Abstract
Human urine has been shown to possess submicrogram per milliliter amounts of DNA. We show here that DNA isolated from human urine resolves into two size categories: the large species, greater than 1 kb, being predominantly cell associated and heterogeneous in size, and the smaller, between 150 to 250 bp, being mostly non-cell associated. We showed that the low molecular weight class of urine DNA is derived from the circulation, by comparing the mutated K-ras sequences present in DNA isolated from tumor, blood, and urine derived from an individual with a colorectal carcinoma (CRC) containing a mutation in codon 12 of the K-ras proto-oncogene. In the urine, mutated K-ras sequences were abundant in the low molecular weight species, but far less abundant in the large molecular weight-derived DNA. Finally, the possibility that detection of mutant K-ras sequences in DNA derived from the urine correlates with the occurrence of a diagnosis of CRC and polyps that contain mutant K-ras was explored in a blinded study. There was an 83% concurrence of mutated DNA detected in urine and its corresponding disease tissue from the same individuals, when paired urine and tissue sections from 20 subjects with either CRC or adenomatous polyps were analyzed for K-ras mutation. The possibility that the source of the trans renal DNA is apoptotic cells, and the potential use of this finding for cancer detection and monitoring is discussed.
Circulating DNA has been detected in serum and plasma of both healthy and diseased individuals.1,2 DNA containing cancer “signatures” (mutations or hypermethylation) has been found in the plasma or serum of patients with small cell lung cancer,3 head and neck cancer,4 clear cell renal cancer,5 pancreatic cancer,6 breast cancer,7 hepatocellular carcinoma,8 non-small cell lung cancer,9 and colorectal carcinoma (CRC).10 In almost all instances where tumor tissue was available, the mutations in the plasma were identical to those detected in the primary tumor, suggesting that the circulating mutant DNA was of tumor origin. Circulating DNA as a source for molecular diagnosis and prognosis is considered promising and was recently reviewed.11
Urine is collected body fluid that has been filtered through the kidney barrier. It has been suggested that small amounts of cell-free circulating DNA in the blood can pass the kidney barrier into urine,12,13 and tumor-specific sequences were detected in DNA isolated from urine. Sample collection of urine is non-invasive, and isolation of DNA from urine is technically easier than from serum or plasma, due to the low protein content of urine. Thus, urine could be a useful source of circulating DNA for molecular diagnosis and prognosis. It was, therefore, of interest to characterize the nature of the nucleic acid recovered from urine, as compared to circulating DNA in the bloodstream. Here, the potential of urine-derived DNA in cancer detection has been explored by using a well- characterized mutation in circulating (plasma and serum) DNA: the K-ras codon 12 mutation in colorectal cancer or adenomatous-polyp patients.
Materials and Methods
Study Subjects and Specimens
Participants were recruited from the Great Lakes New England Clinical Epidemiological Center including University of Michigan, Dana-Farber Harvard Cancer Center, Henry Ford Health System, and University of Toronto. Cancer patients were enrolled from the surgical or oncologic serives (before initiation of chemo- or radiation therapy or surgery). Most “healthy-no known neoplasia” controls were subjects enrolled at the endoscopy suites, but had undergone a negative colonoscopy. Patient samples were obtained under institutional Division of Human Subjects Protection (IRB) approvals.
Urine Collection and Fractionation
Fresh collected urine was immediately mixed with 0.5 mol/L EDTA, pH 8.0, to a final concentration of 10 mmol/L EDTA to inhibit the possible nuclease activity in urine sample, and stored at −70°C. To isolate total urine DNA, frozen urine sample was thawed at room temperature, and then placed immediately in ice before DNA isolation. Thawed urine would be processed for DNA isolation within an hour.
To separate the supernatant and cell debris of freshly collected urine, total urine was centrifuged at 1500 rpm at 4°C for 10 minutes. The supernatant was collected on ice. The cell pellet was then washed briefly, with 1 ml of sodium citrate buffer (pH 3.0). The wash-off solution was collected by centrifugation at 13,000 rpm for 90 seconds and combined with the supernatant fraction. The pellet after centrifugation was collected as the cell debris portion of urine.
Serum Collection
Fifty ml of blood was collected in heparinized containers from informed and consenting subjects. The samples were permitted to sit at room temperature for a minimum of 30 minutes (and a maximum of 60 minutes) to allow the clot to form in the red top tubes, then centrifuged at 1300 × g at 4°C for 20 minutes. The serum was transferred to a polypropylene, capped tube, and frozen on dry ice.
DNA Isolation
Urine samples were mixed with 1.5 volume of 6 mol/L guanidine thiocyanate (Sigma, St. Louis, MO) by inverting eight times. One ml of resin (Wizard DNA Isolation Kit, Promega, Madison, WI) was added into the urine lysate and incubated for 2 hours at room temperature with gentle mixing. Resin-DNA complex was centrifuged, transferred to a mini-column (provided from the kit), washed with a buffer provided by the manufacturer, and DNA was eluted with H2O. Serum DNA was isolated as was the urine DNA, except 2.5 volume of 6 mol/L guanidine thiocyanate was used. DNA from paraffin-embedded tissue sections was isolated using the MasterPure DNA Kit (Epicenter, Madison, WI) per manufacturer’s instruction.
Restriction-Enriched Polymerase Chain Reaction Detecting Codon 12 Mutation of K-ras Gene
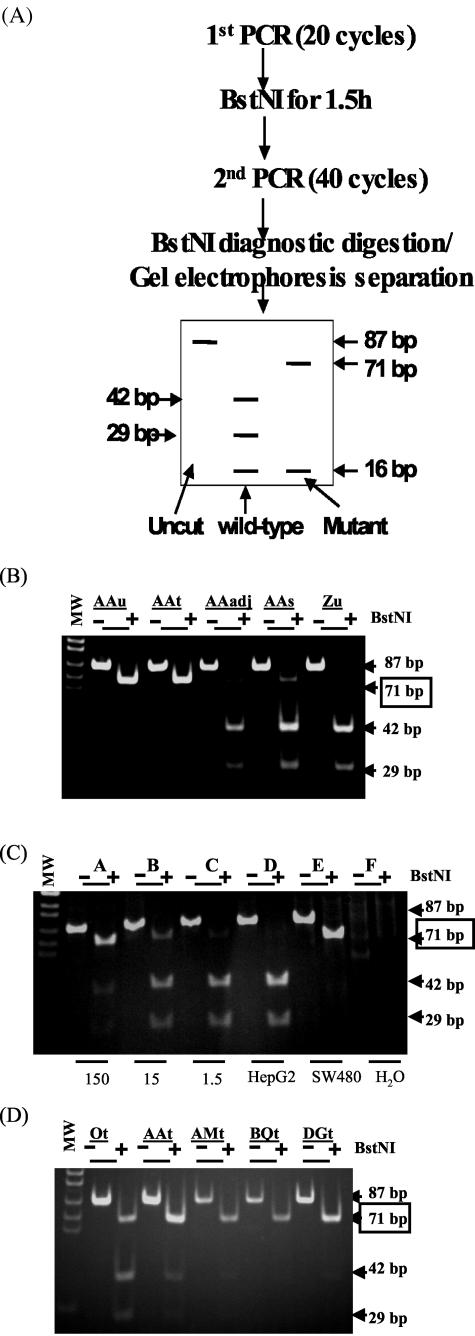
The restriction-enriched polymerase chain reaction (RE-PCR) assay, illustrated in Figure 2A (see Figure 2), distinguishing wild-type and mutant K-ras sequences in DNA isolated from urine, serum, or tissue is described as below. Twenty cycles of Hot-start PCR were performed as following: DNA, equivalent to 0.2 ml of urine, with 1.5 mmol/L MgCl2, 1X PCR buffer (Qiagen, Valencia, CA), 0.5 U Hot-start Taq (Qiagen), 200 μmol/L dNTP, 0.1 μmol/L primers: L-Ext (5′ GCT CTT CGT GGT GTG GTG TCC ATA TAA ACT TGT GGT AGT TGG ACC T 3′) and R-Ext (5′ GCT CTT CGT GGT GTG GTG TCC CGT CCA CAA AAT GAT TCT GA3′) was mixed, denatured at 95°C for 15 minutes and followed by 20 cycles of 94°C for 30 seconds, 52°C for 30 seconds, and 1 cycle of 72°C for 5 minutes. These first 20 cycles of PCR was used to amplify both wild-type and mutated DNA, but only to introduce an artificial BstNI site to the 5′ end of amplified product derived from wild-type DNA. After the first round of PCR, the amplified products were digested with BstNI (New England Biolabs, Beverly, MA) to eliminate amplified products derived from wild-type DNA. One of 20 of the digested product was then subjected to the second Hot-start PCR with the 1 μmol/L primers L (5′ ACT GAA TAT AAA CTT GTG GTA GTT GGA CCT 3′) and R-Bst (5′ GTC CAC AAA ATG ATC CTG GAT TAG C 3′) at the following condition: 95°C for 15 minutes, 40 cycles of 94°C for 30 seconds, 56°C for 30 seconds and followed by 1 cycle of 72°C for 5 minutes. This second set of primers introduced a BstNI site to the 3′ end of amplified product derived from both wild-type and mutated templates serving as internal control for the BstNI diagnostic digestion after the second PCR. The amplified products (87 bp) of the second PCR were digested to completion with BstNI (as shown by disappearance of the 87-bp DNA fragment), and resolved through polyacrylamide gels. The appearance of the 71-bp DNA fragment after BstNI digestion [as illustrated in Figure 1A] is the evidence of the existence of K-ras mutated DNA. Each sample was assayed 2 to 3 times. The samples was scored as “positive” for K-ras mutation when the 71-bp DNA fragments appeared after the second BstNI digestion of the PCR products in 2 of 2 or 2 of 3 repeated assays. As validation controls, the standard reconstitution samples (as in Figure 2C) were included in each assay.
Figure 2.
Wild-type and mutant K-ras sequences detected in human urine and disease tissue by restriction-enriched polymerase chain reactions (RE-PCR). A: The predicted pattern of digested products. B: The results of analysis of DNA derived from the urine of an individual with no diagnosis of cancer (Zu) and from tumor tissue (AAt), non-tumor tissue that was 1 cm apart from the tumor (AAadj), serum (AAs), and urine (AAu) from an individual with CRC. C: The sensitivity and specificity of the assay by analysis of the reconstruction standards (1.5, 15, and 150 copies of SW480 genome per 50 ng of HepG2 DNA), and 50 ng HepG2 DNA and 5 ng SW480 alone, as indicated. D: Detection of mutated K-ras DNA in CRC tissue. CRC disease tissue sections from five CRC patients (Ot, AAt, AMt, BQt, and DGt) were subjected to DNA isolation and the RE-PCR assay for mutated K-ras DNA. MW: DNA MW markers. This represents the data from three independent experiments.
Figure 1.
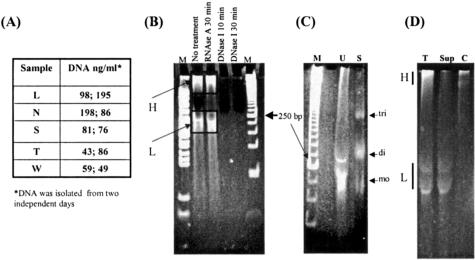
Size and nature of nucleic acid isolated from human urine. Twenty ml of urine was collected from five volunteers (L, N, S, T, and W) on 2 different days. DNA was isolated as described in Materials and Methods, and DNA concentration in each sample was determined by spectrophotometry reading (A). B: DNA from one volunteer was aliquoted. Each aliquot was digested with RNase A, DNase I, or left untreated, and then analyzed in an 8% polyacrylamide gel. The gel was stained with ethidium bromide and photographed. The first and the last lanes labeled “M” are two different DNA molecular weight markers. C: DNA isolated from 15 ml of urine (U) from volunteer W, and 4 ml of human serum (Sigma) (S) was analyzed in an 8% polyacrymide gel, stained with ethidium bromide and photographed. Expected mobilities of 1 to 2 unit sizes of nucleosomal (mononucleosomal (mo) and dinucleosomal (di)) DNA fragments are indicated. D: Recovery of low MW urine DNA (L) in the supernatant of urine. DNA recovered from the total urine (T), the supernatant (Sup), and the cell debris pellet (C), was analyzed in an 8% polyacrymide gel, stained with ethidium bromide and photographed. The high (H) and low (L) MW urine DNA is indicated.
Cloning and Sequencing of Mutated K-ras DNA
The 71-bp fragment of the second BstNI-digested PCR product from the RE-PCR assay was identified by 2% agarose gel electrophoresis, and isolated from the gel by using the Perfectprep Gel Cleanup Kit (Eppendorf, Germany). The purified 71-bp fragment was then cloned into the pPCR-Script Amp SK (+) cloning vector by using the PCR-Script Amp Cloning Kit (Strategene, La Jolla, CA USA). The insert was determined by PCR with the T7 and T3 primers, with the annealing temperature 55°C, 30 cycles. The plasmid DNA containing insert was then isolated by using the Qiaprep Spin Miniprep Kit (Qiagen, Valencia, CA). The plasmid DNA was sequenced at the Nucleic Acid Facility, Thomas Jefferson University, Philadelphia, PA.
Determination of the Ratio of Mutated K-ras DNA to Total K-ras DNA in Low MW and High MW Urine DNA
Total urine DNA was resolved in a 2.5% agarose gel with a molecular weight marker, stained with ethidium bromide. The gel slices corresponding to DNA size at 1 kb and larger (as the high MW urine DNA fraction) or between 100 to 400 bp (as the low MW urine DNA fraction) were excised respectively. DNA was eluted from the gel slices by the MERmaid SPIN Kit (BIO 101, Vista, CA) per manufacturer’s specification.
To measure the ratio of the amplifiable mutated K-ras DNA to total K-ras DNA, a serial of twofold dilutions of eluted high and low MW urine DNA fractions were prepared, and subjected to the PCR with K-ras L-Ext and R-Ext primers to determine the relative amount of amplifiable K-ras templates in each fraction, and the RE-PCR to assay for the K-ras mutation, with the standard reconstitution samples. The relative copy numbers of K-ras amplifiable template was estimated by comparing it to the standard DNA control. The ratio of mutated K-ras DNA, total K-ras DNA, and the fold enrichment of K-ras mutated DNA was then calculated.
Results
DNA Can Be Recovered from Human Urine and Occurs as Two General Size Classes
It has been demonstrated that urine contains DNA and suggested that a portion of this DNA comes from the bloodstream crossing the kidney barrier.12,13 To confirm this observation, nucleic acids from the urine of five individuals, were isolated and examined. The amount of nucleic acids in each sample was determined to be between 40 and 200 ng/ml using spectrophotometry (Figure 1A). The nucleic acids, isolated and concentrated from 15 ml of urine, were resolved by electrophoresis through 8% polyacrylamide gels, stained with ethidium bromide, and shown to resolve into two distinct populations with respect to size: (1) a heterogenous high molecular weight (MW) form that remained near the well (H, 1 kb and larger) and (2) a relatively uniform low molecular weight form (L) between 150 to 250 bp (bp) (Figure 1B).
To verify the nature of nucleic acid recovered from urine, DNase I and RNase A digestions were performed before electrophoresis. The results (Figure 1B) show that both high and low MW forms of the nucleic acid are RNase resistant, but DNase sensitive. They are thus considered to be DNA.
Comparison of DNA Derived from the Blood and Urine
DNA isolated from the circulation (serum) and urine was compared by electrophoresis through an 8% polyacrymide gel. Consistent with other reports,14,15 the serum contained fragmented DNA as indicated in Figure 1C (lane S), as resolved by electrophoresis. Although the relative abundances of the different size populations vary, the low MW (L) urine DNA (lane U) is represented by mostly 1 to 2 nucleosomal size fragments.
To investigate whether the low MW urine DNA is cell-free, total urine was resolved before DNA isolation, by low-speed centrifugation into two fractions: the supernatant and the cell debris, as described in the Materials and Methods section. Figure 1D shows that DNA recovered from total urine (T), the supernatant (Sup), and the cell debris (C), respectively, as resolved in an 8% polyacrymide gel. As predicted, both high and low MW DNA was recovered from the total urine (T). Interestingly, low MW urine DNA was mostly recovered in the supernatant (Sup) of urine. DNA derived from the cell debris pellet (C) contained high MW urine DNA. This result supports the hypothesis that the low MW urine DNA species is enriched for DNA that is cell-free, and the high MW urine DNA is mostly derived from the cells shed from the urinary tract.
Evidence that DNA in Urine Is, in Part, Derived from the Circulation
To further test the hypothesis that urinary DNA was, at least partially, derived from the circulation, the relative amounts of mutated K-ras proto-oncogene in different tissue compartments derived from an individual with a colorectal cancer (CRC) containing a codon 12 mutant K-ras sequence, was determined. DNA derived from malignant cells, as stated, can appear in the circulation.16,17,18 Since the mutant K-ras sequence would be expected to be in the tumor, but not in other tissue, its presence (or relative enrichment) in the small versus large species of urine-derived DNA would be evidence that the small DNA in the urine contained a contribution from the circulation.
To detect the codon 12 mutated K-ras genes present in small DNA fragments, a gel-based restriction-enriched PCR (RE-PCR)10 was modified to permit amplification of DNA of less than 300 bp in length, which includes the size of the smaller species recovered from urine. This approach is graphically outlined in Figure 2A. As validation controls, DNA was prepared from sources known to have either mutant (human adenocarcinoma “12Val” SW480 cells) or wild-type (human hepatoblastoma HepG2 cells) K-ras sequences, and subjected to PCR, with the results shown in Figure 2C.
DNA prepared from the urine of an individual with no diagnosis of cancer (Zu) contained no detectable levels of mutant K-ras (no appearance of a 71-bp fragment after BstNI digestion) under the conditions of amplification used, although wild-type K-ras was readily detected (Figure 2B). DNA was also prepared from tumor (AAt) and surrounding non-tumor tissue (AAadj), serum (AAs), and urine (AAu) obtained from an individual (AA) with a biopsy-confirmed diagnosis of CRC. Figure 2B shows that although DNA derived from the non-malignant tissue surrounding the tumor did not contain detectable mutant K-ras, mutant K-ras sequences are present (the 71-bp fragment after BstNI digestion is detected), above background levels (15 copies/50 ng DNA/reaction) in all other sources (tumor, serum, and urine) from the cancer patient. The absence of mutant K-ras in the non-tumor-derived solid tissue indicates that this individual does not carry the mutant sequence in the germ line, and its presence in the tumor is the evidence for a somatic mutation.
Since mutant K-ras sequences present in the tumor derived from individual AA would be expected to be present in the blood, detection of mutant K-ras in both of these tissue compartments (blood and tumor tissue) is not surprising. However, the presence of mutant K-ras sequences in the urine of samples from individual AA (Figure 2B) is consistent with the theory that DNA from the circulation, containing tumor-derived mutant K-ras, traveled to the urine. If the mutant K-ras sequence present in urine was derived from the tumor in the colon, the type of mutation detected in the urine and in the tumor should be identical. As described in the Materials and Methods section, the 71 bp of DNA fragment after the second BstNI digestion of the PCR products from the RE-PCR of the DNA from urine and tumor was cloned and sequenced to determine whether the mutation detected in urine is the same mutation that was found in the tumor. The DNA sequencing data clearly demonstrate that both the urine and the tumor of patient AA contain the 12 Val mutation in the K-ras sequence (data not shown).
The Small Size Class of DNA in the Urine Comes from, at Least in Part, the Circulation
As stated, the presence of mutant K-ras sequences in the urine could have either been the result of trans-renal passage of DNA from the circulation to the urine, or from cells resident in the urinary tract, which happen to have the same K-ras mutation. Since the larger DNA class detected in the urine might be excluded by kidney filtration, we had reasoned that the most likely population of DNA in the urine, which was derived from the circulation, would be the small size class (150 to 250 bp). In this case, relative to the larger (greater than 1 kb) species of DNA isolated from urine, the small, 150- to 250-bp size class of DNA isolated from the urine of the individual with the mutant K-ras-positive tumor would be expected to be enriched in abundance for mutant K-ras sequences. To test this hypothesis, large- and small-sized classes of DNA isolated from the urine of two CRC patients (AA and O), and one patient with an adenomatous polyps (Q), were recovered from gel slices, following resolution through 2.5% agarose gels. Urine DNA from these three patients had been tested, and found to contain the same K-ras codon 12 mutation as their corresponding tumor or polyp (data not shown). Each DNA fraction recovered from the gel slice was subjected to a determination of the ratio of mutated K-ras DNA to total K-ras DNA as described in Materials and Methods.
Briefly, to determine the copy number of total amplifiable K-ras DNA and mutant K-ras DNA in each fraction, serial twofold dilutions for each fraction were prepared, and then subjected to (1) the PCR amplification for the total amplifiable K-ras DNA, and (2) the RE-PCR assay for the K-ras mutation, as summarized in Table 1. Analysis of the ratio of mutated K-ras DNA to total K-ras DNA in each size fraction (fold enrichment of mutated K-ras detected) shows a clear enrichment (8 to 12-fold) of mutated DNA in low MW DNA fraction for all samples tested. Since the separation of low MW DNA from high MW DNA is not expected to be absolute by agarose gel electrophoresis, it is not surprising that some amount of mutant K-ras would be detected in higher molecular species. However, it is reasoned that the mutated K-ras DNA detected in the low MW species isolated from urine comes from the circulation.
Table 1.
Enrichment of Mutated K-ras DNA in the Low MW Urine DNA Fraction*
| Patient sample† | Ratio of mutated K-ras DNA to total K-ras DNA‡
|
Fold enrichment in low MW fraction§
|
|
|---|---|---|---|
| High MW | Low MW | ||
| Q (adenomatous polyps) | 1/240 | 1/20 | 12 |
| O (CRC) | 1/40 | 1/5 | 8 |
| AA (CRC) | 1/48 | 1/6 | 8 |
The data are represented from two independent experiments.
Diagnosis was biopsy confirmed.
DNA was examined by a RE-PCR, and the amount of mutant and wild type K-ras sequence detected as described in Materials and Methods, is expressed as a ratio, with wild type as the numerator.
The relative enrichment of mutant K-ras in the low molecular weight DNA species, compared with the amount in the high molecular weight species, is shown.
Detection of Mutant K-ras in Urine and Disease Tissue from the Patients with a Diagnosis of Either CRC or Adenomatous Polyps
Having established necessary assay parameters, the comparison between mutant K-ras in colorectal disease tissue (CRC or adenomatous polyps), and corresponding urine from the same individual was investigated. Paraffin-embedded disease tissue sections, as well as corresponding urine from five subjects with a diagnosis of CRC, and 15 subjects with the diagnosis of adenomatous polyps were collected according to the approved IRB protocol. DNA from the collected samples was isolated and subjected to the RE-PCR assay for the mutant K-ras sequence (as in Figure 2). As the control, DNA isolated from urine of 48 subjects with no known neoplasia (healthy) was also subjected to mutant K-ras detection by the RE-PCR assay. As shown in Table 2, 9 of the 48 (19%) urine samples from “healthy” individual contained detectable mutated K-ras sequences. Interestingly, all 9 positive samples were derived from the subjects enrolled at the endoscopy suites, but had undergone a negative colonscopy, as mentioned in Materials and Methods. Significantly, in each of five CRC samples shown to contain mutant K-ras, as shown in Figure 2D, mutant K-ras was detected in the corresponding urine (100% concurrence). Similarly, for 10 of the 13 polyps samples shown to contain mutant K-ras, mutant K-ras was detected in the corresponding urine (77% concurrence).
Table 2.
Correlation between Detection of Mutant K-ras in Tissue and Corresponding Urine
| Tissue specimen and presence (+) or absence (−) of mutant ras (mean age, age range of subjects in the group, in years, and gender)*
|
No. of urine samples
|
No. of urine samples in which mutant K-ras detected
|
No. of corresponding disease tissue†
|
% of samples in which mutant K-ras was detected in both urine and disease tissue
|
|---|---|---|---|---|
| Urine from healthy (no known neoplasia) subjects (n = 48, mean age = 51, range 26–80, 27 females) | 48 | 9 | N/A‡ | (19%)§ |
| CRC “+” for mutant K-ras (n = 5, mean age = 65, range 50–74, 1 female) | 5 | 5 | 5 | 100% |
| Polyps “+” for mutant K-ras (n = 13, mean age = 66, range 54–83, 7 females) | 13 | 10 | 13 | 77% |
| Polyps “−” for mutant K-ras (n = 2, mean age = 60, range 59–62, 1 female) | 2 | 1 | 2 | 50% |
The presence or absence of mutant K-ras (codon 12 missense) was determined by an RE-PCR as described in Materials and Methods. Urine from these individuals was tested by investigators blinded as to the clinical diagnosis, as shown in Table 1.
Number of tissue specimens available in the clinical group.
N/A, not applicable.
Percent of urine samples from “healthy” individuals containing detectable mutant K-ras. No disease tissue available in this “healthy” group.
It is noted that mutated K-ras was detected in urine from one subject with polyp tissues containing no detectable mutant K-ras. Taken together, mutant K-ras sequences were detected in the urine of 15 of the 18 urine samples (83.3%) from individuals with confirmed mutant K-ras containing tumors or polyps, and 19% of those with no diagnosis of colorectal disease.
Discussion
We have shown that DNA isolated from total human urine was comprised of contributions from two sources. The large size class appears to be mainly derived from cells shed into the urine from the urinary tract. The other class is small, between 150 to 250 bp, and following low-speed centrifugation, is mostly recovered from the supernatant, not from the cell and cell debris pellet. By comparing the DNA sequences and the amounts of wild-type and mutated K-ras sequences in tumor, non-tumor tissue, serum, and urine from a patient with a mutated K-ras-positive colorectal tumor, we demonstrated that the low MW urine DNA derives, at least in part, from the circulation.
It is not known how small DNA can cross the kidney barrier. The molecular weight cutoff in the kidney glomeruli is approximately 70 kd,19 and the pore size of the glomerular barrier is about 30Å. Negatively charged molecules in the circulation might face an additional barrier because of the negative charge of the glomerular basement membrane. It is also unknown if DNA crosses the kidney barrier as a protein free molecule, as a nucleoprotein, or even as a part of small apoptotic bodies. Some hypotheses that can explain this phenomenon were discussed earlier.13
The nature of cell-free DNA in blood has been studied extensively.17,20,21 Nucleosome-sized circulating DNA might originate from the internucleosomal cleavage of chromatin, a major hallmark of apoptosis.21 Malignant, benign, and even pre-neoplastic cells often proliferate at abnormal rates that are accompanied by an increase in apoptotic cell death,22,23,24 and this DNA may accumulate in the urine. If the transformed cells possessed somatic mutations, or epigenetic modifications that distinguish them from the germ line, urine could be a good source for detection and monitoring of cancer, even from extra-urinary tract sites.
Presumably, the large size (1 kb and larger) urine DNA is derived from cells shed into the urine from the urinary tract. Consistent with this hypothesis is the fact that the large size class of DNA can be reduced in abundance by low-speed centrifugation before electrophoresis, in which cells are sedimented, and small size class of DNA can only be recovered from the supernatant of urine. It was noted that the amount of this large class of urine DNA varied significantly from sample to sample, even from the same subject, as seen in Figure 1, B-D. This finding might be due to the amount of sloughed off cells, and cell debris in each collection, as well as the degree of cell lysis. Interestingly, the removal of cell debris from urine by centrifugation before DNA isolation increased the mutated K-ras DNA concentration in urine DNA (data not shown). Moreover, it’s unlikely that detected mutated K-ras derived from the cell debris that might come from the urologic tumor of “normal subject,” since the incidence of mutated K-ras in urologic tumors is low. This further suggests that the detected mutated K-ras DNA is present in the cell-free urinary fraction.
Although the comparison between mutant K-ras in tissue and urine was encouraging, the detection of mutant K-ras in 19% (n = 48) of the samples from individuals with no diagnosis of cancer or polyps was disturbing and was greater than we had observed in our previous studies of “healthy” individuals (not shown). The detection of mutant K-ras in the urine of people with no known cancer or polyp is consistent with one of two possibilities: (1) the occurrence of mutant K-ras in this population is truly higher than previously expected, and the assay faithfully detected this or (2) the assay falsely assigned a positive designation for mutant K-ras to samples that did not contain mutant K-ras (assay error).
Explanation (2) seems unlikely, since the samples scored as mutant K-ras positive in repeated assays as described in Materials and Methods, and the presence of the codon 12 mutations was confirmed by the DNA sequencing of the nine sample sets.
Thus, it appears that mutant K-ras can be detected in the urine of as many as 19% of the individuals contributing samples to this study, despite no apparent colorectal disease. Since 38 of 48 had been scheduled patients in the GI clinic receiving endoscopy, and all of the positive urine samples were part of this group, it is possible that they are not representative of the general population, in which the occurrence of mutant K-ras might be lower. Interestingly, similar results were obtained by Kopreski et al,25 in which 28 of 105 subjects without evidence of neoplasia also demonstrated mutated K-ras DNA in their plasma. Nevertheless, the frequency of mutant K-ras detected in the urine of those with a diagnosis of a mutated K-ras-positive colorectal disease was substantially greater (83%) than those without a diagnosis, suggesting that mutant K-ras in the urine itself could be an important molecular “risk” factor. Needless to say, it can be assumed that detection of other mutated oncogenes, tumor suppressors, or hypermethylated CpG islands associated with colorectal diseases and in combination with detection of mutant K-ras, might provide a closer correlation between disease and a positive signal.
Cells shed into urine have been suggested to be a useful source of DNA to detect tumors arising at or near the urinary track.26,27 The study of K-ras mutation in urine and corresponding disease tissues of 20 patients with CRC or adenomatous polyps in this report, strongly suggest that the mutation occurred in sites distant from the urinary tract also can be found in urine. A diagnostic test that is relatively less pleasant, non-invasive and inexpensive, is more likely to be used, especially for screening. For example, there would be improved inpatient comfort, and likely compliance with diagnostic testing, if endoscopic procedures such as colonoscopy could be replaced with serum or simple urine analysis.
Acknowledgments
We thank Dr. Laura Steel and Ms. Melissa Hesley (Thomas Jefferson University, Philadelphia, PA) for the critical reading of the manuscript and Ms. Laurie Fortlage (University of Michigan, Ann Arbor, MI) for coordinating collection and shipment of clinic specimens. The authors acknowledge the Great Lakes New England Clinical Epidemiological Centers (within the EDRN, NCI, Grant CA86400) for providing clinical specimens.
Footnotes
Supported, in part, by The Early Detection Research Network (EDRN) of The National Cancer Institute, The Hepatitis B Foundation of America, and an appropriation from The Commonwealth of Pennsylvania.
References
- Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- Anker P, Lyautey J, Lederrey C, Stroun M. Circulating nucleic acids in plasma or serum. Clin Chim Acta. 2001;313:143–146. doi: 10.1016/s0009-8981(01)00666-0. [DOI] [PubMed] [Google Scholar]
- Chen XQ, Stroun M, Magnenat JL, Nicod LP, Kurt AM, Lyautey J, Lederrey C, Anker P. Microsatellite alterations in plasma DNA of small cell lung cancer pateints. Nat Med. 1996;2:1033–1035. doi: 10.1038/nm0996-1033. [DOI] [PubMed] [Google Scholar]
- Nawroz H, Koch W, Anker P, Stroun M, Sidransky D. Microsatellite alterations in serum DNA of head and neck cancer patients. Nat Med. 1996;2:1035–1037. doi: 10.1038/nm0996-1035. [DOI] [PubMed] [Google Scholar]
- Goessl C, Heicappel R, Munker R, Anker P, Stroun M, Krause H, Muller M, Miller K. Microsatellite analysis of plasma DNA from patients with clear renal carcinoma. Cancer Res. 1998;58:4728–4732. [PubMed] [Google Scholar]
- Mulcahy HE, Lyautey J, Lederrey C, Chen QX, Anker P, Alstead E, Ballinger A, Farthing M, Stroun M. A prospective study of K-ras mutations in plasma of pancreatic cancer patients. Clin Cancer Res. 1998;4:271–275. [PubMed] [Google Scholar]
- Chen XQ, Bonnefoi H, Diebold-Berger C, Lyautey J, Lederrey C, Faltin-Traub E, Stroun M, Anker P. Detecting tumor-related alterations in plasma or serum DNA of patients diagnosed with breast cancer. Clin Cancer Res. 1999;5:2297–2303. [PubMed] [Google Scholar]
- Wong IHN, Lo YMD, Zhang J, Liew C-T, Ng MHL, Wong N, Lai PBS, Lau WY, Hjelm M, Johnson PJ. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res. 1999;59:71–73. [PubMed] [Google Scholar]
- Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999;59:67–70. [PubMed] [Google Scholar]
- Kopreski M, Benko F, Kwee C, Leitzel K, Eskander E, Lipton A, Gocke C. Detection of mutant K-ras DNA in plasma or serum of patients with colorectal cancer. Br J Cancer. 1997;76:1293–1299. doi: 10.1038/bjc.1997.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A, Zangemeister-Wittke U, Stahel RA. Circulating DNA: a new diagnostic gold mine? Cancer Treatment Rev. 2002;28:255–271. doi: 10.1016/s0305-7372(02)00077-4. [DOI] [PubMed] [Google Scholar]
- Botezatu I, Serdyuk O, Potapova G, Shelepov V, Alechina R, Molyaka Y, Anan’ev V, Bazin I, Garin A, Narimanov M, Melkonyan H, Umansky S, Lichtenstein AV. Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin Chem. 2000;46:1078–1084. [PubMed] [Google Scholar]
- Lichtenstein AV, Melkonyan HS, Tomei D, Umansky SR. Circulating nucleic acids and apoptosis. Ann NY Acad Sci. 2001;945:239–249. doi: 10.1111/j.1749-6632.2001.tb03892.x. [DOI] [PubMed] [Google Scholar]
- Stroun M, Anker P, Lyautey J, Lederrey C, Maurice PA. Isolation and characterization of DNA from the plasma of cancer patients. Eur J Cancer Clin Oncol. 1987;23:707–712. doi: 10.1016/0277-5379(87)90266-5. [DOI] [PubMed] [Google Scholar]
- Fournie GJ, Courtin JP, Laval F, Chale JJ, Pourrat JP, Pujazon MC, Lauque D, Carles P. Plasma DNA as a marker of cancerous cell death: investigations in patients suffering from lung cancer and in nude mice bearing human tumours. Cancer Lett. 1995;91:221–227. doi: 10.1016/0304-3835(95)03742-f. [DOI] [PubMed] [Google Scholar]
- Anker P, Stroun M. Tumor-related alterations in circulating DNA, potential for diagnosis, prognosis and detection of minimal residual disease. Leukemia. 2001;15:289–291. doi: 10.1038/sj.leu.2402016. [DOI] [PubMed] [Google Scholar]
- Anker P, Mulcahy H, Chen X, Stroun M. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metatstasis Rev. 1999;18:65–73. doi: 10.1023/a:1006260319913. [DOI] [PubMed] [Google Scholar]
- Mulcahy H, Lyautey J, Lederrey C, Chen X, Lefort F, Vasioukhin V, Anker P, Alstead E, Farthing M, Stroun M. Plasma DNA K-ras mutations in patients with gastrointestinal malignancies. Ann NY Acad Sci. 2000;906:25–28. doi: 10.1111/j.1749-6632.2000.tb06585.x. [DOI] [PubMed] [Google Scholar]
- Lote CJ. London: Chapman & Hall; Principle of Renal Physiology. 1994:33–44. [Google Scholar]
- Stroun M, Anker P, Maurice P, Lyautey J, Lederrey C, Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989;46:318–322. doi: 10.1159/000226740. [DOI] [PubMed] [Google Scholar]
- Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch R, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- Arai N, Mitomi H, Ohtani Y, Igarashi M, Kakita A, Okayasu I. Enhanced epithelial cell turnover associated with p53 accumulation and high p21WAF1/CIP1 expression in ulcerative colitis. Mod Pathol. 1999;12:604–611. [PubMed] [Google Scholar]
- Nakamura T, Nariya S, Sakai T. Cell death in colorectal polyps as evaluated by in situ 3′-tailing reaction and its relationship to BCL-2 expression. Pathol Int. 1995;45:721–728. doi: 10.1111/j.1440-1827.1995.tb03388.x. [DOI] [PubMed] [Google Scholar]
- Schulte-Hermann R, Bursch W, Grasl-Kraupp B, Torok L, Ellinger AML. Role of active cell death (apoptosis) in multi-stage carcinogenesis. Toxicol Lett. 1995;82/83:143–148. doi: 10.1016/0378-4274(95)03550-8. [DOI] [PubMed] [Google Scholar]
- Kopreski M, Benko F, Borys D, Khan A, McGarrity T, Gocke C. Somatic mutation screening: identification of individuals harboring K-ras mutations with the use of plasma DNA. J Natl Cancer Inst. 2000;92:918–923. doi: 10.1093/jnci/92.11.918. [DOI] [PubMed] [Google Scholar]
- Utting M, Werner W, Dahse R, Schubert J, Junker K. Microsatellite analysis of free tumor DNA in urine, serum, and plasma of patients: a minimally invasive method for the detection of bladder cancer. Clin Cancer Res. 2002;8:35–40. [PubMed] [Google Scholar]
- Eisenberger CF, Schoenberg M, Enger C, Hortopan S, Shah S, Chow N, Marshall FF, Sidransky D. Diagnosis of renal cancer by molecular urinalysis. J Natl Cancer Inst. 1999;91:2028–2032. doi: 10.1093/jnci/91.23.2028. [DOI] [PubMed] [Google Scholar]