Abstract
In a recent study, Candida species in clinical blood samples were detected using a real-time PCR-based method (Maaroufi et al, J Clin Microbiol 2003, 41:3293–3298). For the present study, we evaluated the efficiency of this method as an adjunct to the BACTEC blood culture system to early detection of positivity and negativity of simulated low candidemias. We first established an in vitro correlation between the inoculum of the most frequently encountered Candida species and the time to positivity of these microorganisms. Then, aliquots from blood culture bottles infected with a final average candidal inoculum of 3.18 colony-forming units (CFU)/culture bottle (range, 1 to 6 CFU) were collected at increasing incubation times, and DNA was extracted and submitted to the TaqMan-based PCR assay. To optimize this assay, we evaluated the effect of adding 0.5% bovine serum albumin (BSA) to DNA extracts and found that it decreased the effects of inhibitors. Using specific probes for the tested Candida species, the PCR assay was positive on blood culture aliquots collected from the BACTEC system after a minimum culture turnaround time (TAT) of 3.11 ± 1.24 hours. Addition of BSA to PCR reaction mixtures improves the TAT (1.84 ± 0.41 hours). Hence, the combination of DNA “amplification” in the culture bottles by normal growth with an additional DNA amplification by PCR might be a reliable tool facilitating the early diagnosis of low candidemias.
Bloodstream fungal infections constitute a serious health problem because of the excessive hospitalization, added health care costs, and high morbidity and mortality attributed to the diseases.1 Furthermore, this problem is compounded by an increase in resistance to anti-fungal agents.2,3 Candida species now rank fourth among the most commonly isolated organisms from bloodstream infections.4,5 The rates of isolation of the major Candida species causing fungemia have been determined in several studies: C. albicans (50 to 59%), C. tropicalis (11 to 25%), C. glabrata (8 to 18%), C. parapsilosis (7 to 15%), and C. krusei (2 to 4%).2,6,7
One of the most technologically advanced automated blood culture instruments for fungemia detection by the continuous monitoring of microorganisms growth is BACTEC 9240 system (Becton Dickinson Diagnostic Instrument Systems, Sparks, MD). Over the last several decades, significant improvements have been made in culture media of this system in an attempt to shorten the turnaround time (TAT) to detection of a positive or negative result and to increase the yield for a given sample. With the BACTEC system, on average, 5 to 7 days of incubation were required to recover all clinically relevant Candida species8 and definitive identification necessitates further analysis by assimilation and fermentation tests, which can require up to 28 days to complete.9 Similarly, 5 to 7 days must pass before a specimen can be reliably declared to be negative for fungi, and this delay may lead to inappropriate empirical treatment with anti-fungal drugs.
At the molecular level, fungal ribosomal RNA (rRNA) sequence variation offers an alternative to culturing for detection and identification of yeasts. As a result, molecular biology-based diagnostic methods that use rRNA sequences have been used to overcome the problems of sensitivity, specificity, and delay encountered with conventional methods.10,11,12,13,14,15,16 However, the use of PCR-based methods is not always an efficient approach to detect low candidemias, given the significant number of clinical blood samples that did not reach theoretical sensitivity threshold of PCR assay (≥10 colony-forming units (CFU)/ml). Using colony counts obtained directly on blood agar plates by the lysis centrifugation technique, Isenberg17 showed that this may concern more than half clinical specimens; 26.5% of all positive culture for yeast had <1 CFU/ml of blood, and 27.9% had 1 to 10 CFU/ml of blood. It appears that alone neither PCR nor current blood culture methods may detect all patients with candidemias.
We evaluate the efficacy of a recently published real-time PCR-based method for detection of Candida species in clinical blood samples,16 as an adjunct to the BACTEC culture system for early detection of positivity and negativity of simulated low candidemias. The combination of DNA “amplification” in the culture bottles by normal growth of Candida blastoconidies with an additional DNA amplification by PCR was assessed to increase the yield and rate detection of a positive sample. At first, the relationship of initial density of the five commonly encountered Candida species in blood cultures and time to positivity (TTP) in the BACTEC system was evaluated. Then, the PCR assay was performed on aliquots of blood culture fluids collected after the culture media were inoculated with a “low” fungemia colony count (1 to 6 CFU/culture bottle) and incubated in the BACTEC system for increasing times. The Poisson distribution was used to calculate the probability of finding at least one viable CFU per inoculated bottle. Therefore, the purpose of this study was to determine the minimum culture TATs of BACTEC blood cultures for optimal detection and identification of major Candida species by using a TaqMan-based PCR assay.
Materials and Methods
Yeasts and Growth Conditions
Reference strains of Candida albicans ATCC 24433, C. tropicalis ATCC 66029, C. glabrata ATCC 90030, C. parapsilosis ATCC 22019, and C. krusei ATCC 6258 were used. The strains were grown on Sabouraud glucose-agar plates (BBL, Becton Dickinson, Sparks, MD) for 72 hours at 30°C. Species identification was established by using the API 20C kit (BioMérieux, Marcy l’Etoile, France). Cultures were routinely inoculated from single colonies. Yeasts in exponential phase of growth were obtained by inoculation of cells from 24-hour cultures grown in YEPD (10 g of yeast extract, 20 g of peptone, and 20 g of dextrose per L) into 200 ml of YEPD to a starting concentration of 2 × 104 CFU/ml. The cultures were grown overnight at 30°C with agitation.
Inoculation of Blood Culture Bottles
All Candida species from exponential phase cultures were washed twice in phosphate-buffered saline and a microorganism suspension made in sterile distilled water was adjusted to a 0.5 McFarland standard (which is approximately 106 CFU/ml).
Time to Positivity
Yeast suspension (106 CFU/ml) prepared from each Candida species served as a seeded inoculum. Two trials were conducted in duplicate in which initial fungal densities of 1 to 104 CFU/ml of blood were used. To simulate blood cultures, 8 to 10 ml of fresh human blood obtained from healthy volunteers as well as 100 μl of each microorganism suspension were injected aseptically into culture bottles type BACTEC Plus aerobic/F (Becton Dickinson Diagnostic Instrument Systems). All blood cultures were then promptly placed in the BACTEC 9240 Blood Culture system that automatically recorded time to positivity when applicable. Positive bottles were observed microscopically to confirm the presence of fungi.
Minimum Culture TATs of BACTEC Blood Cultures to Detect Low Candidemias by PCR
For each of the five Candida species tested in two independent experiments, 8 to 10 ml of human blood as well as a 100-μl suspension of “low” fungemia colony count (1 to 6 CFU/blood culture) prepared from a dilution of 100 to 101 CFU/ml were inoculated into culture bottles type BACTEC Plus aerobic/F. The yeast concentration was determined by quantitative colony counts on plated media to verify the exact number of CFU in the inoculum. Plates were incubated at 35°C for 48 hours before enumeration. The results of this enumeration showed a final average inoculum of 3.18 CFU/100 μl (range, 1 to 6 CFU/100 μl). For each of the Candida species studied, in all, between 25 and 36 bottles were inoculated concurrently with the same candidal suspension. Thus, a total of 153 blood cultures were tested. After inoculation, blood cultures were immediately placed in the BACTEC system for increasing times or until an alarm signal indicated positivity. After 1 hour of incubation, two blood cultures were withdrawn from BACTEC system and a 10-ml aliquot from each bottle was aseptically collected using a 20-gauge needle to avoid allowing the BACTEC resin particles to pass through.18 The others blood cultures remained undisturbed. The same operation was repeated at each interval of 1 hour. DNA was then extracted from the samples and analyzed.
DNA Extraction and PCR Amplification
The DNA extraction procedure used was a modification of a previously described wash/alkali/heat lysis method;19,20 a larger volume of blood culture (5 ml) and a higher concentration of alkali wash solution (1 mol/L NaOH and 0.1 mol/L sodium citrate) were used. Briefly, after two alkali treatments of blood culture fluid with 10 ml of wash solution, the spheroplasts were directly generated by incubation of fungal cells with 100 μl of lyticase buffer (10 mmol/L Tris [pH 8], 1 mmol/L EDTA, 1 U of recombinant lyticase [ICN Biomedicals, Aurora, OH] per 100 μl) for 1 hour at 37°C. Then, spheroplasts lysis and DNA extraction were accomplished by heating for 1 hour at 100°C, freezing/thawing twice and subsequent centrifugation. The DNA recovered was immediately analyzed or stored at −20°C until testing.
The universal fungal amplification primers ITS86 and ITS411 (Table 1) are complementary to conserved sequences in 5.8S rDNA and 28S rDNA, respectively. Five fluorogenic probes designed by Shin et al10 (Table 1) were used in this study: CA-FAM (where FAM is 6-carboxyfluorescein), CT-TET (where TET is tetrachloro-6-carboxyfluorescein), CG-FAM, CP-HEX (where HEX is hexachloro-6-carboxyfluorescein), and CK-TET for the detection of C. albicans, C. tropicalis, C. glabrata, C. parapsilosis, and C. krusei DNAs, respectively. All of these oligonucleotides were synthesized by Applied Biosystems (Foster City, CA).
Table 1.
Primer and Probe Sequences Used in This Study
| Primer* and probe | Nucleotide sequences (5′-3′) | Reference |
|---|---|---|
| ITS86 (F) | 5′-GTGAATCATCGAATCTTTGAAC-3′ | 11 |
| ITS4 (R) | 5′-TCCTCCGCTTATTGATAGC-3′ | 11 |
| CA-FAM | 5′-FAMATTGCTTGCGGCGGTAACGTCG-3′ | 10 |
| CT-TET | 5′-TETCAAAACGCTTATTTTGCTAGTGGCC-3′ | 10 |
| CP-HEX | 5′-HEXGGTACAAACTCCAAAACTTCTTCCA-3′ | 10 |
| CG-FAM | 5′-FAMTAGGTTTTACCAACTCGGTGTTGAT-3′ | 10 |
| CK-TET | 5′-TETAGTGGCCCGAGCGAACTAGACTTTT-3′ | 10 |
F, forward primer; R, reverse primer.
DNA samples were analyzed by using the GeneAmp 5700 Sequence Detection system (Applied Biosystems) as previously described.16 Briefly, each 25 μl of TaqMan PCR mixture consisted of a 5-μl aliquot of sample DNA, 1X PCR buffer, 3.5 mmol/L MgCl2, 0.2 μmol/L of each primer, each dNTP at a concentration of 0.2 mmol/L, 0.2 μmol/L of a Candida species probe, 2 U of TaqDNA polymerase (Roche Diagnostics), 0.5 μl of ROX passive reference dye (Invitrogen, Groningen, Netherlands), and 2.5 μl of 5% bovine serum albumin (BSA; Sigma, St. Louis, MO). Thermal cycling conditions consisted of heating at 94°C for 10 minutes, which preceded a two-stage temperature profile of 30 seconds at 95°C and 1 minute and 30 seconds at 60°C for 40 cycles. Reactions are characterized by the time during cycling when a threshold of baseline fluorescence (CT) is exceeded.
In each reaction run, Candida species genomic DNA was serially diluted over a range of four logs (1000 to 0.05 pg DNA, corresponding to 105 to 5 CFU) to enable the generation of a standard curve. The latter is generated by plotting the CT values versus log10 (N), where N is the concentration of the standard. The Candida species DNA level in each blood culture sample is determined by locating its CT on the standard curve. Negative controls were tested by using the same PCR reaction mixture under the amplification conditions described above but without template DNA.
Statistical Analysis
Statistical analysis was performed using the Mann-Whitney rank sum test. A P value of less than 0.05 was considered to be significant. The probability of finding at least one viable CFU per inoculated bottle was calculated with the Poisson statistic following the equation 1- e−x, where x is the number of CFU per inoculated blood culture.
Results
Time to Positivity for BACTEC Blood Cultures Inoculated with Candida Species
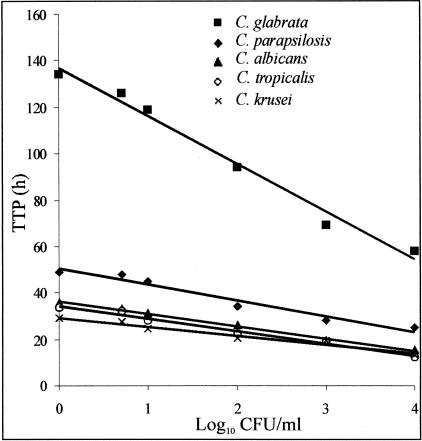
In the preliminary study, a significant inverse correlation was established between TTP and the logarithm of the initial concentration of the inoculum (P < 0.0001) of the five Candida species tested (Figure 1). However, the growth rate varied from species to species. Over the range of initial fungal densities tested, an increase of one log10 in density in the inoculum required different interval of time for detection of positivity. The mean ± SD (SD) values were as follows: for C. albicans, 5.08 ± 1.12 hours; for C. tropicalis, 5.35 ± 1.30 hours; for C. glabrata, 19.00 ± 7.12 hours; for C. parapsilosis, 6.06 ± 3.06 hours; and for C. krusei, 3.91 ± 2.06 hours.
Figure 1.
Time to positivity (TTP) for blood cultures inoculated with increasing numbers (1 to 104 CFU/ml of blood) of exponential-phase Candida species. Incubation time to detection of fungal growth, which is calculated by the BACTEC system, was recorded. Each symbol represents the mean value of the CFU values of two independent experiments. The averaged r = 0.978.
Average Culture Time to Detect Low Candidemias Using the BACTEC System
The average initial colony counts of each of the five Candida species inoculated in the BACTEC blood culture bottles are given in Table 2. However, microorganisms were not detected on all plated media and culture bottles. This was in agreement with the probability of finding at least one viable CFU per inoculated bottle (calculated mean ± SD probability value for the five Candida species, 0.89 ± 0.07). At low fungemia count (range, 1 to 6 CFU/culture bottle), the mean times to detection furnished by the BACTEC system at the end of the incubation period were ranged from 25.59 to 126.49 hours. No growth was signaled by the system when unseeded blood cultures were used as negative controls. On the other hand, the instrument signaled three positive results from the seeded blood cultures that corresponded to a bacterial contamination with Staphylococcus epidermidis.
Table 2.
Incubation Times of BACTEC Blood Cultures to Detect Candida Species
| Number of blood cultures | CFU/blood culture | P* | TTP (h) | |
|---|---|---|---|---|
| C. albicans | 36 | 2.20 (1–3)† | 0.89 (0.63–0.95) | 36.75 (36.40–37.50)‡ |
| C. tropicalis | 34 | 2.67 (2–4) | 0.93 (0.86–0.98) | 27.29 (26.49–28.24) |
| C. glabrata | 25 | 5.67 (5–6) | 1.00 (0.99–1.00) | 126.49 (124.11–131.16) |
| C. parapsilosis | 27 | 3.67 (3–4) | 0.97 (0.95–0.98) | 53.82 (53.13–54.60) |
| C. krusei | 31 | 1.67 (1–2) | 0.81 (0.63–0.86) | 25.59 (25.36–25.87) |
| Mean ± SD | 3.18 ± 1.58 | 0.89 ± 0.07 | 53.99 ± 42.05 |
Probability of having at least one CFU per blood culture bottle.
Three to five replicate 100 μl counts were used for enumeration.
Three to four positive culture bottles were used for calculation.
Range values are in parentheses.
Effect of BSA on PCR
The TaqMan-based PCR assay was carried out as previously described.16 In the absence of any DNA dilution or amplification facilitators, blood culture fluids demonstrated significant inhibition of the PCR assay of tested Candida species and the average rate of positivity was only 62% (Table 3). Moderate DNA dilutions (10-and 100-fold) with water before PCR improve appreciably the DNA amplification (76 and 71%, respectively). When DNA extracts were further diluted at 1:1000, the number of positive samples dramatically decreased to 48%. Addition of BSA to PCR reaction mixtures significantly relieves inhibition of amplification. The optimum concentration of BSA for relief of inhibition was 0.5%; the average rate of positivity was 88%. On the other hand, inclusion of this protein for amplification of different dilutions of DNA extracts did not maintain the sensitivity of PCR as was assessed when the running extracts were not diluted, particularly at much higher dilutions. Of note, higher concentrations of alkali-wash solution used principally to lyse and wash out the human erythrocyte and leukocyte components from blood culture fluid samples before DNA extraction were not sufficient to remove potential inhibitors of PCR (not shown). No signal was obtained in the TaqMan-based PCR assay when genomic DNA extracted from unseeded blood cultures was used as template either in the absence and presence of BSA.
Table 3.
Effect of 0.5% BSA on PCR Amplification of DNA Extracts from Blood Culture Samples
| At dilution of | Number of PCR-positive samples (%)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| In the absence of BSA
|
In the presence of BSA
|
|||||||
| Undiluted | 1:10 | 1:100 | 1:1000 | Undiluted | 1:10 | 1:100 | 1:1000 | |
| C. albicans | 23 (64) | 32 (89) | 28 (78) | 27 (75) | 32 (89) | 32 (89) | 31 (86) | 23 (64) |
| C. tropicalis | 23 (68) | 30 (88) | 25 (74) | 18 (53) | 31 (91) | 29 (85) | 25 (74) | 20 (59) |
| C. glabrata | 13 (54) | 15 (60) | 17 (68) | 12 (48) | 19 (76) | 18 (72) | 18 (72) | 13 (52) |
| C. parapsilosis | 18 (67) | 20 (74) | 20 (74) | 13 (48) | 25 (93) | 25 (93) | 23 (85) | 14 (52) |
| C. krusei | 18 (58) | 19 (61) | 18 (58) | 04 (13) | 28 (90) | 28 (90) | 24 (77) | 07 (23) |
| Total | 95 (62) | 116 (76) | 108 (71) | 74 (48) | 135 (88) | 132 (86) | 121 (79) | 77 (50) |
Minimum Incubation TATs of BACTEC Blood Cultures to Detect Candida Species by Using TaqMan-Based PCR Assay
To determine the minimum culture TATs to PCR-positive, blood cultures were incubated in the BACTEC instrument for increasing times. After each interval of 1 hour, aliquots were subjected to DNA extraction and submitted to PCR detection in the absence and presence of 0.5% BSA. As shown in Table 4, the TaqMan-based PCR assay, using specific probes for the five Candida species, was positive on blood culture aliquots collected from 95 (when PCR was performed in the absence of BSA) or 135 (when PCR was performed in the presence of BSA) of 153 simulated bottles. On average, decreasing the BACTEC incubation time to 3.11 hours resulted on a faster TAT to detect Candida species. Addition of BSA to PCR reaction mixtures improves this detection (1.84 ± 0.41 hours). The differences between the TTPs (Table 2) and the BACTEC minimum incubation TATs to detect a PCR-positive signal in the presence of BSA were roughly more than 24 hours for all Candida species tested: C. albicans, 35.00 hours; C. tropicalis, 24.79 hours; C. glabrata, 124.99 hours; C. parapsilosis, 52.07 hours; and C. krusei, 24.09 hours.
Table 4.
Minimum Incubation TATs of BACTEC Blood Cultures to Detect Candida Species from Undiluted DNA Extracts by Using TaqMan-Based PCR Assay
| Number of PCR-positive (%) blood cultures
|
BACTEC incubation TATs (h) for PCR detection
|
|||
|---|---|---|---|---|
| Minus BSA | Plus BSA | Minus BSA | Plus BSA | |
| C. albicans | 23 (64) | 32 (89) | 2.50 (2–3)* | 1.75 (1–2)* |
| C. tropicalis | 23 (68) | 31 (91) | 2.75 (2–4) | 2.50 (2–3) |
| C. glabrata | 13 (54) | 19 (76) | 5.50 (5–6) | 1.50 (1–2) |
| C. parapsilosis | 18 (67) | 25 (93) | 3.00 (2–4) | 1.75 (1–2) |
| C. krusei | 18 (58) | 28 (90) | 2.75 (2–4) | 1.50 (1–2) |
| Total | 95 (62) | 135 (88) | 3.11 ± 1.24 | 1.84 ± 0.41 |
Minimum culture times values obtained in two independent experiments to detect a PCR-positive signal.
The monitoring of Candida species multiplication in blood culture bottles was also evaluated with the TaqMan-based PCR assay. As expected, with blood culture aliquots collected at each interval of 1 hour, the fungal burden steadily increased with longer time following incubation, reaching a peak which correlated with the onset of BACTEC alarm signal indicating positivity. For instance, Figure 2 shows the C. albicans load in blood cultures when measured at increasing incubation times following infection. C. albicans load increased following an exponential way to reach a peak of 250,100 ± 32,400 CFU/blood culture bottle at hour 31, when the alarm signal is set off.
Figure 2.
Determination of C. albicans load in blood culture fluids inoculated with a low colony count. Culture bottles inoculated with 3.67 CFU of exponential-phase C. albicans were placed in the BACTEC system for increasing times or until an alarm signal indicated positivity. At each interval of 1 hour, an aliquot was withdrawn, DNA extracted and submitted to PCR amplification in the presence of 0.5% BSA. The mean value of the CFU values of two independent experiments was calculated. Each point in each experiment was tested in duplicate. The detection time indicating positivity was determined to be 31 hours by the BACTEC system.
Discussion
Blood culture systems remain the cornerstone in the detection of microbiological agents responsible for bloodstream fungal infections. In many instances, these systems failed to provide a positive result. Even in autopsy-verified candidiasis, negative outcome of blood cultures is reportedly as high as 56%.21 Kelly et al, 22 in a study comparing BACTEC resin bottles with lysis centrifugation, found that the first method detected 67% (20 of 30) of positive blood cultures for Candida spp. and the second 70% (21 of 30). Moreover, with highly automated blood culture systems, 5 to 7 days were required to recover all clinically relevant yeasts. However, with this longer incubation time, more microorganisms considered to be skin contaminants may be isolated and final results of negative blood cultures will be more delayed.23 The availability of blood culture material which exists in most clinical microbiological laboratories offers the opportunity to make a molecular diagnosis concurrently to the culture methods. The sensitivity of detection of a positive result may thus be improved and the identification of the infecting microorganisms may be made earlier.
However, investigators have demonstrated the presence of PCR-inhibitory substances in both blood culture aerobic and anaerobic bottles.19,24 Hemoglobin, lactoferrin, and sodium polyanetholesulfonate were found to be major PCR inhibitors in erythrocytes, leukocytes, and blood culture media, respectively.24,25 To deal with this problem, various additives and proteins facilitating PCR, such as BSA, have been widely included to the PCR mixture for relieving interference in amplification.25,26,27 In the present study, we evaluated DNA extracted from blood culture samples and found that it will contain varying amounts of PCR-inhibiting factors. In fact, PCR inhibition was not fully dilutable, and is presumably caused by compounds that tightly bind to the DNA-like hemoglobin degradation products.25 Moreover, it should be noted that level of DNA target in some samples was too low to detect after diluting to reduce inhibition. Even when the BSA was used, TaqMan-based PCR assay sensitivity performed on blood culture material was not 100%. Thus, factors other than inhibition must have caused negative results. However, we showed that blood culture aliquots collected at very early incubation times during the culture-negative period remained also PCR-negative. This could be explained by the much lower fungemia levels used for simulation which have been “missed” by the PCR assay. Sample-to-sample variation due to low numbers of viable organisms was also a source of these negative results.
The efficacy of the TaqMan-based PCR assay as an adjunct to the BACTEC blood culture system to shorten the time of detection of low candidemias was assessed on the most frequently encountered Candida species (C. albicans, C. tropicalis, C. glabrata, C. parapsilosis, and C. krusei) that represent about 94% of the total yeasts recovered from blood cultures.2,7,28 Our data indicate that, under our experimental conditions, almost all tested yeast species were detected more than 24 hours earlier with the PCR assay (performed on 2.50 to 5.50 hours-blood culture aliquots) than with the BACTEC system (TTP range, 25.59 to 126.49 hours). From the clinical point of view, this study shows that, on average, if a blood culture bottle is inoculated with a fungal suspension of 3.18 ± 1.58 CFU, there is a probability of 89 ± 7% to detect a positive result by using the TaqMan-based PCR assay following an incubation of 3.11 ± 1.24 hours in the BACTEC system. Addition of BSA to PCR reaction mixtures improves this detection (1.84 ± 0.41 hours). Hence, the combination of DNA “amplification” in the culture bottles by normal growth with an additional DNA amplification by PCR speeds up the detection of low fungal concentrations.
In a recent study, Selvarangan et al29 reported the development of a real-time PCR-based method for the rapid identification (≤ 2 hours) of most common invasive Candida species. However, the authors waited to have positive fungal blood culture bottles before PCR identification, therefore losing the real-time advantage for identification. By combining DNA “amplification” in the culture bottles with a real-time PCR-based assay, the speed with which we started to detect candidal species in simulated cultures was remarkable compared to that for the BACTEC system. This rapidity of detection is of major importance especially when identification of the Candida species was simultaneously made. This information could improve the survival of patients at risk for invasive fungal infections by allowing the selection of appropriate treatment while the fungal concentration is still low.
In summary, this study demonstrated that the PCR-enhanced culture method, using a simulated candidemia model, is effective for the recovery of most Candida species. Performing the TaqMan-based PCR assay after an average incubation time of 3.11 hours decreased the total TAT. The critical time of 3.11 hours of BACTEC incubation may allow at least two runs per day on carefully selected samples from high-risk patients. In practice, blood cultures arriving up to 4 a.m. can be tested by PCR at 8 a.m. The second run could be performed during the day for blood cultures arriving later than 4 a.m. Shorter TAT allows early initiation of anti-fungal treatment, which is critical in reducing the high mortality in immunocompromised patients; particularly in neutropenic patients treated for cancer or lymphoproliferative disorders, in patients suffering from infectious complications after serious surgery, and in burn and neonatal intensive care patients. However, because of its relatively high cost of screening, the TaqMan-based PCR method performed on blood culture bottles after 3 to 4 hours of incubation did not seem to be justifiable for all patients. Nevertheless, as we suggested, this approach can be mainly dedicated for analysis of highly selected blood cultures from immunocompromised and postoperative patients most at risk of developing fungal infections. An automated system for the fungal DNA extraction (ie, the MagNA Pure LC Instrument, Roche Diagnostics) able to recover minute amounts of DNA in a rapid and efficient manner30 would be useful for accelerating the analysis. A prospective clinical study to validate our findings is underway. An all Candida genus probe10 would be useful for verification of the presence of the main clinically significant Candida species included in the present study.
Acknowledgments
We thank Dr. H. Rodriguez-Villalobos for helpful suggestions. We are also grateful for assistance provided by A.-M. Bogaerts-Bourguignon.
References
- Weinstein MP, Towns ML, Quartey SM, Mirrett S, Reimer LG, Parmigiani G, Reller LB. The clinical significance of positive blood cultures in the 1990s: a prospective comprehensive evaluation of microbiology, epidemiology, and outcome of bacteremia and fungemia in adults. Clin Infect Dis. 1997;24:584–602. doi: 10.1093/clind/24.4.584. [DOI] [PubMed] [Google Scholar]
- Nguyen MH, Peacock JE, Morris AJ, Tanner DC, Nguyen ML, Snydman DR, Wagener MM, Rinaldi MG, Yu VL. The changing face of candidemia: emergence of non-Candida albicans species and anti-fungal resistance. Am J Med. 1996;100:617–623. doi: 10.1016/s0002-9343(95)00010-0. [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Jones RN, Messer SA, Edmond MB, Wenzel RP. National surveillance of nosocomial blood stream infection due to species of Candida other than Candida albicans: frequency of occurrence and anti-fungal susceptibility in the SCOPE program. Diagn Microbiol Infect Dis. 1998;30:121–129. doi: 10.1016/s0732-8893(97)00240-x. [DOI] [PubMed] [Google Scholar]
- Banerjee SN, Emori TG, Culver DH, Gaynes RP, Jarvis WR, Horan T, Edwards JR, Tolson J, Martone WJ, the National Nosocomial Infections Surveillance System Secular trends in nosocomial primary bloodstream infections in the United States, 1980–1990. Am J Med. 1991;91(Suppl 3B):86B–89B. doi: 10.1016/0002-9343(91)90349-3. [DOI] [PubMed] [Google Scholar]
- Pittet D, Li N, Woolson RF, Wenzel RP. Microbiological factors influencing the outcome of nosocomial bloodstream infections: a 6-year validated, population-based model. Clin Infect Dis. 1997;24:1068–1078. doi: 10.1086/513640. [DOI] [PubMed] [Google Scholar]
- Beck-Sague C, Jarvis WR. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980–1990: National Nosocomial Infections Surveillance System. J Infect Dis. 1993;167:1247–1251. doi: 10.1093/infdis/167.5.1247. [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Messer SA, Hollis RJ, Jones RN, Doern GV, Brandt ME, Hajjeh RA. Trends in species distribution and susceptibility to fluconazole among blood stream isolates of Candida species in the United States. Mycology. 1999;33:217–222. doi: 10.1016/s0732-8893(98)00160-6. [DOI] [PubMed] [Google Scholar]
- Reimer LG, Wilson ML, Weinstein MP. Update on detection of bacteremia and fungemia. Clin Microbiol Rev. 1997;10:444–465. doi: 10.1128/cmr.10.3.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren NG, Hazen KC. Manual of Clinical Microbiology. Candida, Cryptococcus, and Yeasts of Medical Importance. Murray PR, Barton EJ, Pfaller MA, Tenover FC, Yolken RH, editors. Washington, DC: American Society of Microbiology; 1995:723–737. [Google Scholar]
- Shin JH, Nolte FS, Holloway BP, Morrison CJ. Rapid identification of up to three Candida species in a single reaction tube by a 5′ exonuclease assay using fluorescent DNA probes. J Clin Microbiol. 1999;37:165–170. doi: 10.1128/jcm.37.1.165-170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turenne CY, Sanche SE, Hoban DJ, Karlowsky JA, Kabani AM. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J Clin Microbiol. 1999;37:1846–1851. doi: 10.1128/jcm.37.6.1846-1851.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einsele H, Hebart H, Roller G, Löeffler J, Rothenhöfer I, Muller CA, Bowden A, van Burik JA, Engelhard D, Kanz L, Schumacher U. Detection and identification of fungal pathogens in blood by using molecular probes. J Clin Microbiol. 1997;35:1353–1360. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand KH, Houck H, Wolff M. Detection of candidemia by polymerase chain reaction. Mol Cell Probes. 1994;8:215–221. doi: 10.1006/mcpr.1994.1030. [DOI] [PubMed] [Google Scholar]
- Shin JH, Nolte FS, Morrison CJ. Rapid identification of Candida species in blood cultures by a clinically useful PCR method. J Clin Microbiol. 1997;35:1454–1459. doi: 10.1128/jcm.35.6.1454-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löeffler J, Henke N, Hebart H, Schmidt K, Hagmeyer L, Schumacher U, Einsele H. Quantification of fungal DNA by using fluorescence resonance energy transfer and the Light Cycler system. J Clin Microbiol. 2000;38:586–590. doi: 10.1128/jcm.38.2.586-590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaroufi Y, Heymans C, De Bruyne JM, Duchateau V, Rodriguez-Villalobos H, Aoun M, Crokaert F. Rapid detection of Candida albicans in clinical blood samples by using a TaqMan-based PCR assay. J Clin Microbiol. 2003;41:3293–3298. doi: 10.1128/JCM.41.7.3293-3298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg HD. Washington DC: American Society of Microbiologists; Clinical Microbiology Procedures HandbookYeast Identification Using Morphology, 6.9.1. (ed 1.) 1992:399. [Google Scholar]
- Jorgensen JH, Mirrett S, McDonald LC, Murray PR, Weinstein MP, Fune J, Trippy CW, Materson M, Reller LB. Controlled clinical laboratory comparison of BACTEC Plus aerobic/F resin medium with BacT/Alert aerobic FAN medium for detection of bacteremia and fungemia. J Clin Microbiol. 1997;35:53–58. doi: 10.1128/jcm.35.1.53-58.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulski JK, Pryce T. Preparation of mycobacterial DNA from blood culture fluids by simple alkali wash and heat lysis method for PCR detection. J Clin Microbiol. 1996;34:1985–1991. doi: 10.1128/jcm.34.8.1985-1991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar BC, Jiru X, Moore JE, Earle JAP. A simple and sensitive method to extract bacterial, yeast, and fungal DNA from blood culture material. J Microbiol Methods. 2000;42:139–147. doi: 10.1016/s0167-7012(00)00174-3. [DOI] [PubMed] [Google Scholar]
- de Repentigny L, Reiss E. Current trends in immunodiagnosis of candidiasis and aspergillosis. Rev Infect Dis. 1984;6:301–312. doi: 10.1093/clinids/6.3.301. [DOI] [PubMed] [Google Scholar]
- Kelly MT, Roberts FJ, Henry D, Geere I, Smith JA. Clinical comparison of Isolator and BACTEC 660 resin media for blood culture. J Clin Microbiol. 1990;28:1925–1927. doi: 10.1128/jcm.28.9.1925-1927.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisner B, Woods GL. Times to detection of bacteria and yeast in BACTEC 9240 blood culture bottles. J Clin Microbiol. 1999;37:2024–2026. doi: 10.1128/jcm.37.6.2024-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredricks DN, Relman DA. Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate. J Clin Microbiol. 1998;36:2810–2816. doi: 10.1128/jcm.36.10.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu Al-Soud W, Rådström P. Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol. 2001;39:485–493. doi: 10.1128/JCM.39.2.485-493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreader CA. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol. 1996;62:1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeke T, Adams RP. The effects of plant polysaccharides and buffer additives on PCR. BioTechniques. 1992;12:332–334. [PubMed] [Google Scholar]
- Rex JH, Bennett JE, Sugar AM, Pappas PGC, van der Horst M, Edwards JE, Washburn RG, Scheld WM, Karchmer AW, Dine AP, Levenstein MJ, Webb CD. A randomized trial comparing fluconazole with amphotericin B for the treatment of candidemia in patients without neutropenia. N Engl J Med. 1994;331:1325–1330. doi: 10.1056/NEJM199411173312001. [DOI] [PubMed] [Google Scholar]
- Selvarangan R, Bui U, Limaye A, Cookson B: Real-time identification of commonly encountered Candida species from blood culture bottles. 42nd Interscience Conference on Antimicrobial Agents and Chemotherapy, San Diego, CA, 2002, Abstract number M-914 [Google Scholar]
- Löeffler J, Schmidt K, Hebart H, Schumacher U, Einsele H. Automated extraction of genomic DNA from medically important yeasts species and filamentous fungi by using the MagNA Pure LC system. J Clin Microbiol. 2002;40:2240–2243. doi: 10.1128/JCM.40.6.2240-2243.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]