CHS.192 The CHS1 protein is predicted to contain three to four defined domains including a weak ARM/HEAT repeat domain, a perilipin domain, a BEACH domain, and a series of WD-40 repeats. These motifs are thought to be important in mediating membrane associations, vesicular transport, and lipid association. The exact biochemical function of LYST/CHS1 is unknown however, yeast two-hybrid studies demonstrate that LYST interacts with proteins important for vesicular transport and signal transduction including SNAR-complex protein HRS, 14–3-3, and casein kinase II.193 Many cell types have large intracytoplasmic granulations194 and deficient cytotoxic lymphocyte activity as a result of defective vesicular trafficking of the endosome/lysosome compartment. Defective T cells and natural killer cell cytotoxic function develop due to defective exocytosis and delivery of lytic proteins.195 Macrophages and neutrophils show decreased chemotaxis with normal phagocytic function. The defective lysosomal function results in a delay in fusion of phagosomes with lysosomes. Recent data suggest the presence of genotype-phenotypic correlation in childhood and adult forms of CHS. Most of the mutations in patients with severe childhood presentations result in null alleles, frame-shifts, nonsense mutations, or gene deletions that abolish the expression of the full-length polypeptide.196 About 10 to 15% of patients exhibit a much milder clinical phenotype and survive to adulthood. In these patients, missense mutant alleles that encode peptides with partial function are found197
Griscelli Syndrome
Griscelli syndrome is a rare autosomal recessive disorder that results in hypopigmentation of hair and skin, the presence of large clumps of pigment in hair shafts, and an accumulation of melanosomes in melanocytes.198 Most patients develop an uncontrolled T lymphocyte and macrophage activation syndrome, known as hemophagocytic syndrome, potentially leading to death.199 Some Griscelli syndrome patients, early in life, show severe neurological impairment without apparent immune abnormalities. Despite an adequate number of T and B lymphocytes, the patients are hypogammaglobulinemic, deficient in antibody production, and incapable of delayed skin hypersensitivity and skin graft rejection. Griscelli syndrome resembles Chediak-Higashi syndrome, but differs in that the polymorphonuclear leukocytes in GS are morphologically normal, and lack the abnormal giant granules that are characteristic of CH syndrome.
Molecular and Cellular Defects
Two closely linked genes located on human 15q21 region have been found to be responsible for the disease. Nucleotide substitutions of myosin 5A (MYO5A) gene have been reported200,201 while others have reported the involvement of RAB27A.199 Myosin 5A, a member of the unconventional myosin family thought to participate in organelle-transport is important for lymphocyte cytotoxic function. RAB27A encodes a RAB guanosine triphosphate (GTP)-binding protein required for prenylation and activation of RAB GTPases which are critical regulators of vesicular transport. Studies of the mouse model of Rab27A mutant ashen homozygotes show that it is important for the regulated secretion of granzyme A and hexosaminidase of cytotxic T cells.202 Griscelli disease presents with a heterogenous clinical picture that seem to reflect the involved gene defect. The form due to myosin 5A mutation is associated with a primary neurological impairment, and immune features such as susceptibility to infections and occurrence of hemophagocytic syndrome are absent. The form due to RAB27A mutations has no neurological features but is associated with an uncontrolled T cell and macrophage activation syndrome, the hemophagocytic syndrome. T cells of RAB27A-deficient individuals had normal granule content in perforin and granzymes but showed defective granule release. Indeed, bone marrow transplantation with peripheral blood stem cell transplant in a 6-month-old girl with Griscelli syndrome due to deletion of the RAB27A has been beneficial.203
Familial Hemophagocytic Lymphohistiocytosis
Familial hemophagocytic lymphohistiocytosis (FHL) is inherited as an autosomal recessive disorder with a rapidly fatal clinical presentation.204 It is characterized by the overwhelming activation of T cells and macrophages which results in fever, hepatosplenomegaly, pancytopenia, coagulation abnormality, and liver dysfunction. Multiple organs including liver, spleen, bone marrow, and central nervous system become infiltrated by activated T cells and macrophages that become engulfed by phagocytes, a process called hemophagocytosis.205 In addition, neurological involvement develops during the latter course of the syndrome with manifestations ranging from confusion to severe seizures and neurological deficits. These clinical features are associated with the overproduction by T lymphocytes and macrophages of cytokines including interferon-γ, tumor necrosis factor-α, IL-1, and IL-6.206 Combination of chemotherapy and bone marrow transplantation can cure many patients.207
Molecular and Cellular Defects
Both homozygous and heterozygous mutations208 involving the perforin gene have been identified in patients with familial hemophagocytic lymphohistiocytosis. Lymphocytes obtained from FHL patients demonstrated significantly reduced CD3-dependent perforin-mediated cytotoxic activity of CD8+ T and natural killer cells than from normal control patients. Clonal expansion of αβ-T cells with restricted Jβ1 usage of T cells was seen in these patients209 which could be detected at relapse after chemotherapy.210 The level of perforin-mediated cytotoxic activity is correlated with perforin immunoreactivity. More recently, a comprehensive survey of 34 patients for mutations in the coding region of the perforin gene have been reported.211 Homozygous and heterozygous mutations of the perforin gene were detected in 20% (7 of 34) of all FHL patients investigated, with a somewhat higher prevalence, approximately 30% (6 of 20), in children whose parents originated from Turkey. Prenatal diagnosis of perforin gene mutations can identify a subset of patients with FHL.212
X-Linked Lymphoproliferative Disease
In X-linked lymphoproliferative disease (XLP) or Duncan’s disease, patients are exceptionally susceptible to Epstein-Barr virus (EBV)213,214 leading to uncontrolled expansion of EBV-infected B cells and cytotoxic T cells. The average age of onset is 2.5 years, with 100% mortality by the age of 40 years.215 Patients are totally asymptomatic before EBV infection however, following infection with EBV, patients mount a vigorous, uncontrolled polyclonal expansion of T and B cells. The subtle, progressive combined variable immunodeficiency disease is characterized by benign or malignant proliferation of lymphocytes that may be polyclonal or overly malignant with monoclonal populations. Proliferation of histiocytes and alterations in concentrations of serum immunoglobulins are also characteristic. In many patients, a differential diagnosis of lymphoma is operative as proliferating lymphocytes infiltrate the lymph nodes, spleen, and liver. Patients’ symptoms are fever, pharyngitis, lymphadenopathy, hepatosplenomegaly, atypical lymphocytosis, and a spectrum of deficiencies ranging from agammaglobulinemia to polyclonal hypergammaglobulinemia. The primary cause of death is hepatic necrosis and bone marrow failure.
Molecular and Cellular Defects
The genetic abnormality associated with XLP involves a gene designated SH2 domain protein-1A (SH2D1A)214 also called SAP (SLAM-associated protein). SLAM (signaling lymphocyte activation molecule), also known as CDw150, appears on the surface of T cells, where it has a crucial function in cell stimulation. SAP is physiologically expressed in T cells and NK cells and is an SH2 domain-containing protein. SAP is associated with the SLAM protein that is expressed on the surface of activated T-cells216 and 2B4 molecule on NK cells and cytotoxic T cells.217 It is thought to act as an adaptor molecule involved in transducing activation signals in T cells following T-B cell contact. It is expressed in activated T and NK cells but not in activated B cells.218 Mutations in SAP affect the interaction between T and B cells and leads into uncontrolled B cell proliferation in EBV infection.213 The identification of the SAP gene and its association with XLP has led to molecular diagnoses of related disorders although the mutation detection rate in several series is only 55 to 60%. Disease causing mutations of the SAP gene have been reported in male patients initially diagnosed as affected by CVID,219 suggesting the possibility that a subgroup of patients with CVID may represent phenotypic variants of XLP. As the molecular genetic basis of CVID remains elusive, the diagnosis involves the exclusion of other molecularly defined disorders.
Immunodeficiency, Centromeric Instability, Facial Anomaly Syndrome
Immunodeficiency, centromeric instability, facial anomaly (ICF) syndrome is a rare autosomal recessive disorder with variable immune deficiency. The most common diagnostic feature is the branching of chromosomes due to centromeric instability of chromosomes 1, 9, 16, and rarely, 2, with an increased frequency of somatic recombination of the arms of these chromosomes.220 The affected children suffer serious variable immunodeficiency, mild developmental delay, and facial abnormalities with hypertelorism, a flat nasal bridge, epicanthal folds, protrusion of the tongue, and mild micrognathia. The severity of the disorder is indicated by the fact that most die during childhood.
Molecular and Cellular Defects
Positional cloning has revealed the de novo DNA methyltransferase 3B (DNMT3B) as the candidate gene in a large percentage of ICF patients.221,222 ICF patients show marked hypomethylation of their DNA. Mutations involving the DNA methyltransferase 3B (DNMT3B) gene were identified in four patients from three families. There is evidence however for genetic heterogeneity, since two ICF patients from consanguineous families who did not show autozygosity (homozygosity by descent) for the DNMT3B locus also did not reveal DNMT3B mutations. Mutations including missense, nonsense, and splice-site involvement223 have been documented. No homozygous mutations have been found suggesting that the absence of the enzyme is incompatible with life. Genomic methylation is critical for maintenance of structure and expression and undermethylation of certain chromosomal segments is associated with centromere instability and abnormal expression of key immunoregulatory genes. Microarray expression analysis of B cell lymphoblastoid cell lines from ICF patients with diverse DNMT3B mutations demonstrated dysregulation of numerous genes associated with lymphocyte signaling, maturation, and migration compared to normal lymphoblastoid cell lines224
Nijmegen Breakage Syndrome
Nijmegen breakage syndrome (NBS) is a rare autosomal disorder characterized by microcephaly, stunted growth, mental retardation, cafe-au-lait spots, and immunodeficiency with impaired DNA repair and hypersensitivity to ionizing radiation.225,226 Most patients suffer from recurrent respiratory tract infections and others have repeated urinary tract infections. There is a high predisposition for malignancy, occuring in 40% of patients before the age of 21. Hypogammaglobulinemia is typically present involving the levels of IgA, IgG2, IgG4, and IgE.226 NBS patients lack the classical signs and symptoms of AT such as cerebellar ataxia, oculocutaneous telangiectasia, and increased α-fetoprotein (AFP).
Molecular and Cellular Defects
Most patients have a truncating deletion of the NBS-1 gene, which is located on chromosome 8q21. The NBS-1 gene encodes a protein nibrin, a member of the hMre11/hRad50 nuclease complex involved in DNA double-strand break repair.227 The defect in DNA repair mechanism is thought to contribute to decreased immunoglobulins as a result of defective class switching.228 Such findings as recurrent infections, immunodeficiency, chromosomal instability with multiple 7 and/or 14 rearrangements, cellular and chromosomal hypersensitivity to ionizing radiation and bleomycin, and radioresistance of DNA replication, are common features of both NBS and AT. More recent studies demonstrate a functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. The phosphorylation and function of NBS1 protein induced by ionizing radiation requires the catalytically active kinase function of ATM.229 These results demonstrate a biochemical link between cell-cycle checkpoints activated by DNA damage and DNA repair in two genetic diseases with overlapping phenotypes.
Job Syndrome
Job syndrome of hyperimmunoglobulinemia E and recurrent infections (hyper-IgE syndrome) was described in 1966.230 The disorder is characterized by markedly elevated serum IgE levels and recurrent cutaneous and sinopulmonary infections.230 Many also present with eczematoid rashes, mucocutaneous candidiasis, subcutaneous abscesses, bony abnormalities, and osteoporotic fractures.231
Molecular and Cellular Defects
The genetic basis for Job syndrome is not known and the central immunological defect is largely undefined. Reduced neutrophil chemotaxis is often described, and variable T cell defects have been demonstrated in some patients. It has been hypothesized that hyper-IgE is associated with a Th1/Th2 imbalance.232 It is inherited as a single-locus, autosomal dominant trait with variable expressivity.54
Autoimmune Lymphoproliferative Syndrome
Autoimmune lymphoproliferative syndrome (ALPS) is a disorder of defective lymphocyte apoptosis due to genetic aberrations of several members of the Fas-signaling pathway. Patients with ALPS present with peripheral lymphocytosis, diffuse lymphadenopathy, hepatosplenomegaly, hypergammaglobulinemia, autoimmune cytopenias and rarely autoimmune glomerulonephritis and hepatitis.233 The peripheral blood is characterized by a markedly expanded population of CD5+ B cells, NK cells, and TCRαβ CD4-CD8- (double-negative) HLA-DR+ naive T cells.234 Many patients display a dysregulated cytokine pattern with dysfunctional T cells, suggesting that Fas defects may alter pathways of T cell activation235 and differentiation. In most patients, this population constitutes between 15 to 70% of the peripheral blood T cells. The T cells have impaired proliferative and cytokine reponses to T cell activation.
Molecular and Cellular Defects
ALPS result from mutations in several genes critical in the apoptosis pathway, including Fas,236 Fas ligand, Caspase 8, and 10.237 The genetics and pathogenesis of ALPS is complex. The disease can be inherited as an autosomal dominant trait, but development of full phenotype depends on additional factors suggesting that other host factors or genes play a role in modulating the disease phenotype. Moreover, the observation of ALPS in individuals without mutations of the above mentioned genes suggest a role for other apoptosis pathway genes in the predisposition to this disorder.
Conclusions
Recent developments in molecular diagnostics and gene-based therapies have led to fundamental shifts in the diagnosis, treatment, and management of families affected by primary immunodeficiencies. The discovery of specific gene abnormalities important in lymphocyte biology and their link to these rare diseases has led to significant insights into the complexity of the immune system and provided new insights regarding the spectrum of clinical severity seen in a particular disorder and the phenotypic overlap resulting from mutations of different genes.
Immunophenotypic and molecular genetic studies are very useful in the evaluation of these processes. However, the results should be interpreted with caution and correlation with clinical features is imperative. Despite the increased susceptibility of primary immunodeficiency patients to the development of lymphoma, the most frequent cause of mortality in these patients continues to be infection. Further delineation of the molecular and cellular defects in these diseases may provide the framework for the development of therapies targeted at the specific defects which may prevent the occurrence of devastating secondary complications such as infection and malignancy.
Table 2.
(Continued)
| Predominantly antibody deficiencies
| |||
|---|---|---|---|
| Designation | Pathogenesis/defect | Inheritance | Diagnostic tests |
| 8. Chronic granulomatous disease | |||
| (a) X-linked CGD (deficiency of 91 kDa chain of cytochrome b) | Killing (faulty production of superoxide metabolites), CYBB | AR | Mutation analysis |
| (b) Autosomal recessive-CGD (deficiencies of 22 kDa chain of cytochrome b or of p47 or p67 cytosol factors) | Killing-as above | AR or XL | Mutation analysis |
| 1. p22 phox | Mutations in LYBA | XL | Mutation analysis |
| 2. p47 phox | Mutations in NCF1 | AR | Mutation analysis |
| 3. p67 phox | Mutations in NCF2 | XL | Mutation analysis |
| 9. Myeloperoxidase deficiency | Killing, mutations in MPO gene | AR | Mutation analysis |
| 10. Leukocyte mycobactericidal defect | Killing, mutations | AR | |
| (a) TNF-γ receptor 1 deficiency | Killing defect | AR or AD | Mutation analysis |
| (b) TNF-γ receptor 2 deficiency | Killing defect | AR or AD | Mutation analysis |
| (c) Interleukin-12 receptor deficiency | Killing defect | AR or AD | Mutation analysis |
| (d) Interleukin-12 deficiency | Killing defect | AR or AD | Mutation analysis |
| Other well-defined immunodeficiency | syndromes | |||
|---|---|---|---|---|
| 1. Wiskott-Aldrich syndrome | Mutations in WAS gene; cytoskeletal defect affecting haematopoietic stem cell derivatives | XL | Mutation analysis | |
| Detection of WASP mRNA or protein | ||||
| 2. Ataxia-telangiectasia | Mutation in A-T gene (ATM); disorder of cell cycle checkpoint pathway leading to chromosomal instability | AR | Mutation analysis | |
| DNA radiation sensitivity | ||||
| 3. Nijmegen breakage syndrome | Defect in NBS1 (Nibrin); disorder of cell cycle check point and DNA double strand break repair | AR | Mutation analysis | |
| DNA radiation sensitivity | ||||
| 4. DiGeorge anomaly | Contiguous gene defect in 90% affecting thymic development | De novo defect or AD | FISH, mutation analysis | |
| 5. Immunodeficiency with albinism | Mutation analysis | |||
| (a) Chediak-Higashi syndrome | Defect in Lyst | AR | Mutation analysis | |
| (b) Griscelli syndrome | Defect in myosin 5a | AR | Mutation analysis | |
| Defect in RAB27A | ||||
| 6. X-linked lymphoproliferative syndrome | Defect in SAP | XL | Mutation analysis | |
| 7. Familial hemophagocytic lymphohistiocytosis | Perforin mutation (10q21–22) | AR | Mutation analysis | |
| 8. ICF Syndrome | DNMT3B mutation (20q11.2) | AR | Mutation analysis | |
References
- Rosen FS, Cooper MD, Wedgwood RJ. The primary immunodeficiencies (1). N Engl J Med. 1984;311:235–242. doi: 10.1056/NEJM198407263110406. [DOI] [PubMed] [Google Scholar]
- Rosen FS, Cooper MD, Wedgwood RJ. The primary immunodeficiencies. (2). N Engl J Med. 1984;311:300–310. doi: 10.1056/NEJM198408023110506. [DOI] [PubMed] [Google Scholar]
- Rosen FS, Cooper MD, Wedgwood RJ. The primary immunodeficiencies. N Engl J Med. 1995;333:431–440. doi: 10.1056/NEJM199508173330707. [DOI] [PubMed] [Google Scholar]
- International Union of Immunological Societies Primary immunodeficiency diseases: report of an IUIS Scientific Committee. Clin Exp Immunol. 1999;118 (Suppl 1):1–28. doi: 10.1046/j.1365-2249.1999.00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicherer SH, Winkelstein JA. Primary immunodeficiency diseases in adults. JAMA. 1998;279:58–61. doi: 10.1001/jama.279.1.58. [DOI] [PubMed] [Google Scholar]
- Korpi M, Valiaho J, Vihinen M. Structure-function effects in primary immunodeficiencies. Scand J Immunol. 2000;52:226–232. doi: 10.1046/j.1365-3083.2000.00799.x. [DOI] [PubMed] [Google Scholar]
- Puck JM. Primary immunodeficiency diseases. JAMA. 1997;278:1835–1841. [PubMed] [Google Scholar]
- Lappalainen I, Ollila J, Smith CI, Vihinen M. Registries of immunodeficiency patients and mutations. Hum Mutat. 1997;10:261–267. doi: 10.1002/(SICI)1098-1004(1997)10:4<261::AID-HUMU1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Tangsinmankong N, Bahna SL, Good RA. The immunologic workup of the child suspected of immunodeficiency. Ann Allergy Asthma Immunol. 2001;87:362–369. doi: 10.1016/S1081-1206(10)62915-8. quiz 370, 423. [DOI] [PubMed] [Google Scholar]
- Conley ME, Notarangelo LD, Etzioni A. Diagnostic criteria for primary immunodeficiencies: representing PAGID (Pan-American Group for Immunodeficiency) and ESID (European Society for Immunodeficiencies). Clin Immunol. 1999;93:190–197. doi: 10.1006/clim.1999.4799. [DOI] [PubMed] [Google Scholar]
- Lederman HM, Winkelstein JA. X-linked agammaglobulinemia: an analysis of 96 patients. Medicine (Baltimore) 1985;64:145–156. [PubMed] [Google Scholar]
- Stewart DM, Tian L, Nelson DL. A case of X-linked agammaglobulinemia diagnosed in adulthood. Clin Immunol. 2001;99:94–99. doi: 10.1006/clim.2001.5024. [DOI] [PubMed] [Google Scholar]
- Bruton OC. Agammaglobulinemia. Pediatrics. 1952;9:722–727. [PubMed] [Google Scholar]
- Tsukada S, Saffran DC, Rawlings DJ, Parolini O, Allen RC, Klisak I, Sparkes RS, Kubagawa H, Mohandas T, Quan S, Belmont JW, Cooper MD, Conley ME, Witte ON. Deficient expression of a B cell cytoplasmic tyrosine kinase in human X- linked agammaglobulinemia. Cell. 1993;72:279–290. doi: 10.1016/0092-8674(93)90667-f. [DOI] [PubMed] [Google Scholar]
- Rawlings DJ, Witte ON. Bruton’s tyrosine kinase is a key regulator in B-cell development. Immunol Rev. 1994;138:105–119. doi: 10.1111/j.1600-065x.1994.tb00849.x. [DOI] [PubMed] [Google Scholar]
- Islam TC, Smith CI. The cellular phenotype conditions Btk for cell survival or apoptosis signaling. Immunol Rev. 2000;178:49–63. doi: 10.1034/j.1600-065x.2000.17811.x. [DOI] [PubMed] [Google Scholar]
- Futatani T, Miyawaki T, Tsukada S, Hashimoto S, Kunikata T, Arai S, Kurimoto M, Niida Y, Matsuoka H, Sakiyama Y, Iwata T, Tsuchiya S, Tatsuzawa O, Yoshizaki K, Kishimoto T. Deficient expression of Bruton’s tyrosine kinase in monocytes from X- linked agammaglobulinemia as evaluated by a flow cytometric analysis and its clinical application to carrier detection. Blood. 1998;91:595–602. [PubMed] [Google Scholar]
- Vihinen M, Arredondo-Vega FX, Casanova JL, Etzioni A, Giliani S, Hammarstrom L, Hershfield MS, Heyworth PG, Hsu AP, Lahdesmaki A, Lappalainen I, Notarangelo LD, Puck JM, Reith W, Roos D, Schumacher RF, Schwarz K, Vezzoni P, Villa A, Valiaho J, Smith CI. Primary immunodeficiency mutation databases. Adv Genet. 2001;43:103–188. doi: 10.1016/s0065-2660(01)43005-7. [DOI] [PubMed] [Google Scholar]
- Yel L, Minegishi Y, Coustan-Smith E, Buckley RH, Trubel H, Pachman LM, Kitchingman GR, Campana D, Rohrer J, Conley ME. Mutations in the μ heavy-chain gene in patients with agammaglobulinemia. N Engl J Med. 1996;335:1486–1493. doi: 10.1056/NEJM199611143352003. [DOI] [PubMed] [Google Scholar]
- Minegishi Y, Coustan-Smith E, Wang YH, Cooper MD, Campana D, Conley ME. Mutations in the human λ5/14.1 gene result in B cell deficiency and agammaglobulinemia. J Exp Med. 1998;187:71–77. doi: 10.1084/jem.187.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi Y, Rohrer J, Coustan-Smith E, Lederman HM, Pappu R, Campana D, Chan AC, Conley ME. An essential role for BLNK in human B cell development. Science. 1999;286:1954–1957. doi: 10.1126/science.286.5446.1954. [DOI] [PubMed] [Google Scholar]
- Minegishi Y, Coustan-Smith E, Rapalus L, Ersoy F, Campana D, Conley ME. Mutations in Igα (CD79a) result in a complete block in B-cell development. J Clin Invest. 1999;104:1115–1121. doi: 10.1172/JCI7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notarangelo LD, Hayward AR. X-linked immunodeficiency with hyper-IgM (XHIM). Clin Exp Immunol. 2000;120:399–405. doi: 10.1046/j.1365-2249.2000.01142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantman HJ, Stiehm ER, Stevens RH, Saxon A, Seeger RC. Abnormal B cell differentiation and variable increased T cell suppression in immunodeficiency with hyper-IgM. Clin Exp Immunol. 1980;40:147–156. [PMC free article] [PubMed] [Google Scholar]
- Pascual-Salcedo D, de la Concha EG, Garcia-Rodriguez MC, Zabay JM, Sainz T, Fontan G. Cellular basis of hyper IgM immunodeficiency. J Clin Lab Immunol. 1983;10:29–34. [PubMed] [Google Scholar]
- Hayward AR, Levy J, Facchetti F, Notarangelo L, Ochs HD, Etzioni A, Bonnefoy JY, Cosyns M, Weinberg A. Cholangiopathy and tumors of the pancreas, liver, and biliary tree in boys with X-linked immunodeficiency with hyper-IgM. J Immunol. 1997;158:977–983. [PubMed] [Google Scholar]
- Levy J, Espanol-Boren T, Thomas C, Fischer A, Tovo P, Bordigoni P, Resnick I, Fasth A, Baer M, Gomez L, Sanders EA, Tabone MD, Plantaz D, Etzioni A, Monafo V, Abinun M, Hammarstrom L, Abrabamsen T, Jones A, Finn A, Klemola T, DeVries E, Sanal O, Peitsch MC, Notarangelo LD. Clinical spectrum of X-linked hyper-IgM syndrome. J Pediatr. 1997;131:47–54. doi: 10.1016/s0022-3476(97)70123-9. [DOI] [PubMed] [Google Scholar]
- Thomas C, de Saint Basile G, Le Deist F, Theophile D, Benkerrou M, Haddad E, Blanche S, Fischer A. Brief report: correction of X-linked hyper-IgM syndrome by allogeneic bone marrow transplantation. N Engl J Med. 1995;333:426–429. doi: 10.1056/NEJM199508173330705. [DOI] [PubMed] [Google Scholar]
- Hadzic N, Pagliuca A, Rela M, Portmann B, Jones A, Veys P, Heaton ND, Mufti GJ, Mieli-Vergani G. Correction of the hyper-IgM syndrome after liver and bone marrow transplantation. N Engl J Med. 2000;342:320–324. doi: 10.1056/NEJM200002033420504. [DOI] [PubMed] [Google Scholar]
- Aruffo A, Farrington M, Hollenbaugh D, Li X, Milatovich A, Nonoyama S, Bajorath J, Grosmaire LS, Stenkamp R, Neubauer M, Roberts RL, Noelle RJ, Ledsetter JA, Francke U, Ochs HD. The CD40 ligand, gp39, is defective in activated T cells from patients with X-linked hyper-IgM syndrome. Cell. 1993;72:291–300. doi: 10.1016/0092-8674(93)90668-g. [DOI] [PubMed] [Google Scholar]
- Seyama K, Nonoyama S, Gangsaas I, Hollenbaugh D, Pabst HF, Aruffo A, Ochs HD. Mutations of the CD40 ligand gene and its effect on CD40 ligand expression in patients with X-linked hyper-IgM syndrome. Blood. 1998;92:2421–2434. [PubMed] [Google Scholar]
- Jain A, Atkinson TP, Lipsky PE, Slater JE, Nelson DL, Strober W. Defects of T-cell effector function and post-thymic maturation in X-linked hyper-IgM syndrome. J Clin Invest. 1999;103:1151–1158. doi: 10.1172/JCI5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Ma CA, Liu S, Brown M, Cohen J, Strober W. Specific missense mutations in NEMO result in hyper-IgM syndrome with hypohydrotic ectodermal dysplasia. Nat Immunol. 2001;2:223–228. doi: 10.1038/85277. [DOI] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Ferrari S, Giliani S, Insalaco A, Al-Ghonaium A, Soresina AR, Loubser M, Avanzini MA, Marconi M, Badolato R, Ugazio AG, Levy Y, Catalan N, Durandy A, Tbakhi A, Notarangelo LD, Plebani A. Mutations of CD40 gene cause an autosomal recessive form of immunodeficiency with hyper IgM. Proc Natl Acad Sci USA. 2001;98:12614–12619. doi: 10.1073/pnas.221456898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneller MC, Strober W, Eisenstein E, Jaffe JS, Cunningham-Rundles C. NIH conference: new insights into common variable immunodeficiency. Ann Intern Med. 1993;118:720–730. doi: 10.7326/0003-4819-118-9-199305010-00011. [DOI] [PubMed] [Google Scholar]
- Nonoyama S, Farrington M, Ishida H, Howard M, Ochs HD. Activated B cells from patients with common variable immunodeficiency proliferate and synthesize immunoglobulin. J Clin Invest. 1993;92:1282–1287. doi: 10.1172/JCI116701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agematsu K, Futatani T, Hokibara S, Kobayashi N, Takamoto M, Tsukada S, Suzuki H, Koyasu S, Miyawaki T, Sugane K, Komiyama A, Ochs HD. Absence of memory B cells in patients with common variable immunodeficiency. Clin Immunol. 2002;103:34–42. doi: 10.1006/clim.2001.5197. [DOI] [PubMed] [Google Scholar]
- Warnatz K, Denz A, Drager R, Braun M, Groth C, Wolff-Vorbeck G, Eibel H, Schlesier M, Peter HH. Severe deficiency of switched memory B cells (CD27(+)IgM(−)IgD(−)) in subgroups of patients with common variable immunodeficiency: a new approach to classify a heterogeneous disease. Blood. 2002;99:1544–1551. doi: 10.1182/blood.v99.5.1544. [DOI] [PubMed] [Google Scholar]
- Kaneko H, Kondo N, Motoyoshi F, Mori S, Kobayashi Y, Inoue Y, Orii T. Expression of immunoglobulin genes in common variable immunodeficiency. J Clin Immunol. 1991;11:262–267. doi: 10.1007/BF00918184. [DOI] [PubMed] [Google Scholar]
- Spickett GP, Farrant J, North ME, Zhang JG, Morgan L, Webster AD. Common variable immunodeficiency: how many diseases? Immunol Today. 1997;18:325–328. doi: 10.1016/s0167-5699(97)01086-4. [DOI] [PubMed] [Google Scholar]
- Spickett GP, Matamoros N, Farrant J. Lymphocyte surface phenotype in common variable immunodeficiency. Dis Markers. 1992;10:67–80. [PubMed] [Google Scholar]
- Iglesias J, Matamoros N, Raga S, Ferrer JM, Mila J. CD95 expression and function on lymphocyte subpopulations in common variable immunodeficiency (CVID): related to increased apoptosis. Clin Exp Immunol. 1999;117:138–146. doi: 10.1046/j.1365-2249.1999.00946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawabe T, Horiuchi T, Nakamura M, Tsukamoto H, Nakahara K, Harashima SI, Tsuchiya T, Nakano S. Defect of lck in a patient with common variable immunodeficiency. Int J Mol Med. 2001;7:609–614. doi: 10.3892/ijmm.7.6.609. [DOI] [PubMed] [Google Scholar]
- Sneller MC, Strober W. Abnormalities of lymphokine gene expression in patients with common variable immunodeficiency. J Immunol. 1990;144:3762–3769. [PubMed] [Google Scholar]
- Morra M, Silander O, Calpe S, Choi M, Oettgen H, Myers L, Etzioni A, Buckley R, Terhorst C. Alterations of the X-linked lymphoproliferative disease gene SH2D1A in common variable immunodeficiency syndrome. Blood. 2001;98:1321–1325. doi: 10.1182/blood.v98.5.1321. [DOI] [PubMed] [Google Scholar]
- Abbas A, Lichtman AH, Pober JS. Abbas A, Lichtman AH, Pober JS, editors. Philadelphia: WB Saunders; Congenital and acquired immunodeficiencies. Cellular and Molecular Immunology. 1997:439–462. [Google Scholar]
- Strober W, Sneller MC. IgA deficiency. Ann Allergy. 1991;66:363–375. [PubMed] [Google Scholar]
- Arulanandam BP, Raeder RH, Nedrud JG, Bucher DJ, Le J, Metzger DW. IgA Immunodeficiency leads to inadequate Th cell priming and increased susceptibility to influenza virus infection. J Immunol. 2001;166:226–231. doi: 10.4049/jimmunol.166.1.226. [DOI] [PubMed] [Google Scholar]
- Alper CA, Marcus-Bagley D, Awdeh Z, Kruskall MS, Eisenbarth GS, Brink SJ, Katz AJ, Stein R, Bing DH, Yunis EJ, Schur PH. Prospective analysis suggests susceptibility genes for deficiencies of IgA and several other immunoglobulins on the [HLA-B8, SC01, DR3] conserved extended haplotype. Tissue Antigens. 2000;56:207–216. doi: 10.1034/j.1399-0039.2000.560302.x. [DOI] [PubMed] [Google Scholar]
- De la Concha EG, Fernandez-Arquero M, Vigil P, Lazaro F, Ferreira A, Garcia-Rodriguez MC, Fontan G. Tumor necrosis factor genomic polymorphism in Spanish IGA deficiency patients. Tissue Antigens. 2000;55:359–363. doi: 10.1034/j.1399-0039.2000.550410.x. [DOI] [PubMed] [Google Scholar]
- Hammarstrom L, Grubb R, Smith CI. Gm allotypes in IgA deficiency. J Immunogenet. 1985;12:125–130. doi: 10.1111/j.1744-313x.1985.tb00839.x. [DOI] [PubMed] [Google Scholar]
- Grimbacher B, Holland SM, Gallin JI, Greenberg F, Hill SC, Malech HL, Miller JA, O’Connell AC, Puck JM. Hyper-IgE syndrome with recurrent infections: an autosomal dominant multi-system disorder. N Engl J Med. 1999;340:692–702. doi: 10.1056/NEJM199903043400904. [DOI] [PubMed] [Google Scholar]
- Grimbacher B, Schaffer AA, Holland SM, Davis J, Gallin JI, Malech HL, Atkinson TP, Belohradsky BH, Buckley RH, Cossu F, Espanol T, Garty BZ, Matamoros N, Myers LA, Nelson RP, Ochs HD, Renner ED, Wellinghausen N, Puck JM. Genetic linkage of hyper-IgE syndrome to chromosome 4. Am J Hum Genet. 1999;65:735–744. doi: 10.1086/302547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey GK, Friedrich MF, Esswein LA, Thomas ML, Chatila TA. The association of atopy with a gain-of-function mutation in the α subunit of the interleukin-4 receptor. N Engl J Med. 1997;337:1720–1725. doi: 10.1056/NEJM199712113372403. [DOI] [PubMed] [Google Scholar]
- Giblett ER, Ammann AJ, Wara DW, Sandman R, Diamond LK. Nucleoside-phosphorylase deficiency in a child with severely defective T-cell immunity and normal B-cell immunity. Lancet. 1975;1:1010–1013. doi: 10.1016/s0140-6736(75)91950-9. [DOI] [PubMed] [Google Scholar]
- Simmonds HA, Fairbanks LD, Morris GS, Morgan G, Watson AR, Timms P, Singh B. Central nervous system dysfunction and erythrocyte guanosine triphosphate depletion in purine nucleoside phosphorylase deficiency. Arch Dis Child. 1987;62:385–391. doi: 10.1136/adc.62.4.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddeo A, Fairbanks LD, Simmonds HA, Duley JA, Morris GS. Deoxy GTP accumulates in thymocytes, but not in T or B lymphocytes in simulated PNP deficiency. Adv Exp Med Biol. 1989:275–280. doi: 10.1007/978-1-4684-5676-9_40. [DOI] [PubMed] [Google Scholar]
- Dalal I, Grunebaum E, Cohen A, Roifman CM. Two novel mutations in a purine nucleoside phosphorylase (PNP)-deficient patient. Clin Genet. 2001;59:430–437. doi: 10.1034/j.1399-0004.2001.590608.x. [DOI] [PubMed] [Google Scholar]
- Arpaia E, Benveniste P, Di Cristofano A, Gu Y, Dalal I, Kelly S, Hershfield M, Pandolfi PP, Roifman CM, Cohen A. Mitochondrial basis for immune deficiency: evidence from purine nucleoside phosphorylase-deficient mice. J Exp Med. 2000;191:2197–2208. doi: 10.1084/jem.191.12.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder ME, Lin D, Clever J, Chan AC, Hope TJ, Weiss A, Parslow TG. Human severe combined immunodeficiency due to a defect in ZAP-70, a T cell tyrosine kinase. Science. 1994;264:1596–1599. doi: 10.1126/science.8202712. [DOI] [PubMed] [Google Scholar]
- Weil R, Cloutier JF, Fournel M, Veillette A. Regulation of Zap-70 by Src family tyrosine protein kinases in an antigen-specific T-cell line. J Biol Chem. 1995;270:2791–2799. doi: 10.1074/jbc.270.6.2791. [DOI] [PubMed] [Google Scholar]
- Arnaiz-Villena A, Timon M, Corell A, Perez-Aciego P, Martin-Villa JM, Regueiro JR. Brief report: primary immunodeficiency caused by mutations in the gene encoding the CD3-γ subunit of the T-lymphocyte receptor. N Engl J Med. 1992;327:529–533. doi: 10.1056/NEJM199208203270805. [DOI] [PubMed] [Google Scholar]
- Soudais C, De Villartay JP, Le Deist F, Fischer A, Lisowska-Grospierre B. Genetic analysis of the human CD3-ε gene in a T cell receptor/CD3 immunodeficiency. Immunodeficiency. 1993;4:117–119. [PubMed] [Google Scholar]
- Schwarz K, Gauss GH, Ludwig L, Pannicke U, Li Z, Lindner D, Friedrich W, Seger RA, Hansen-Hagge TE, Desiderio S, Lieber MR, Bartram CR. RAG mutations in human B cell-negative SCID. Science. 1996;274:97–99. doi: 10.1126/science.274.5284.97. [DOI] [PubMed] [Google Scholar]
- Villa A, Santagata S, Bozzi F, Giliani S, Frattini A, Imberti L, Gatta LB, Ochs HD, Schwarz K, Notarangelo LD, Vezzoni P, Spanopoulou E. Partial V(D)J recombination activity leads to Omenn syndrome. Cell. 1998;93:885–896. doi: 10.1016/s0092-8674(00)81448-8. [DOI] [PubMed] [Google Scholar]
- Villa A, Sobacchi C, Notarangelo LD, Bozzi F, Abinun M, Abrahamsen TG, Arkwright PD, Baniyash M, Brooks EG, Conley ME, Cortes P, Duse M, Fasth A, Filipovich AM, Infante AJ, Jones A, Mazzolari E, Muller SM, Pasic S, Rechavi G, Sacco MG, Santagata S, Schroeder ML, Seger R, Strina D, Ugazio A, Valiaho J, Vihinen M, Vogler LB, Ochs H, Vezzoni P, Friedrich W, Schwarz K. V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood. 2001;97:81–88. doi: 10.1182/blood.v97.1.81. [DOI] [PubMed] [Google Scholar]
- Omenn GS. Familial reticuloendotheliosis with eosinophilia. N Engl J Med. 1965;273:427. doi: 10.1056/NEJM196508192730806. [DOI] [PubMed] [Google Scholar]
- Corneo B, Moshous D, Gungor T, Wulffraat N, Philippet P, Le Deist FL, Fischer A, de Villartay JP. Identical mutations in RAG1 or RAG2 genes leading to defective V(D)J recombinase activity can cause either T-B-severe combined immune deficiency or Omenn syndrome. Blood. 2001;97:2772–2776. doi: 10.1182/blood.v97.9.2772. [DOI] [PubMed] [Google Scholar]
- Brooks EG, Filipovich AH, Padgett JW, Mamlock R, Goldblum RM. T-cell receptor analysis in Omenn’s syndrome: evidence for defects in gene rearrangement and assembly. Blood. 1999;93:242–250. [PubMed] [Google Scholar]
- Hershfield MS. Adenosine deaminase deficiency: clinical expression, molecular basis, and therapy. Semin Hematol. 1998;35:291–298. [PubMed] [Google Scholar]
- Shovlin CL, Simmonds HA, Fairbanks LD, Deacock SJ, Hughes JM, Lechler RI, Webster AD, Sun XM, Webb JC, Soutar AK. Adult onset immunodeficiency caused by inherited adenosine deaminase deficiency. J Immunol. 1994;153:2331–2339. [PubMed] [Google Scholar]
- Ozsahin H, Arredondo-Vega FX, Santisteban I, Fuhrer H, Tuchschmid P, Jochum W, Aguzzi A, Lederman HM, Fleischman A, Winkelstein JA, Seger RA, Hershfield MS. Adenosine deaminase deficiency in adults. Blood. 1997;89:2849–2855. [PubMed] [Google Scholar]
- Santisteban I, Arredondo-Vega FX, Kelly S, Mary A, Fischer A, Hummell DS, Lawton A, Sorensen RU, Stiehm ER, Uribe L, Weinberg K, Hershfield MS. Novel splicing, missense, and deletion mutations in seven adenosine deaminase-deficient patients with late/delayed onset of combined immunodeficiency disease: contribution of genotype to phenotype. J Clin Invest. 1993;92:2291–2302. doi: 10.1172/JCI116833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apasov SG, Blackburn MR, Kellems RE, Smith PT, Sitkovsky MV. Adenosine deaminase deficiency increases thymic apoptosis and causes defective T cell receptor signaling. J Clin Invest. 2001;108:131–141. doi: 10.1172/JCI10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Hacein-Bey S, Le Deist F, De Saint Basile G, Cavazzana-Calvo M. Gene therapy of severe combined immunodeficiencies. Adv Exp Med Biol. 2001;495:199–204. doi: 10.1007/978-1-4615-0685-0_27. [DOI] [PubMed] [Google Scholar]
- Otsu M, Hershfield MS, Tuschong LM, Muul LM, Onodera M, Ariga T, Sakiyama Y, Candotti F. Flow cytometry analysis of adenosine deaminase (ADA) expression: a simple and reliable tool for the assessment of ADA-deficient patients before and after gene therapy. Hum Gene Ther. 2002;13:425–432. doi: 10.1089/10430340252792558. [DOI] [PubMed] [Google Scholar]
- Puck JM, Conley ME, Bailey LC. Refinement of linkage of human severe combined immunodeficiency (SCIDX1) to polymorphic markers in Xq13. Am J Hum Genet. 1993;53:176–184. [PMC free article] [PubMed] [Google Scholar]
- Russell SM, Tayebi N, Nakajima H, Riedy MC, Roberts JL, Aman MJ, Migone TS, Noguchi M, Markert ML, Buckley RH, O’Shea JJ, Leonard WJ. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science. 1995;270:797–800. doi: 10.1126/science.270.5237.797. [DOI] [PubMed] [Google Scholar]
- Russell SM, Johnston JA, Noguchi M, Kawamura M, Bacon CM, Friedmann M, Berg M, McVicar DW, Witthuhn BA, Silvennoinen O, Golman AS, Schmalstieg FC, Ihle ON, O’Shea JJ, Leonard WJ. Interaction of IL-2R β and γ c chains with Jak1 and Jak3: implications for XSCID and XCID. Science. 1994;266:1042–1045. doi: 10.1126/science.7973658. [DOI] [PubMed] [Google Scholar]
- Goldman AS, Palkowetz KH, Rudloff HE, Dallas DV, Schmalstieg FC. Genesis of progressive T-cell deficiency owing to a single missense mutation in the common γ chain gene. Scand J Immunol. 2001;54:582–591. doi: 10.1046/j.1365-3083.2001.01006.x. [DOI] [PubMed] [Google Scholar]
- Puck JM, Middelton L, Pepper AE. Carrier and prenatal diagnosis of X-linked severe combined immunodeficiency: mutation detection methods and utilization. Hum Genet. 1997;99:628–633. doi: 10.1007/s004390050418. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Le Deist F, Carlier F, Bouneaud C, Hue C, De Villartay JP, Thrasher AJ, Wulffraat N, Sorensen R, Dupuis-Girod S, Fischer A, Davies EG, Kuis W, Leiva L, Cavazzana-Calvo M. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- Macchi P, Villa A, Giliani S, Sacco MG, Frattini A, Porta F, Ugazio AG, Johnston JA, Candotti F, O’Shea JJ, Vezzoni P, Notorangelo LD. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID). Nature. 1995;377:65–68. doi: 10.1038/377065a0. [DOI] [PubMed] [Google Scholar]
- Notarangelo LD, Giliani S, Mella P, Schumacher RF, Mazza C, Savoldi G, Rodriguez-Perez C, Badolato R, Mazzolari E, Porta F, Candotti F, Ugazio AG. Combined immunodeficiencies due to defects in signal transduction: defects of the γ-JAK3 signaling pathway as a model. Immunobiology. 2000;202:106–119. doi: 10.1016/s0171-2985(00)80058-3. [DOI] [PubMed] [Google Scholar]
- Schumacher RF, Mella P, Badolato R, Fiorini M, Savoldi G, Giliani S, Villa A, Candotti F, Tampalini A, O’Shea JJ, Notarangelo LD. Complete genomic organization of the human JAK3 gene and mutation analysis in severe combined immunodeficiency by single-strand conformation polymorphism. Hum Genet. 2000;106:73–79. doi: 10.1007/s004390051012. [DOI] [PubMed] [Google Scholar]
- Schumacher RF, Mella P, Lalatta F, Fiorini M, Giliani S, Villa A, Candotti F, Notarangelo LD. Prenatal diagnosis of JAK3-deficient SCID. Prenat Diagn. 1999;19:653–656. [PubMed] [Google Scholar]
- Candotti F, Oakes SA, Johnston JA, Giliani S, Schumacher RF, Mella P, Fiorini M, Ugazio AG, Badolato R, Notarangelo LD, Bozzi F, Macchi P, Strina D, Vezzoni P, Blaese RM, O’Shea JJ, Villa A. Structural and functional basis for JAK3-deficient severe combined immunodeficiency. Blood. 1997;90:3996–4003. [PubMed] [Google Scholar]
- Candotti F, Oakes SA, Johnston JA, Notarangelo LD, O’Shea JJ, Blaese RM. In vitro correction of JAK3-deficient severe combined immunodeficiency by retroviral-mediated gene transduction. J Exp Med. 1996;183:2687–2692. doi: 10.1084/jem.183.6.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Salle H, Zimmer J, Fricker D, Angenieux C, Cazenave J-P, Okubo M, Maeda H, Plebani A, Tongio M-M, Dormoy A, Hanau D. HLA class I deficiencies due to mutations in subunit 1 of the peptide transporter TAP1. J Clin Invest. 1999;103:R9–13. doi: 10.1172/JCI5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Salle H, Hanau D, Fricker D, Urlacher A, Kelly A, Salamero J, Powis SH, Donato L, Bausinger H, Laforet M, Jeras M, Spehner D, Bieber T, Falkenrodt A, Cavenae J-P, Trowsdale J, Tongio M-T. Homozygous human TAP peptide transporter mutation in HLA class I deficiency. Science. 1994;265:237–241. doi: 10.1126/science.7517574. [DOI] [PubMed] [Google Scholar]
- Yabe T, Kawamura S, Sato M, Kashiwase K, Tanaka H, Ishikawa Y, Asao Y, Oyama J, Tsuruta K, Tokunaga K, Tadokoro K, Juji T. A subject with a novel type I bare lymphocyte syndrome has tapasin deficiency due to deletion of 4 exons by Alu-mediated recombination. Blood. 2002;100:1496–1498. doi: 10.1182/blood-2001-12-0252. [DOI] [PubMed] [Google Scholar]
- Elhasid R, Etzioni A. Major histocompatibility complex class II deficiency: a clinical review. Blood Rev. 1996;10:242–248. doi: 10.1016/s0268-960x(96)90008-9. [DOI] [PubMed] [Google Scholar]
- Klein C, Lisowska-Grospierre B, LeDeist F, Fischer A, Griscelli C. Major histocompatibility complex class II deficiency: clinical manifestations, immunologic features, and outcome. J Pediatr. 1993;123:921–928. doi: 10.1016/s0022-3476(05)80388-9. [DOI] [PubMed] [Google Scholar]
- Benoist C, Mathis D. Regulation of major histocompatibility complex class-II genes: x, y, and other letters of the alphabet. Annu Rev Immunol. 1990;8:681–715. doi: 10.1146/annurev.iy.08.040190.003341. [DOI] [PubMed] [Google Scholar]
- Mach B, Steimle V, Martinez-Soria E, Reith W. Regulation of MHC class II genes: lessons from a disease. Annu Rev Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- Villard J, Masternak K, Lisowska-Grospierre B, Fischer A, Reith W. MHC class II deficiency: a disease of gene regulation. Medicine (Baltimore) 2001;80:405–418. doi: 10.1097/00005792-200111000-00006. [DOI] [PubMed] [Google Scholar]
- Tchilian EZ, Wallace DL, Wells RS, Flower DR, Morgan G, Beverley PC. A deletion in the gene encoding the CD45 antigen in a patient with SCID. J Immunol. 2001;166:1308–1313. doi: 10.4049/jimmunol.166.2.1308. [DOI] [PubMed] [Google Scholar]
- Kung C, Pingel JT, Heikinheimo M, Klemola T, Varkila K, Yoo LI, Vuopala K, Poyhonen M, Uhari M, Rogers M, Speck SH, Chatila T, Thomas ML. Mutations in the tyrosine phosphatase CD45 gene in a child with severe combined immunodeficiency disease. Nat Med. 2000;6:343–345. doi: 10.1038/73208. [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, Selz F, Hue C, Certain S, Casanova JL, Bousso P, Deist FL, Fischer A. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- Dale DC, Guerry Dt, Wewerka JR, Bull JM, Chusid MJ. Chronic neutropenia. Medicine (Baltimore) 1979;58:128–144. doi: 10.1097/00005792-197903000-00002. [DOI] [PubMed] [Google Scholar]
- Horwitz M, Benson KF, Person RE, Aprikyan AG, Dale DC. Mutations in ELA2, encoding neutrophil elastase, define a 21-day biological clock in cyclic haematopoiesis. Nat Genet. 1999;23:433–436. doi: 10.1038/70544. [DOI] [PubMed] [Google Scholar]
- Bode W, Meyer E, Jr, Powers JC. Human leukocyte and porcine pancreatic elastase: x-ray crystal structures, mechanism, substrate specificity, and mechanism-based inhibitors. Biochemistry. 1989;28:1951–1963. doi: 10.1021/bi00431a001. [DOI] [PubMed] [Google Scholar]
- Haurie C, Dale DC, Mackey MC. Cyclical neutropenia and other periodic hematological disorders: a review of mechanisms and mathematical models. Blood. 1998;92:2629–2640. [PubMed] [Google Scholar]
- Dale DC, Person RE, Bolyard AA, Aprikyan AG, Bos C, Bonilla MA, Boxer LA, Kannourakis G, Zeidler C, Welte K, Benson KF, Horwitz M. Mutations in the gene encoding neutrophil elastase in congenital and cyclic neutropenia. Blood. 2000;96:2317–2322. [PubMed] [Google Scholar]
- Dale DC, Liles WC, Garwicz D, Aprikyan AG. Clinical implications of mutations of neutrophil elastase in congenital and cyclic neutropenia. J Pediatr Hematol Oncol. 2001;23:208–210. doi: 10.1097/00043426-200105000-00005. [DOI] [PubMed] [Google Scholar]
- Welte K, Dale D. Pathophysiology and treatment of severe chronic neutropenia. Ann Hematol. 1996;72:158–165. doi: 10.1007/s002770050156. [DOI] [PubMed] [Google Scholar]
- Bernini JC. Diagnosis and management of chronic neutropenia during childhood. Pediatr Clin North Am. 1996;43:773–792. doi: 10.1016/s0031-3955(05)70432-6. [DOI] [PubMed] [Google Scholar]
- Sigurdsson F, Khanna-Gupta A, Lawson N, Berliner N. Control of late neutrophil-specific gene expression: insights into regulation of myeloid differentiation. Semin Hematol. 1997;34:303–310. [PubMed] [Google Scholar]
- Aprikyan AA, Liles WC, Rodger E, Jonas M, Chi EY, Dale DC. Impaired survival of bone marrow hematopoietic progenitor cells in cyclic neutropenia. Blood. 2001;97:147–153. doi: 10.1182/blood.v97.1.147. [DOI] [PubMed] [Google Scholar]
- Germeshausen M, Schulze H, Ballmaier M, Zeidler C, Welte K. Mutations in the gene encoding neutrophil elastase (ELA2) are not sufficient to cause the phenotype of congenital neutropenia. Br J Haematol. 2001;115:222–224. doi: 10.1046/j.1365-2141.2001.03069.x. [DOI] [PubMed] [Google Scholar]
- Welte K, Boxer LA. Severe chronic neutropenia: pathophysiology and therapy. Semin Hematol. 1997;34:267–278. [PubMed] [Google Scholar]
- Devriendt K, Kim AS, Mathijs G, Frints SG, Schwartz M, Van Den Oord JJ, Verhoef GE, Boogaerts MA, Fryns JP, You D, Rosen MK, Vandenberghe P. Constitutively activating mutation in WASP causes X-linked severe congenital neutropenia. Nat Genet. 2001;27:313–317. doi: 10.1038/85886. [DOI] [PubMed] [Google Scholar]
- Smith OP, Hann IM, Chessells JM, Reeves BR, Milla P. Haematological abnormalities in Shwachman-Diamond syndrome. Br J Haematol. 1996;94:279–284. doi: 10.1046/j.1365-2141.1996.d01-1788.x. [DOI] [PubMed] [Google Scholar]
- Dokal I, Rule S, Chen F, Potter M, Goldman J. Adult onset of acute myeloid leukaemia (M6) in patients with Shwachman-Diamond syndrome. Br J Haematol. 1997;99:171–173. doi: 10.1046/j.1365-2141.1997.3673181.x. [DOI] [PubMed] [Google Scholar]
- Dror Y, Ginzberg H, Dalal I, Cherepanov V, Downey G, Durie P, Roifman CM, Freedman MH. Immune function in patients with Shwachman-Diamond syndrome. Br J Haematol. 2001;114:712–717. doi: 10.1046/j.1365-2141.2001.02996.x. [DOI] [PubMed] [Google Scholar]
- Dror Y, Freedman MH. Shwachman-Diamond syndrome marrow cells show abnormally increased apoptosis mediated through the Fas pathway. Blood. 2001;97:3011–3016. doi: 10.1182/blood.v97.10.3011. [DOI] [PubMed] [Google Scholar]
- Goobie S, Popovic M, Morrison J, Ellis L, Ginzberg H, Boocock GR, Ehtesham N, Betard C, Brewer CG, Roslin NM, Hudson TJ, Morgan K, Fujiwara TM, Durie PR, Rommens JM. Shwachman-Diamond syndrome with exocrine pancreatic dysfunction and bone marrow failure maps to the centromeric region of chromosome 7. Am J Hum Genet. 2001;68:1048–1054. doi: 10.1086/319505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F, Mertens F, Johansson B. A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nat Genet. 1997;15(Spec No):417–474. doi: 10.1038/ng0497supp-417. [DOI] [PubMed] [Google Scholar]
- Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, Rommens JM. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33:97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- Brown E. Neutrophil adhesion and the therapy of inflammation. Semin Hematol. 1997;34:319–326. [PubMed] [Google Scholar]
- Larson RS, Springer TA. Structure and function of leukocyte integrins. Immunol Rev. 1990;114:181–217. doi: 10.1111/j.1600-065x.1990.tb00565.x. [DOI] [PubMed] [Google Scholar]
- Etzioni A. Integrins: the glue of life. Lancet. 1999;353:341–343. doi: 10.1016/S0140-6736(05)74944-8. [DOI] [PubMed] [Google Scholar]
- Phillips ML, Schwartz BR, Etzioni A, Bayer R, Ochs HD, Paulson JC, Harlan JM. Neutrophil adhesion in leukocyte adhesion deficiency syndrome type 2. J Clin Invest. 1995;96:2898–2906. doi: 10.1172/JCI118361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T, Brune T, Luhn K, Zimmer KP, Korner C, Fabritz L, van der Werft N, Vormoor J, Freeze HH, Louwen F, Biermann B, Harms E, von Figura K, Vestweber D, Koch HG. Leukocyte adhesion deficiency II syndrome, a generalized defect in fucose metabolism. J Pediatr. 1999;134:681–688. doi: 10.1016/S0022-3476(99)70281-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambruso DR, Knall C, Abell AN, Panepinto J, Kurkchubasche A, Thurman G, Gonzalez-Aller C, Hiester A, deBoer M, Harbeck RJ, Oyer R, Johnson GL, Roos D. Human neutrophil immunodeficiency syndrome is associated with an inhibitory Rac2 mutation. Proc Natl Acad Sci USA. 2000;97:4654–4659. doi: 10.1073/pnas.080074897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold BA, Bokoch GM. Molecular basis for Rac2 regulation of phagocyte NADPH oxidase. Nat Immunol. 2001;2:211–215. doi: 10.1038/85259. [DOI] [PubMed] [Google Scholar]
- Williams DA, Tao W, Yang F, Kim C, Gu Y, Mansfield P, Levine JE, Petryniak B, Derrow CW, Harris C, Jia B, Zheng Y, Ambruso DR, Lowe JB, Atkinson SJ, Dinauer MC, Boxer L. Dominant-negative mutation of the hematopoietic-specific Rho GTPase, Rac2, is associated with a human phagocyte immunodeficiency. Blood. 2000;96:1646–1654. [PubMed] [Google Scholar]
- Gu Y, Jia B, Yang F-C, D’Souza M, Harris CE, Derrow CW, Zheng Y, Williams DA. Biochemical and biological characterization of a human Rac2 GTPase mutant associated with phagocytic immunodeficiency. J Biol Chem. 2001;276:15929–15938. doi: 10.1074/jbc.M010445200. [DOI] [PubMed] [Google Scholar]
- Dorman SE, Holland SM. Interferon-γ and interleukin-12 pathway defects and human disease. Cytokine Growth Factor Rev. 2000;11:321–333. doi: 10.1016/s1359-6101(00)00010-1. [DOI] [PubMed] [Google Scholar]
- Gallin JI, Farber JM, Holland SM, Nutman TB. Interferon-γ in the management of infectious diseases. Ann Intern Med. 1995;123:216–224. doi: 10.7326/0003-4819-123-3-199508010-00009. [DOI] [PubMed] [Google Scholar]
- Jouanguy E, Altare F, Lamhamedi S, Revy P, Emile JF, Newport M, Levin M, Blanche S, Seboun E, Fischer A, Casanova JL. Interferon-γ-receptor deficiency in an infant with fatal bacille Calmette-Guerin infection. N Engl J Med. 1996;335:1956–1961. doi: 10.1056/NEJM199612263352604. [DOI] [PubMed] [Google Scholar]
- Komiyama A, Morosawa H, Nakahata T, Miyagawa Y, Akabane T. Abnormal neutrophil maturation in a neutrophil defect with morphologic abnormality and impaired function. J Pediatr. 1979;94:19–25. doi: 10.1016/s0022-3476(79)80343-1. [DOI] [PubMed] [Google Scholar]
- Gallin JI. Neutrophil-specific granule deficiency. Annu Rev Med. 1985;36:263–274. doi: 10.1146/annurev.me.36.020185.001403. [DOI] [PubMed] [Google Scholar]
- Lekstrom-Himes JA, Gallin JI. Immunodeficiency diseases caused by defects in phagocytes. N Engl J Med. 2000;343:1703–1714. doi: 10.1056/NEJM200012073432307. [DOI] [PubMed] [Google Scholar]
- Gombart AF, Koeffler HP. Neutrophil-specific granule deficiency and mutations in the gene encoding transcription factor C/EBPvarε. Curr Opin Hematol. 2002;9:36–42. doi: 10.1097/00062752-200201000-00007. [DOI] [PubMed] [Google Scholar]
- Clark RA, Malech HL, Gallin JI, Nunoi H, Volpp BD, Pearson DW, Nauseef WM, Curnutte JT. Genetic variants of chronic granulomatous disease: prevalence of deficiencies of two cytosolic components of the NADPH oxidase system. N Engl J Med. 1989;321:647–652. doi: 10.1056/NEJM198909073211005. [DOI] [PubMed] [Google Scholar]
- Baehner Rl MLK. Deficiency of reduced nicotinamide-adenine dinucleotide oxidase in chronic granulomatous disease. Science. 1968;162:1277–1279. doi: 10.1126/science.162.3859.1277. [DOI] [PubMed] [Google Scholar]
- Dinauer MC, Pierce EA, Bruns GA, Curnutte JT, Orkin SH. Human neutrophil cytochrome b light chain (p22-phox): gene structure, chromosomal location, and mutations in cytochrome-negative autosomal recessive chronic granulomatous disease. J Clin Invest. 1990;86:1729–1737. doi: 10.1172/JCI114898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae J, Noack D, Heyworth PG, Ellis BA, Curnutte JT, Cross AR. Molecular analysis of 9 new families with chronic granulomatous disease caused by mutations in CYBA, the gene encoding p22(phox). Blood. 2000;96:1106–1112. [PubMed] [Google Scholar]
- Yamada M, Ariga T, Kawamura N, Ohtsu M, Imajoh-Ohmi S, Ohshika E, Tatsuzawa O, Kobayashi K, Sakiyama Y. Genetic studies of three Japanese patients with p22-phox-deficient chronic granulomatous disease: detection of a possible common mutant CYBA allele in Japan and a genotype-phenotype correlation in these patients. Br J Haematol. 2000;108:511–517. doi: 10.1046/j.1365-2141.2000.01857.x. [DOI] [PubMed] [Google Scholar]
- Stasia J, Bordigoni P, Martel C, Morel F. A novel and unusual case of chronic granulomatous disease in a child with a homozygous 36-bp deletion in the CYBA gene (A22(0)) leading to the activation of a cryptic splice site in intron 4. Hum Genet. 2002;110:444–450. doi: 10.1007/s00439-002-0720-8. [DOI] [PubMed] [Google Scholar]
- Vazquez N, Lehrnbecher T, Chen R, Christensen BL, Gallin JI, Malech H, Holland S, Zhu S, Chanock SJ. Mutational analysis of patients with p47-phox-deficient chronic granulomatous disease: the significance of recombination events between the p47-phox gene (NCF1) and its highly homologous pseudogenes. Exp Hematol. 2001;29:234–243. doi: 10.1016/s0301-472x(00)00646-9. [DOI] [PubMed] [Google Scholar]
- Dekker J, de Boer M, Roos D. Gene-scan method for the recognition of carriers and patients with p47(phox)-deficient autosomal recessive chronic granulomatous disease. Exp Hematol. 2001;29:1319–1325. doi: 10.1016/s0301-472x(01)00731-7. [DOI] [PubMed] [Google Scholar]
- de Boer M, Singh V, Dekker J, Di Rocco M, Goldblatt D, Roos D. Prenatal diagnosis in two families with autosomal, p47(phox)-deficient chronic granulomatous disease due to a novel point mutation in NCF1. Prenat Diagn. 2002;22:235–240. doi: 10.1002/pd.296. [DOI] [PubMed] [Google Scholar]
- Lomax KJ, Leto TL, Nunoi H, Gallin JI, Malech HL. Recombinant 47-kilodalton cytosol factor restores NADPH oxidase in chronic granulomatous disease. Science. 1989;245:409–412. doi: 10.1126/science.2547247. [DOI] [PubMed] [Google Scholar]
- Leto TL, Lomax KJ, Volpp BD, Nunoi H, Sechler JM, Nauseef WM, Clark RA, Gallin JI, Malech HL. Cloning of a 67-kD neutrophil oxidase factor with similarity to a noncatalytic region of p60c-src. Science. 1990;248:727–730. doi: 10.1126/science.1692159. [DOI] [PubMed] [Google Scholar]
- Nauseef WM, Cogley M, Bock S, Petrides PE. Pattern of inheritance in hereditary myeloperoxidase deficiency associated with the R569W missense mutation. J Leukoc Biol. 1998;63:264–269. doi: 10.1002/jlb.63.2.264. [DOI] [PubMed] [Google Scholar]
- Nauseef WM, Brigham S, Cogley M. Hereditary myeloperoxidase deficiency due to a missense mutation of arginine 569 to tryptophan. J Biol Chem. 1994;269:1212–1216. [PubMed] [Google Scholar]
- Aldrich RA, steinberg AG, Campbell DC. Pedigree demonstrating a sex-linked condition characterized by draining ears, eczematoid dermatitis, and bloody diarrhea. Pediatrics. 1954;13:133–139. [PubMed] [Google Scholar]
- Ochs HD, Slichter SJ, Harker LA, Von Behrens WE, Clark RA, Wedgwood RJ. The Wiskott-Aldrich syndrome: studies of lymphocytes, granulocytes, and platelets. Blood. 1980;55:243–252. [PubMed] [Google Scholar]
- Molina IJ, Sancho J, Terhorst C, Rosen FS, Remold-O’Donnell E. T cells of patients with the Wiskott-Aldrich syndrome have a restricted defect in proliferative responses. J Immunol. 1993;151:4383–4390. [PubMed] [Google Scholar]
- Cooper MD, Chae HP, Lowman JT, Krivit W, Good RA. Wiskott-Aldrich syndrome: an immunologic deficiency disease involving the afferent limb of immunity. Am J Med. 1968;44:499–513. doi: 10.1016/0002-9343(68)90051-x. [DOI] [PubMed] [Google Scholar]
- Sullivan KE, Mullen CA, Blaese RM, Winkelstein JA. A multi-institutional survey of the Wiskott-Aldrich syndrome. J Pediatr. 1994;125:876–885. doi: 10.1016/s0022-3476(05)82002-5. [DOI] [PubMed] [Google Scholar]
- Simon HU, Mills GB, Hashimoto S, Siminovitch KA. Evidence for defective transmembrane signaling in B cells from patients with Wiskott-Aldrich syndrome. J Clin Invest. 1992;90:1396–1405. doi: 10.1172/JCI116006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan SP, Hagemann TL, Radtke BE, Blaese RM, Rosen FS. Identification of mutations in the Wiskott-Aldrich syndrome gene and characterization of a polymorphic dinucleotide repeat at DXS6940, adjacent to the disease gene. Proc Natl Acad Sci USA. 1995;92:4706–4710. doi: 10.1073/pnas.92.10.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remold-O’Donnell E, Rosen FS, Kenney DM. Defects in Wiskott-Aldrich syndrome blood cells. Blood. 1996;87:2621–2631. [PubMed] [Google Scholar]
- Snapper SB, Rosen FS. The Wiskott-Aldrich syndrome protein (WASP): roles in signaling and cytoskeletal organization. Annu Rev Immunol. 1999;17:905–929. doi: 10.1146/annurev.immunol.17.1.905. [DOI] [PubMed] [Google Scholar]
- Altman LC, Snyderman R, Blaese RM. Abnormalities of chemotactic lymphokine synthesis and mononuclear leukocyte chemotaxis in Wiskott-Aldrich syndrome. J Clin Invest. 1974;54:486–493. doi: 10.1172/JCI107784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Ohtsu M, Kobayashi I, Kawamura N, Kobayashi K, Ariga T, Sakiyama Y, Nelson DL, Tsuruta S, Anakura M, Ishikawa N. Flow cytometric analysis of Wiskott-Aldrich Syndrome (WAS) protein in lymphocytes from WAS patients and their familial carriers. Blood. 1999;93:756–758. [PubMed] [Google Scholar]
- Kawai S, Minegishi M, Ohashi Y, Sasahara Y, Kumaki S, Konno T, Miki H, Derry J, Nonoyama S, Miyawaki T, Horibe K, Tachibana N, Kudoh E, Yoshimura Y, Izumikawa Y, Sako M, Tsuchiya S. Flow cytometric determination of intracytoplasmic Wiskott-Aldrich syndrome protein in peripheral blood lymphocyte subpopulations. J Immunol Methods. 2002;260:195–205. doi: 10.1016/s0022-1759(01)00549-x. [DOI] [PubMed] [Google Scholar]
- Giliani S, Fiorini M, Mella P, Candotti F, Schumacher RF, Wengler GS, Lalatta F, Fasth A, Badolato R, Ugazio AG, Albertini A, Notarangelo LD. Prenatal molecular diagnosis of Wiskott-Aldrich syndrome by direct mutation analysis. Prenat Diagn. 1999;19:36–40. [PubMed] [Google Scholar]
- Shcherbina A, Rosen FS, Remold-O’Donnell E. WASP levels in platelets and lymphocytes of Wiskott-Aldrich syndrome patients correlate with cell dysfunction. J Immunol. 1999;163:6314–6320. [PubMed] [Google Scholar]
- Notarangelo LD, Mazza C, Giliani S, D’Aria C, Gandellini F, Ravelli C, Locatelli MG, Nelson DL, Ochs HD. Missense mutations of the WASP gene cause intermittent X-linked thrombocytopenia. Blood. 2002;99:2268–2269. doi: 10.1182/blood.v99.6.2268. [DOI] [PubMed] [Google Scholar]
- Wada T, Jagadeesh GJ, Nelson DL, Candotti F. Retrovirus-mediated WASP gene transfer corrects Wiskott-Aldrich syndrome T-cell dysfunction. Hum Gene Ther. 2002;13:1039–1046. doi: 10.1089/104303402753812449. [DOI] [PubMed] [Google Scholar]
- Goldmuntz E, Emanuel BS. Genetic disorders of cardiac morphogenesis: the DiGeorge and velocardiofacial syndromes. Circ Res. 1997;80:437–443. doi: 10.1161/01.res.80.4.437. [DOI] [PubMed] [Google Scholar]
- Jawad AF, McDonald-Mcginn DM, Zackai E, Sullivan KE. Immunologic features of chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome). J Pediatr. 2001;139:715–723. doi: 10.1067/mpd.2001.118534. [DOI] [PubMed] [Google Scholar]
- van den Bosch MA, Wittebol S, van Dijk H, Kramer MH. Hypocalcemic tetany as an early sign of DiGeorge syndrome in an adult woman. Am J Med. 2002;112:161–162. doi: 10.1016/s0002-9343(01)00955-x. [DOI] [PubMed] [Google Scholar]
- Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S, Tokooya K, Jore BS, Lopez M, Pandita RK, Lia M, Carrion D, Xu H, Schorle H, Kobler JB, Scambler P, Wynshaw-Boris A, Skoultchi AI, Morrow BE, Kucherlapati R. TBX1 is responsible for cardiovascular defects in velo-cardio- facial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- Maynard TM, Haskell GT, Bhasin N, Lee JM, Gassman AA, Lieberman JA, LaMantia AS. RanBP1, a velocardiofacial/DiGeorge syndrome candidate gene, is expressed at sites of mesenchymal/epithelial induction. Mech Dev. 2002;111:177–180. doi: 10.1016/s0925-4773(01)00616-5. [DOI] [PubMed] [Google Scholar]
- Guris DL, Fantes J, Tara D, Druker BJ, Imamoto A. Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat Genet. 2001;27:293–298. doi: 10.1038/85855. [DOI] [PubMed] [Google Scholar]
- Driscoll DA. Prenatal diagnosis of the 22q11.2 deletion syndrome. Genet Med. 2001;3:14–18. doi: 10.1097/00125817-200101000-00004. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Metcalfe JA, Thick J, Mak YF. Leukemia and lymphoma in ataxia telangiectasia. Blood. 1996;87:423–438. [PubMed] [Google Scholar]
- Loeb DM, Lederman HM, Winkelstein JA. Lymphoid malignancy as a presenting sign of ataxia-telangiectasia. J Pediatr Hematol Oncol. 2000;22:464–467. doi: 10.1097/00043426-200009000-00017. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Harnden DG, Arlett CF, Harcourt SA, Lehmann AR, Stevens S, Bridges BA. Ataxia telangiectasia: a human mutation with abnormal radiation sensitivity. Nature. 1975;258:427–429. doi: 10.1038/258427a0. [DOI] [PubMed] [Google Scholar]
- Gatti RA, Berkel I, Boder E, Braedt G, Charmley P, Concannon P, Ersoy F, Foroud T, Jaspers NG, Lange K, Lathrop GM, Leppert M, Nakamura Y, O’Connell P, Paterson M, Salser W, Sanal O, Silver J, Sparkes RS, Susi E, Weeks DE, Wei S, White R, Yoder F. Localization of an ataxia-telangiectasia gene to chromosome 11q22–23. Nature. 1988;336:577–580. doi: 10.1038/336577a0. [DOI] [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, Ashkenazi M, Pecker I, Frydman M, Harnick R, Patanjali SR, Simmons A, Clines GA, Sartiel A, Gatti RA, Chessa L, Sanal O, Lavin MF, Jaspers NGJ, Taylor AMR, Arlett CF, Miri T, Weissman SM, Lovett M, Collins FS, Shiloh Y. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- Hartley KO, Gell D, Smith GC, Zhang H, Divecha N, Connelly MA, Admon A, Lees-Miller SP, Anderson CW, Jackson SP. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH, Kastan MB. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- Xu Y, Baltimore D. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev. 1996;10:2401–2410. doi: 10.1101/gad.10.19.2401. [DOI] [PubMed] [Google Scholar]
- Dork T, Bendix R, Bremer M, Rades D, Klopper K, Nicke M, Skawran B, Hector A, Yamini P, Steinmann D, Weise S, Stuhrmann M, Karstens JH. Spectrum of ATM gene mutations in a hospital-based series of unselected breast cancer patients. Cancer Res. 2001;61:7608–7615. [PubMed] [Google Scholar]
- Stankovi T, Stewart GS, Byrd P, Fegan C, Moss PA, Taylor AM. ATM mutations in sporadic lymphoid tumours. Leuk Lymphoma. 2002;43:1563–1571. doi: 10.1080/1042819021000002884. [DOI] [PubMed] [Google Scholar]
- Fang NY, Greiner TC, Weisenburger DD, Chan WC, Vose JM, Smith LM, Armitage JO, Mayer RA, Pike BL, Collins FS, Hacia JG. Oligonucleotide microarrays demonstrate the highest frequency of ATM mutations in the mantle cell subtype of lymphoma. Proc Natl Acad Sci USA. 2003;100:5372–5377. doi: 10.1073/pnas.0831102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring K, Ahangari F, Scott SP, Waring P, Purdie DM, Chen PC, Hourigan K, Ramsay J, McKinnon PJ, Swift M, Lavin MF. Mice heterozygous for mutation in Atm, the gene involved in ataxia-telangiectasia, have heightened susceptibility to cancer. Nat Genet. 2002;32:185–190. doi: 10.1038/ng958. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST, Lane WS, Kastan MB, D’Andrea AD. Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- Blume RS, Wolff SM. The Chediak-Higashi syndrome: studies in four patients and a review of the literature. Medicine (Baltimore) 1972;51:247–280. [PubMed] [Google Scholar]
- Bejaoui M, Veber F, Girault D, Gaud C, Blanche S, Griscelli C, Fischer A. The accelerated phase of Chediak-Higashi syndrome. Arch Fr Pediatr. 1989;46:733–736. [PubMed] [Google Scholar]
- Perou CM, Moore KJ, Nagle DL, Misumi DJ, Woolf EA, McGrail SH, Holmgren L, Brody TH, Dussault BJ, Jr, Monroe CA, Duyk GM, Pryor RJ, Li L, Justice MJ, Kaplan J. Identification of the murine beige gene by YAC complementation and positional cloning. Nat Genet. 1996;13:303–308. doi: 10.1038/ng0796-303. [DOI] [PubMed] [Google Scholar]
- Tchernev VT, Mansfield TA, Giot L, Kumar AM, Nandabalan K, Li Y, Mishra VS, Detter JC, Rothberg JM, Wallace MR, Southwick FS, Kingsmore SF. The Chediak-Higashi protein interacts with SNARE complex and signal transduction proteins. Mol Med. 2002;8:56–64. [PMC free article] [PubMed] [Google Scholar]
- Burkhardt JK, Wiebel FA, Hester S, Argon Y. The giant organelles in beige and Chediak-Higashi fibroblasts are derived from late endosomes and mature lysosomes. J Exp Med. 1993;178:1845–1856. doi: 10.1084/jem.178.6.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetz K, Isaaz S, Griffiths GM. Loss of cytotoxic T lymphocyte function in Chediak-Higashi syndrome arises from a secretory defect that prevents lytic granule exocytosis. J Immunol. 1995;154:6122–6131. [PubMed] [Google Scholar]
- Nagle DL, Karim MA, Woolf EA, Holmgren L, Bork P, Misumi DJ, McGrail SH, Dussault BJ, Jr, Perou CM, Boissy RE, Duyk GM, Spritz RA, Moore KJ. Identification and mutation analysis of the complete gene for Chediak-Higashi syndrome. Nat Genet. 1996;14:307–311. doi: 10.1038/ng1196-307. [DOI] [PubMed] [Google Scholar]
- Karim MA, Suzuki K, Fukai K, Oh J, Nagle DL, Moore KJ, Barbosa E, Falik-Borenstein T, Filipovich A, Ishida Y, Kivrikko S, Klein C, Kreuz F, Levin A, Miyajima H, Regueiro J, Russo C, Uyama E, Vierimaa O, Spritz RA. Apparent genotype-phenotype correlation in childhood, adolescent, and adult Chediak-Higashi syndrome. Am J Med Genet. 2002;108:16–22. [PubMed] [Google Scholar]
- Griscelli C, Durandy A, Guy-Grand D, Daguillard F, Herzog C, Prunieras M. A syndrome associating partial albinism and immunodeficiency. Am J Med. 1978;65:691–702. doi: 10.1016/0002-9343(78)90858-6. [DOI] [PubMed] [Google Scholar]
- Menasche G, Pastural E, Feldmann J, Certain S, Ersoy F, Dupuis S, Wulffraat N, Bianchi D, Fischer A, Le Deist F, de Saint Basile G. Mutations in RAB27A cause Griscelli syndrome associated with haemophagocytic syndrome. Nat Genet. 2000;25:173–176. doi: 10.1038/76024. [DOI] [PubMed] [Google Scholar]
- Pastural E, Barrat FJ, Dufourcq-Lagelouse R, Certain S, Sanal O, Jabado N, Seger R, Griscelli C, Fischer A, de Saint Basile G. Griscelli disease maps to chromosome 15q21 and is associated with mutations in the myosin-Va gene. Nat Genet. 1997;16:289–292. doi: 10.1038/ng0797-289. [DOI] [PubMed] [Google Scholar]
- Sanal O, Ersoy F, Tezcan I, Metin A, Yel L, Menasche G, Gurgey A, Berkel I, de Saint Basile G. Griscelli disease: genotype-phenotype correlation in an array of clinical heterogeneity. J Clin Immunol. 2002;22:237–243. doi: 10.1023/a:1016045026204. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JC, Barral DC, Mules EH, Booth S, Hume AN, Machesky LM, Seabra MC, Griffiths GM. Rab27a is required for regulated secretion in cytotoxic T lymphocytes. J Cell Biol. 2001;152:825–834. doi: 10.1083/jcb.152.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster F, Stachel DK, Schmid I, Baumeister FA, Graubner UB, Weiss M, Haas RJ, Belohradsky BH. Griscelli syndrome: report of the first peripheral blood stem cell transplant and the role of mutations in the RAB27A gene as an indication for BMT. Bone Marrow Transplant. 2001;28:409–412. doi: 10.1038/sj.bmt.1703114. [DOI] [PubMed] [Google Scholar]
- Janka GE. Familial hemophagocytic lymphohistiocytosis. Eur J Pediatr. 1983;140:221–230. doi: 10.1007/BF00443367. [DOI] [PubMed] [Google Scholar]
- Henter JI, Arico M, Elinder G, Imashuku S, Janka G. Familial hemophagocytic lymphohistiocytosis: primary hemophagocytic lymphohistiocytosis. Hematol Oncol Clin North Am. 1998;12:417–433. doi: 10.1016/s0889-8588(05)70520-7. [DOI] [PubMed] [Google Scholar]
- Henter JI, Elinder G, Soder O, Hansson M, Andersson B, Andersson U. Hypercytokinemia in familial hemophagocytic lymphohistiocytosis. Blood. 1991;78:2918–2922. [PubMed] [Google Scholar]
- Jabado N, de Graeff-Meeder ER, Cavazzana-Calvo M, Haddad E, Le Deist F, Benkerrou M, Dufourcq R, Caillat S, Blanche S, Fischer A. Treatment of familial hemophagocytic lymphohistiocytosis with bone marrow transplantation from HLA genetically nonidentical donors. Blood. 1997;90:4743–4748. [PubMed] [Google Scholar]
- Stepp SE, Dufourcq-Lagelouse R, Le Deist F, Bhawan S, Certain S, Mathew PA, Henter JI, Bennett M, Fischer A, de Saint Basile G, Kumar V. Perforin gene defects in familial hemophagocytic lymphohistiocytosis. Science. 1999;286:1957–1959. doi: 10.1126/science.286.5446.1957. [DOI] [PubMed] [Google Scholar]
- Nagano M, Kimura N, Ishii E, Yoshida N, Yoshida T, Sako M, Hibi S, Imashuku S, Miyazaki S, Hara T, Mizutani S. Clonal expansion of αβ-T lymphocytes with inverted Jβ1 bias in familial hemophagocytic lymphohistiocytosis. Blood. 1999;94:2374–2382. [PubMed] [Google Scholar]
- Kimura N, Ishii E, Sako M, Yoshida T, Nagano M, Takada H, Imashuku S, Tamura K. Effect of chemotherapy and stem cell transplantation on T lymphocyte clones in familial haemophagocytic lymphohistiocytosis. Br J Haematol. 2001;113:822–831. doi: 10.1046/j.1365-2141.2001.02784.x. [DOI] [PubMed] [Google Scholar]
- Goransdotter Ericson K, Fadeel B, Nilsson-Ardnor S, Soderhall C, Samuelsson A, Janka G, Schneider M, Gurgey A, Yalman N, Revesz T, Egeler R, Jahnukainen K, Storm-Mathiesen I, Haraldsson A, Poole J, de Saint Basile G, Nordenskjold M, Henter J. Spectrum of perforin gene mutations in familial hemophagocytic lymphohistiocytosis. Am J Hum Genet. 2001;68:590–597. doi: 10.1086/318796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Stadt U, Pruggmayer M, Jung H, Henter JI, Schneider M, Kabisch H, Janka G. Prenatal diagnosis of perforin gene mutations in familial hemophagocytic lymphohistiocytosis (FHLH). Prenat Diagn. 2002;22:80–81. doi: 10.1002/pd.231. [DOI] [PubMed] [Google Scholar]
- Coffey AJ, Brooksbank RA, Brandau O, Oohashi T, Howell GR, Bye JM, Cahn AP, Durham J, Heath P, Wray P, Pavitt R, Wilkinson J, Leversha M, Huckle E, Shaw-Smith CJ, Dunham A, Rhodes S, Schuster V, Porta G, Yin L, Serafini P, Sylla B, Zollo M, Franco B, Bolino A, Seri M, Lanyi A, Davis JR, Webster D, Harris A, Lenoir G, de St. Basile G, Jones A, Beloradsky BH, Achatz H, Murken J, Fassler R, Sumegi J, Romeo G, Vaudin M, Ross MT, Meindl A, Bentley DR. Host response to EBV infection in X-linked lymphoproliferative disease results from mutations in an SH2-domain encoding gene. Nat Genet. 1998;20:129–135. doi: 10.1038/2424. [DOI] [PubMed] [Google Scholar]
- Bolino A, Yin L, Seri M, Cusano R, Cinti R, Coffey A, Brooksbank R, Howell G, Bentley D, Davis JR, Lanyi A, Huang D, Stark M, Creaven M, Bjorkhaug L, Heitzmann F, Lamartine J, Gaudi S, Sylla BS, Lenoir GM, Castagnola E, Giacchino R, Porta G, Franco B, Romeo G. A new candidate region for the positional cloning of the XLP gene. Eur J Hum Genet. 1998;6:509–517. doi: 10.1038/sj.ejhg.5200249. [DOI] [PubMed] [Google Scholar]
- Nelson DL, Terhorst C. X-linked lymphoproliferative syndrome. Clin Exp Immunol. 2000;122:291–295. doi: 10.1046/j.1365-2249.2000.01400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayos J, Wu C, Morra M, Wang N, Zhang X, Allen D, van Schaik S, Notarangelo L, Geha R, Roncarolo MG, Oettgen H, De Vries JE, Aversa G, Terhorst C. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature. 1998;395:462–469. doi: 10.1038/26683. [DOI] [PubMed] [Google Scholar]
- Parolini S, Bottino C, Falco M, Augugliaro R, Giliani S, Franceschini R, Ochs HD, Wolf H, Bonnefoy JY, Biassoni R, Moretta L, Notarangelo LD, Moretta A. X-linked lymphoproliferative disease: 2B4 molecules displaying inhibitory rather than activating function are responsible for the inability of natural killer cells to kill Epstein-Barr virus-infected cells. J Exp Med. 2000;192:337–346. doi: 10.1084/jem.192.3.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy N, Mattsson K, Maeda A, Liu A, Szekely L, Klein E. The X-linked lymphoproliferative disease gene product SAP is expressed in activated T and NK cells. Immunol Lett. 2002;82:141–147. doi: 10.1016/s0165-2478(02)00029-9. [DOI] [PubMed] [Google Scholar]
- Nistala K, Gilmour KC, Cranston T, Davies EG, Goldblatt D, Gaspar HB, Jones AM. X-linked lymphoproliferative disease: three atypical cases. Clin Exp Immunol. 2001;126:126–130. doi: 10.1046/j.1365-2249.2001.01599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiepolo L, Maraschio P, Gimelli G, Cuoco C, Gargani GF, Romano C. Multi-branched chromosomes 1, 9, and 16 in a patient with combined IgA and IgE deficiency. Hum Genet. 1979;51:127–137. doi: 10.1007/BF00287166. [DOI] [PubMed] [Google Scholar]
- Xu GL, Bestor TH, Bourc’his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Pequignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijmenga C, Hansen RS, Gimelli G, Bjorck EJ, Davies EG, Valentine D, Belohradsky BH, van Dongen JJ, Smeets DF, van den Heuvel LP, Luyten JA, Strengman E, Weemaes C, Pearson PL. Genetic variation in ICF syndrome: evidence for genetic heterogeneity. Hum Mutat. 2000;16:509–517. doi: 10.1002/1098-1004(200012)16:6<509::AID-HUMU8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Buchanan KL, Tsien F, Jiang G, Sun B, Uicker W, Weemaes CM, Smeets D, Sperling K, Belohradsky BH, Tommerup N, Misek DE, Rouillard JM, Kuick R, Hanash SM. DNA methyltransferase 3B mutations linked to the ICF syndrome cause dysregulation of lymphogenesis genes. Hum Mol Genet. 2001;10:2917–2931. doi: 10.1093/hmg/10.25.2917. [DOI] [PubMed] [Google Scholar]
- Weemaes CM, Hustinx TW, Scheres JM, van Munster PJ, Bakkeren JA, Taalman RD. A new chromosomal instability disorder: the Nijmegen breakage syndrome. Acta Paediatr Scand. 1981;70:557–564. doi: 10.1111/j.1651-2227.1981.tb05740.x. [DOI] [PubMed] [Google Scholar]
- Weemaes CM, Smeets DF, van der Burgt CJ. Nijmegen breakage syndrome: a progress report. Int J Radiat Biol. 1994;66:S185–188. [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- van Engelen BG, Hiel JA, Gabreels FJ, van den Heuvel LP, van Gent DC, Weemaes CM. Decreased immunoglobulin class switching in Nijmegen breakage syndrome due to the DNA repair defect. Hum Immunol. 2001;62:1324–1327. doi: 10.1016/s0198-8859(01)00345-7. [DOI] [PubMed] [Google Scholar]
- Zhao S, Weng YC, Yuan SS, Lin YT, Hsu HC, Lin SC, Gerbino E, Song MH, Zdzienicka MZ, Gatti RA, Shay JW, Ziv Y, Shiloh Y, Lee EY. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature. 2000;405:473–477. doi: 10.1038/35013083. [DOI] [PubMed] [Google Scholar]
- Davis SD, Schaller J, Wedgwood RJ. Job’s syndrome: recurrent, “cold,” staphylococcal abscesses. Lancet. 1966;1:1013–1015. doi: 10.1016/s0140-6736(66)90119-x. [DOI] [PubMed] [Google Scholar]
- Hill HR, Estensen RD, Hogan NA, Quie PG. Severe staphylococcal disease associated with allergic manifestations, hyperimmunoglobulinemia E, and defective neutrophil chemotaxis. J Lab Clin Med. 1976;88:796–806. [PubMed] [Google Scholar]
- Chehimi J, Elder M, Greene J, Noroski L, Stiehm ER, Winkelstein JA, Sullivan KE. Cytokine and chemokine dysregulation in hyper-IgE syndrome. Clin Immunol. 2001;100:49–56. doi: 10.1006/clim.2001.5039. [DOI] [PubMed] [Google Scholar]
- Fleisher TA, Puck JM, Strober W, Dale JK, Lenardo MJ, Siegel RM, Straus SE, Bleesing JJ. The autoimmune lymphoproliferative syndrome: a disorder of human lymphocyte apoptosis. Clin Rev Allergy Immunol. 2001;20:109–120. doi: 10.1385/criai:20:1:109. [DOI] [PubMed] [Google Scholar]
- Sneller MC, Wang J, Dale JK, Strober W, Middelton LA, Choi Y, Fleisher TA, Lim MS, Jaffe ES, Puck JM, Lenardo MJ, Straus SE. Clincial, immunologic, and genetic features of an autoimmune lymphoproliferative syndrome associated with abnormal lymphocyte apoptosis. Blood. 1997;89:1341–1348. [PubMed] [Google Scholar]
- Goldman FD, Vibhakar R, Puck JM, Straus SE, Ballas ZK, Hollenback C, Loew T, Thompson A, Song K, Cook RT. Aberrant T-cell antigen receptor-mediated responses in autoimmune lymphoproliferative syndrome. Clin Immunol. 2002;104:31–39. doi: 10.1006/clim.2002.5249. [DOI] [PubMed] [Google Scholar]
- Fisher GH, Rosenberg FJ, Straus SE, Dale JK, Middleton LA, Lin AY, Strober W, Lenardo MJ, Puck JM. Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell. 1995;81:935–946. doi: 10.1016/0092-8674(95)90013-6. [DOI] [PubMed] [Google Scholar]
- Wang J, Zheng L, Lobito A, Chan FK, Dale J, Sneller M, Yao X, Puck JM, Straus SE, Lenardo MJ. Inherited human caspase 10 mutations underlie defective lymphocyte and dendritic cell apoptosis in autoimmune lymphoproliferative syndrome type II. Cell. 1999;98:47–58. doi: 10.1016/S0092-8674(00)80605-4. [DOI] [PubMed] [Google Scholar]