Abstract
Many tumors have large homozygous deletions of the CDKN2A locus (encoding p14ARF and p16) and of CDKN2B (p15). Our aim was to determine which gene is the major target in bladder cancer. We used quantitative real-time PCR (RTQ-PCR) to determine copy number of p15, of p14ARF exon 1β, and p16 exon 2 in 22 tumor cell lines and 83 bladder tumors, some of which had been assessed previously by duplex PCR. Titration experiments showed that homozygous deletion could be detected in the presence of up to 30% normal DNA. Results for cell lines were compatible with previous cytogenetic analyses. Ten cell lines and 32 tumors (38.5%) had homozygous deletion of at least one target. Thirteen tumors (15.7%) had deletion of all three targets. Two tumors had deletion of p14ARF exon 1β alone and four of p16 exon 2 alone. RTQ-PCR detected more homozygous deletions than duplex PCR. Finally we used a multiplex ligation-dependent probe amplification kit to provide independent confirmation of results. We conclude that with appropriate controls RTQ-PCR is a sensitive and robust method to detect copy number changes in tumors even in the presence of contaminating normal cell DNA.
More than 60% of transitional cell carcinomas (TCC) of the bladder of all stages and grades show loss of heterozygosity (LOH) of chromosome 9.1,2,3,4,5,6 Although many tumors have LOH on both arms of the chromosome, a significant number (∼10% of tumors overall) have a region of interstitial LOH of 9p.7 Many such deletions are small and this has allowed the identification of a critical region at 9p21 that contains CDKN2B and CDKN2A/ARF. These two loci encode three tumor suppressor genes, p15, p16, and p14ARF, all of which are cyclin-dependent kinase inhibitors with key functions in cell cycle regulation.8,9
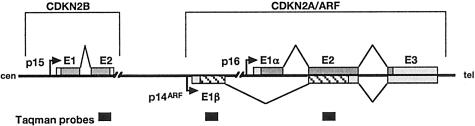
The CDKN2A locus encodes p16 and p14ARF, both of which are capable of inducing cell cycle arrest.10,11 p16 and p14ARF have different first exons (1α and 1β, respectively) approximately 13 kb apart,12 giving rise to products in alternate reading frames with no homology at the protein level13,14 (Figure 1). Exons 1α, 2, and 3 encode p16, which induces G1 cell cycle arrest via the Rb pathway. Exons 1β, 2, and 3 encode p14ARF, which inhibits p53 degradation via binding to mdm2.
Figure 1.
Genomic organization of the CDKN2A locus showing positions of the Taqman probes.
The mechanism of CDKN2A/ARF inactivation in human cancers is somewhat tumor specific. Homozygous deletions at CDKN2A/9p21 are common in bladder tumors6,15,16,17 and have been described in a variety of other sporadic tumors, including melanomas18 and gliomas.19 Pancreatic adenocarcinomas show inactivation of CDKN2A by either homozygous deletion or point mutation,20 whereas esophageal tumors commonly show inactivation by point mutation.21 A third mechanism of inactivation, transcriptional silencing by promoter hypermethylation, is commonly found in colorectal carcinoma.22 Point mutations have only rarely been identified in bladder tumors15,23,24 and promoter methylation, only infrequently.25
Since the discovery of p14ARF,14 much interest has centered on the relative involvement of p16 and p14ARF as tumor suppressor genes in both mouse models and in human cancer. The tumor suppressor function of p16 and p14ARF (p19 in the mouse) has been confirmed by many studies including mouse knockout experiments,26,27,28,29 experiments on cells derived from such knockouts eg,30 and gene transfer studies in cultured cells eg31. Introduction of wild-type p16 into bladder tumor cell lines lacking functional p16, induced cell cycle arrest and suppression of tumorigenicity.32 Further evidence for a suppressor function of p16 in the urothelium has come from studies of cultured primary bladder tumor biopsies where the majority of superficial papillary tumors showed limited in vitro lifespan, which was associated with elevated levels of p16 expression at senescence. In contrast, cultures derived from muscle invasive tumors commonly bypassed senescence and this was associated with loss of either p16 or pRb expression.33
The majority of human tumors with homozygous deletion of this genomic region have large deletions of the whole of the CDKN2A/ARF locus often including the gene encoding p15 (CDKN2B). Homozygous deletion may represent an efficient way of inactivating two or more closely linked genes. It has been suggested that in tumors such as TCC where large homozygous deletions are found and only a small proportion of tumors show inactivation by point mutation or methylation of 1α and 1β promoters, more than one gene in the region may be functionally important.
Few studies have systematically searched for homozygous deletions targeting only p15 or exon 1β of p14ARF in tumor samples. We have previously identified two bladder tumor cell lines in which p16 exon 2 is deleted but a marker R2.734 now known to be between p16 and CDKN2A exon 1β is retained, suggesting that exon 1β is also retained.15 A germline deletion of p14ARF but not CDKN2A exon 1α has been reported in a melanoma-neural system syndrome family.35 Similarly, deletion of exon 1β has been detected in a sporadic melanoma patient36 in two melanoma cell lines37 and three acute lymphocytic leukemia samples.38
The rare detection of p14ARF-specific deletions in tumor samples could be due to technical difficulties, since homozygous deletions are difficult to detect by conventional PCR due to the presence of contaminating normal cells.6,15,24 Real-time quantitative PCR (RTQ-PCR) potentially offers a more accurate approach to measure gene dosage in tumor samples. This technique has been successfully used to measure DNA deletion and amplification at microsatellite loci39 and was recently applied to a study of the 9p21 region in 105 primary gliomas, where no selective deletion of p14ARF was found.40
The relative frequency of homozygous deletion of CDKN2A/ARF and CDKN2B has not previously been examined in bladder cancer. Our aim was to determine whether p14ARF or p16 is the major deletion target by using RTQ-PCR to map homozygous deletion boundaries in tumor samples. We have analyzed the copy number of p15, p14ARF, and p16 exon 2 (an exon common to both p16 and p14ARF) relative to two reference genes (ALDOB on 9q and PFKL on 21q) in bladder cancer cell lines and primary TCC samples, many of which have been previously analyzed by conventional duplex PCR.15 We found RTQ-PCR to be superior to standard multiplex PCR for detecting homozygous deletions in tumor samples. Cell lines with known homozygous deletions and known copy numbers of 9p and 9q gave the expected gene dosage ratios. Thirty-eight percent of tumors had a homozygous deletion of one or more genes, many of which encompassed all three genes. A small number of tumors had homozygous deletion of p16 exon 2 or p14ARF only, indicating that, at least in some cases, only one of these two genes may be involved. Some tumors without 9p21 homozygous deletion nevertheless showed under-representation of 9p relative to 9q. However, under-representation of 9q relative to 9p was much more common. To differentiate clearly between the DNA targets assessed, throughout the text we refer to CDKN2A exon 2 as p16 exon 2, CDKN2A exon 1β as p14ARF, and CDKN2B as p15.
Materials and Methods
Tissues
Primary bladder tumor samples were obtained at cystoscopy and immediately frozen in liquid nitrogen or at −20°C.Tumors were graded according to World Health Organization (WHO) recommendations41 and staged according to the Tumor Nodes Metastasis (T.N.M.) classification.42 A venous blood sample was obtained from each patient in EDTA tubes as a source of constitutional DNA. High molecular weight DNA was prepared from tissue samples and blood leukocytes as described previously.43 The LOH status and an assessment of CDKN2A status for some of the tumors has been reported previously.7,15 To compare the sensitivity of real-time quantitative PCR with the duplex PCR method used previously on some of this tumor series,15 we used the same DNA samples and did not microdissect the tumors. All of these tumors were previously assessed for LOH at multiple loci and judged to contain no more than 30% contaminating normal cell DNA.
Cell Lines
Twenty-two bladder tumor-derived cell lines were used: RT4, RT112, T24, SD, JO’N, 5637, 253J, J82, UMUC3, VM-CUB-II, A1698, DSH1, SW1710, HT1376, 97–7, 97–18, 97–24, 96–1, 92–1, 97–6, 97–29, and 97–21. The latter eight cell lines were kindly provided by Dr. C. Reznikoff.33 DNA was prepared using standard phenol:chloroform extraction and ethanol precipitation.
Real-Time Quantitative PCR
The Taqman PCR reaction includes a dual-labeled fluorogenic probe (5′ reporter dye and 3′ quencher dye), which anneals to the template between the forward and reverse PCR primers. When the probe is intact the reporter dye is quenched and no fluorescence from the reporter is detected. Once polymerization from the 5′ end proceeds, the newly synthesized DNA displaces the quencher and the reporter is cleaved and can fluoresce. The amount of fluorescence detected is directly proportional to the amount of DNA synthesized.44,45,46,47
Taqman probes and primers were designed using Primer Express software (Applied Biosystems, Warrington, U.K.) and were optimized according to the manufacturer’s guidelines. Target gene probes (p15, p14ARF, and p16 exon 2) and reference gene probes [aldolase B, ALDOB (9q22.3) and liver phosphofructokinase, PFKL (21q22.3)] contained a TAMRA dye at the 3′ end and FAM as reporter dye at the 5′ end. Primer and probe sequences are shown in Table 1.
Table 1.
Primers and Probes
| Gene | Primer/probe sequence |
|---|---|
| p15 | forward primer: CCCTCGACACTCACCATGAA |
| reverse primer CGACCCCTGGAATGTCACAC | |
| probe AAATATCCCTGGAAATCCGCTTCTCTGTGTT-FAM | |
| p14ARF | forward primer TGATGCTACTGAGGAGCCAGC |
| reverse primer AGGGCCTTTCCTACCTGGTC | |
| probe TCTAGGGCAGCAGCCGCTTCCTAGA-FAM | |
| p16 exon 2 | forward primer TTTCCGTCATGCCGGC |
| reverse primer TCATCATGACCTGCCAGAGAGA | |
| probe CCCACCCTGGCTCTGACCATTCTG-FAM | |
| ALDOB | forward primer TGACAGGAAAGCCCTGGC |
| reverse primer TTCCCCATGGTACCTATGGTG | |
| probe CTCCTTATGCTGCCCTTGGCCCTC-FAM | |
| PFKL | forward primer GATGCTGGCACAATACCGC |
| reverse primer GGGTCACGTGCTCCAGCT | |
| probe TCAGTATGGCCGCCTACGTGTCAGG-FAM |
Monoplex real-time quantitative PCR was carried out on 83 TCCs and 23 TCC cell lines, using 10-ng template. Reactions were carried out in a total volume of 25 μl containing 1X Taqman master mix (ABI) and 10-ng DNA template. Primer and probe concentrations were optimized for each target according to the manufacturer’s instructions. The PCR program consisted of 50°C for 2 minutes and 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute.
A standard curve was included on each 96-well plate for reference and target genes, consisting of serially diluted normal diploid DNA (Sigma D-3035), where 1 ng DNA = 330 diploid gene copies. A no-template control was also included. In each run, all templates were assayed in triplicate and for the majority of samples, each run was repeated at least twice.
Data analysis was carried out using ABI Prism 7700 Sequence Detection Software which calculates threshold cycle numbers (Ct). The Ct value is the cycle at which the fluorescent signal from a sample crosses a threshold as it enters the exponential phase in the PCR reaction. A standard curve is formulated from the starting copy number (x-axis) against Ct (y-axis) value. The Ct values of unknown samples can then be used to determine copy number from the standard curve. Only standard curves with correlation coefficients of 0.98 and above were used. The gene dosage ratio was calculated as: average copy number of target gene ÷ average copy number of the reference gene. All gene dosage ratios were then normalized against the normal diploid control DNA to give a normalized gene dosage ratio (GDR).
Homozygous deletion was scored when a sample had a GDR less than or equal to 0.36. This allows for the presence of up to 30% contaminating normal cell DNA in the template. A GDR between 0.36 and 0.63 was interpreted as having under-representation of the test gene relative to the reference gene. A GDR between 0.64 and 1.36 was interpreted as having equal copy numbers of target and reference gene and any sample with a GDR above 1.36 was interpreted as over-representation relative to the reference gene.
Multiplex Ligation-Dependent Probe Amplification
Multiplex ligation-dependent probe amplification (MLPA) is a novel method designed to measure the relative copy number of many different DNA sequences in a single reaction.48 Probes are added to genomic DNA and amplification of these probes depends on the presence of target sequences in the sample. Each probe consists of two oligonucleotides that anneal to adjacent sites in the target DNA. Following ligation, only ligated sequences can be amplified in the subsequent PCR reaction. The end sequences of all probes are identical, allowing amplification with a single pair of primers. Probe sequences contain stuffer fragments of different lengths to generate a series of small products of unique size that can be resolved simultaneously on a sequencing gel and quantified.
MLPA was carried out using 50 ng of bladder tumor DNA and a blood control DNA, with the P024 kit (MRC-Holland, Amsterdam, The Netherlands). The kit contains 38 probes, 11 probes covering the CDKN2A/2B genes, three MTAP probes, and seven probes from the genomic region surrounding these genes (www.mrc-holland.com). 9p21 probes from cen-tel are: 1, TEK; 2, ELAVL2; 3, CDKN2B promoter/exon 1; 4, CDKN2B exon 1; 5, CDKN2B intron; 6, CDKN2A 0.5 kb upstream of arf; 7, CDKN2A/ARF exon 1; 8, between arf and CDKN2A exon 1; 9, between arf and CDKN2A exon 1 probe 2; 10, CDKN2A exon 1; 11, CDKN2A exon 2; 12, CDKN2A exon 3; 13, MTAP3; 14, MTAP2; 15, MTAP1; 16, KIAA; 17, IFNW1; 18, IFNB1; 19, MLLT3 2; 20, MLLT3 1; 21, FLJ00026.
Probe hybridization, ligation, and PCR reactions were carried out according to the manufacturer’s protocol. The PCR products were analyzed on an Applied Biosystems 3100 Genetic Analyzer using Genescan software. Dosage quotients were calculated as described by MRC-Holland48 using peak heights rather than peak areas.
Results
Assessment of the Sensitivity of RTQ-PCR
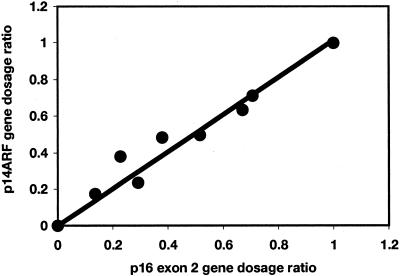
To test the sensitivity of the technique, RTQ-PCR was carried out on a series of templates containing control diploid DNA diluted with DNA from the bladder tumor cell line RT112. RT112 is diploid and known to have a homozygous deletion of all three genes at the CDKN2A locus. Dilutions (control:RT112) were 100:0, 70:30, 60:40, 50:50, 40:60, 30:70, 20:80, 10:90, and 0:100. A plot of p14ARF GDR against p16 exon 2 GDR relative to the control gene ALDOB gave a linear relationship and showed that the technique is sensitive enough to detect a homozygous deletion in the presence of 30% normal DNA contamination (Figure 2). Since it was known that the tumor DNA samples contain contaminating normal cell DNA derived from blood vessels and inflammatory cells (minimum tumor cell content, 70%) we used a cut-off value of 0.36 as the GDR below which we scored homozygous deletion. This represents 0.3 + 2 SD to allow for the measured variability of the assay. A GDR between 0.64 and 1.36 was scored as having equal copies of the target and reference gene and any GDR above 1.36 was scored as 9p over-representation relative to the reference gene(s).
Figure 2.
p14ARF and p16 gene dosage ratios relative to ALDOB for a DNA series containing 0 to 100% normal DNA and 0 to 100% DNA from a cell line with homozygous deletion of both targets.
9p21 Copy Number Changes in Bladder Tumor Cell Lines
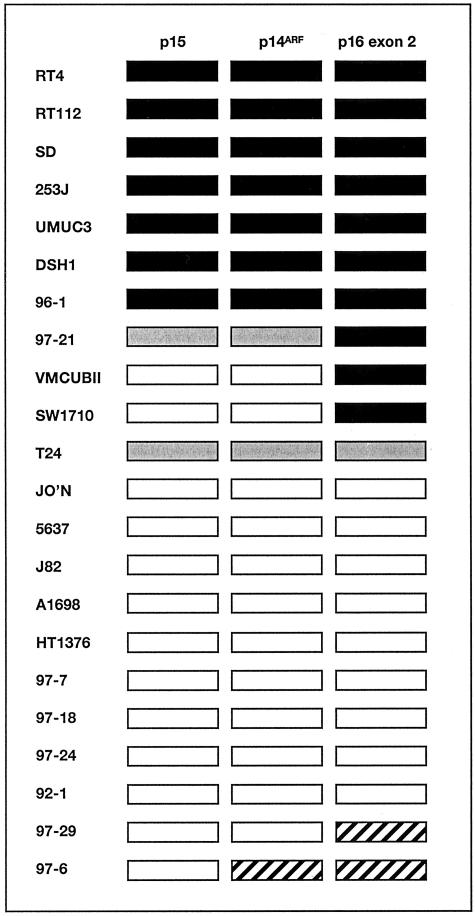
Initially we investigated the status of the three targets in a panel of bladder tumor-derived cell lines including some for which the 9p21 status had been studied in detail (15,49 and unpublished results) to assess the ability of the assay to detect both homozygous deletions and copy number changes relative to the control probe. For these analyses we used the 9q control, ALDOB to see differences in 9p and 9q representation and to compare these with known information. We have not previously characterized chromosome 21 where the PFKL gene maps in all of these cell lines so this control was not used in these validation experiments. As previously determined by conventional PCR and fluorescence in situ hybridization (FISH), RT4, RT112, SD, 253J, UMUC3, DSH1, and 96–1 have homozygous deletion of all three genes.15,33 VMCUB-II, SW1710, and 97–21 have homozygous deletion of p16 exon 2 only and retain p15 and p14ARF. These results were confirmed by RTQ-PCR (Figure 3). The cell line 97–21 showed homozygous deletion of p16 exon 2 and retained p14ARF and p15 but at lower copy number than the 9q reference gene ALDOB. This difference in relative copy number is not apparent by conventional PCR (our unpublished results). Similarly, T24 showed under-representation of all three targets relative to ALDOB as expected from previous karyotypic findings.49 Two cell lines, 97–6 and 97–29, showed over-representation of one or two 9p21 targets relative to ALDOB and the other 9p21 target(s) indicating a likely breakpoint at 9p21 with deletion extending into 9q and including ALDOB. The remaining cell lines had equal copies of 9p21 target to ALDOB (ie, a GDR of approximately 1.0), despite some cell lines being aneuploid. Two of these (J82 and 5637) have been studied extensively by FISH49 and 24-color FISH (Williams S, Adams J, Coulter J, Summersgill B, Shipley J, Knowles M, submitted) and contain only apparently normal chromosomes.
Figure 3.
Pattern of DNA copy number alterations in bladder tumor cell lines. Gene dosage ratios were calculated relative to ALDOB. ▪, homozygous deletion; ░⃞, under-representation of 9p21 gene; ▨, over-representation of 9p21 gene; □, no copy number change.
9p21 Copy Number Changes in Bladder Tumors
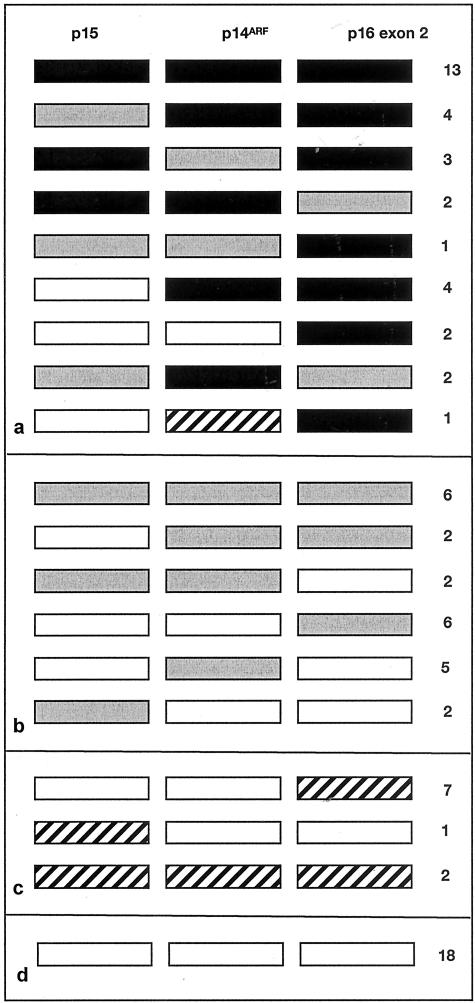
We then searched for homozygous deletions of 9p21 in 83 TCC samples. We used PFKL as control to detect 9p homozygous deletions with higher sensitivity than can be achieved using the 9q control gene that itself is commonly under-represented. PFKL maps to chromosome 21, a chromosome that is rarely altered numerically in TCC. Thirty-two of 83 tumors (38.5%) had apparent homozygous deletion of at least one gene and 13 of these (15.6%) had homozygous deletion of all three targets. Eight tumors (9.6%) had homozygous deletion of both p14ARF and p16 exon 2 but not of p15. Two tumors (2.4%) had homozygous deletion of p14ARF alone and five (6%) showed under-representation of p14ARF but not of p15 and p16 exon 2, indicating that this may represent an independent target for deletion. Similarly, four tumors (4.8%) had homozygous deletion of p16 exon 2 alone and six (7.2%) showed under-representation of p16 exon 2 but not of the other two targets. No tumors showed selective homozygous deletion of p15 though two showed selective under-representation. Three tumors showed apparent under-representation of p14ARF and homozygous deletion of the flanking p15 and p16 exon 2. These latter are considered likely to have equal copy number of all three targets as GDR values for p14ARF were just above the cut-off for scoring as homozygous deletion (GDRs 0.38, 0.40, and 0.42). The patterns of deletion for p16 exon 2 and p14ARF are illustrated in Figure 4. Information on tumor grade and stage was available for 79 and 61 tumors respectively. No associations with homozygous deletion were found by the χ2 test (P > 0.1).
Figure 4.
Pattern of DNA copy number changes in primary bladder tumors. Gene dosage ratios were calculated relative to PFKL. ▪, homozygous deletion; ░⃞, under-representation of 9p21 gene; ▨, over-representation of 9p21 gene; □, no copy number change. Patterns of deletion are shown for 32 tumors with homozygous deletion of one or more genes (a), 23 tumors with under-representation of one or more genes (b), 10 tumors with over-representation of one or more genes (c), 18 tumors with no copy number change (d).
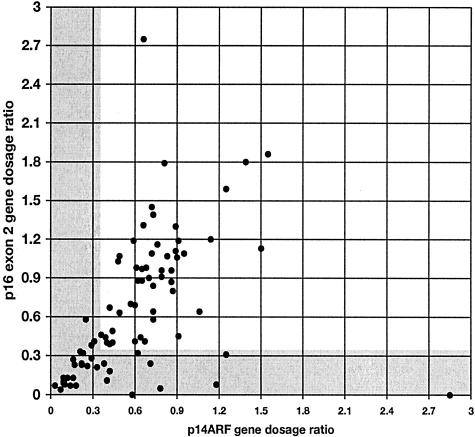
A scatter plot of GDR of p16 exon 2 versus p14ARF with PFKL as control is shown in Figure 5. Many samples yielded values that lay close to the line x = y indicating that p16 exon 2 and p14ARF copy number was equivalent. Values for some tumors lay above this line, indicating that these tumors had fewer copies of p14ARF than p16 exon 2 and similarly values below the line x = y indicate under-representation of p16 exon 2 relative to p14ARF. A population of tumors with values in the bottom left of the scatter plot (x and y ≤0.36) were those with homozygous deletions of both p14ARF and p16 exon 2.
Figure 5.
Relative gene dosage ratios for p16 exon 2 and p14ARF in 83 primary bladder tumors. Gray areas are those scored as homozygous deletion for each gene (ie, GDR ≤0.36). Values are all relative to PFKL control.
Subsequently, we assessed tumors for copy number of p14ARF, p16 exon 2, and p15 relative to the ALDOB control (9q). As expected, in light of the known frequent 9q copy number reduction in TCC, using a GDR of 0.36 as cut-off, we identified fewer apparent homozygous deletions. Nevertheless, 13 of the 83 tumors (15.6%) showed clear homozygous deletion of at least one target relative to the ALDOB control. Where only one or two of the targets showed homozygous deletion, the results were compatible with those obtained using PFKL as control. Eleven of the samples shown to have homozygous deletion relative to the chromosome 21 control had apparent under-representation of 9p relative to 9q when the ALDOB control was used but did not show sufficient reduction in GDR to be scored as homozygous deletion. Using the ALDOB control, a reduction in 9q copy number relative to 9p (9p:ALDOB GDR >1.3) was identified in 49 tumors (59%). An additional measure of 9q under-representation was obtained by comparing GDR for ALDOB and PFKL. This indicated that 9q was under-represented relative to 21q (GDR PFKL:ALDOB ≥1.36) in 70 of 83 of samples (84%) and over-represented (PFKL:ALDOB ≤0.63) in one sample (1.2%) which showed a consistent GDR of >100 for PFKL:ALDOB. These results suggest that 21 tumors had under-representation of both 9p and 9q relative to 21q, which may indicate loss of an entire chromosome (hemizygous loss).
Fifty-two of the samples had been assessed previously for deletions of p16 by duplex PCR.15 Of these, 13 had been scored as homozygous deletion, 9 as under-represented, and 30 as retained. More homozygous deletions were identified by RTQ-PCR. These included all six of the cases that had previously been confirmed by Southern blotting and 11 additional homozygous deletions that had previously been scored as under-represented or retained. Interestingly, three cases that had been previously assessed as homozygous deletion by duplex PCR were scored as under-representation rather than homozygous deletion by RTQ-PCR.
Confirmation of Deletions by Multiplex Ligation-Dependent Probe Amplification
Multiplex ligation-dependent probe amplification (MLPA) is a novel method for relative quantitation of multiple DNA sequences in a single reaction.48 After our RTQ-PCR study had been completed, an MLPA kit was developed to measure DNA copy number of 21 sequences from 9p21. To compare the efficiency of this new method and to provide independent confirmation of the deletions we had detected, we applied MLPA to a subset of 32 samples for which DNA was still available.
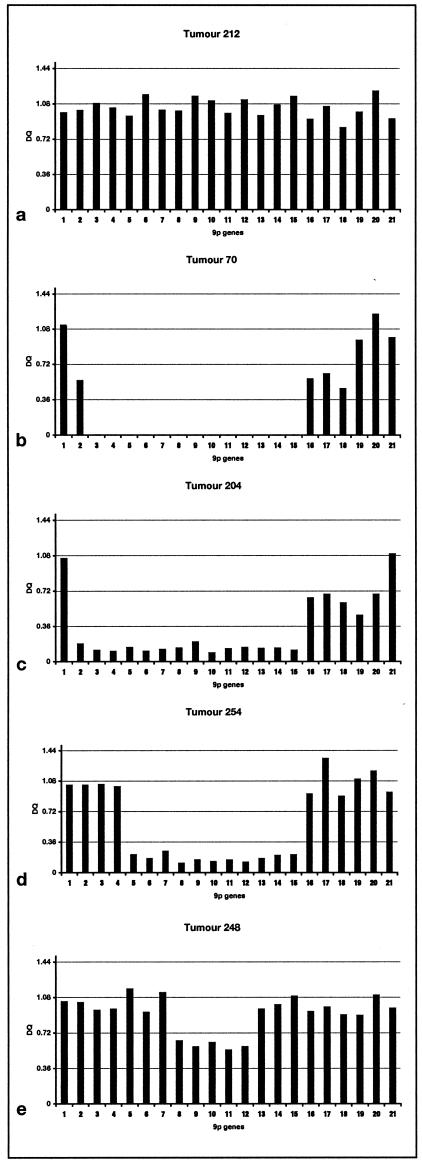
Figure 6 shows examples of the results. Homozygous deletions were detected with ease and the inclusion of multiple probes allowed breakpoints to be defined with high resolution. For example, the deletions in tumors 70 and 204 that encompass all three tumor suppressor gene targets have clearly defined and different centromeric breakpoints. Tumor 70 retains probe 1, has hemizygous deletion of probe 2 and homozygous deletion of probes 3 to 15 whereas the breakpoint on both alleles in tumor 204 lies between probes 1 and 2 (Figure 6, b and c). The results for many tumors including 70 and 204 indicated that homozygous deletions did not involve identical breakpoints on both alleles and that regions of reduction in copy number (which may represent hemizygosity) were commonly found flanking regions of homozygous deletion. The assay confirmed deletion of p14ARF and p16 exon 2 but not p15 in tumor 254 (Figure 6d). Interestingly, the deletion of p16 exon 2 alone in tumor 248 which had been scored as homozygous deletion by RTQ-PCR (GDR = 0.24) was detected as under-representation but not homozygous loss by MLPA (Figure 6e).
Figure 6.
MLPA results for 21 probes on 9p21 for 5 TCC samples. a: Tumor with no copy number change in region. b: Tumor with minimal normal DNA contamination showing clear homozygous deletion with no products for probes encompassing p15, p14ARF, and p16. c: Tumor with homozygous deletion of all three genes but distinct centromeric breakpoint. d: Tumor with homozygous deletion encompassing p14ARF and p16 but not p15. e: Tumor scored as having homozygous deletion of p16 exon 2 alone by RTQ-PCR using PFKL as control. MLPA result indicates under-representation rather than homozygous deletion but confirms small region of deletion involving p16 only.
Discussion
It is clear that deletions of 9p21 including CDKN2A/ARF and CDKN2B play an important role in the development of bladder tumors. Many studies have reported frequent LOH and homozygous deletion in the region as a clonal event eg,6,16,25,50,51,52 However, the relative importance of the three genes within the region of deletion has not been determined previously.
Assessment of homozygous deletion is technically difficult in tumor samples due to the presence of contaminating normal cell DNA. Various methods can be used to detect homozygous deletion including Southern blotting and FISH to tumor cells, and various PCR-based techniques. Southern blotting provides an accurate estimate of copy number but requires a large amount of DNA. FISH is relatively expensive and labor-intensive method for screening large numbers of clinical tissue samples, though in the case of bladder cancer FISH is ideal for examining cells in urine sediments. PCR-based methods are suitable for high-throughput analysis and easy to use but are particularly vulnerable to the problem of normal DNA contamination. Here we have successfully used RTQ-PCR to examine both the frequency of homozygous deletion on 9p21 in bladder tumors and to assess the relative deletion frequency of three targets within the region.
Our results indicate that the technique is robust but that attention to several key parameters is required to generate reliable data. It is desirable that minimal amounts of contaminating normal cell DNA are present and in the future this can be achieved using microdissection. In the present study we wished to compare RTQ-PCR with duplex PCR and for this reason used historical samples extracted from grossly dissected tissue. None contained more than 30% normal cell contamination. A recent study of p14ARF gene dosage in bladder tumors53 used samples with similar levels of contamination and defined “gray” zones in the data that were difficult to interpret and that most probably reflect the variable level of normal tissue contamination. We found a similar group of samples that gave results close to the threshold values we had defined and that would have benefited from microdissection to confirm results if tissue had been available. Alternatively, it would be possible to correct for the amount of contamination in individual samples and this has recently been described in a study of p16 deletion in childhood acute lymphoblastic leukemia.54
We used a cut-off GDR value of 0.36 to allow for a maximum of +/− 6% sample variation (calculated from the GDR variation of the normal control) and 30% normal cells in the tumor sample. Since RTQ-PCR is quantitative we can be confident that any tumor with a GDR of 0.36 or less almost certainly has a homozygous deletion. However, in tumor samples with most normal tissue contamination the effects of gross aneuploidy may prevent the detection of homozygous deletion. Similarly tumors with subclones of cells containing homozygous deletion are less likely to be detected in the presence of normal cell contamination.
Our results highlight the need for appropriate choice of controls. We used one control on 21q, a chromosome that is not reported to undergo frequent copy number changes in TCC. Thus the copy number of PFKL usually reflects the overall ploidy of the cell. This may not be the case for all tumors and in the future we recommend the use of at least two control probes derived from chromosomes that are not expected to show copy number alterations. We also used a control probe derived from 9q and our results demonstrated very clearly the loss of sensitivity for homozygous deletion detection when using a control derived from a region of common under-representation.
When the present results were compared with our previous results on 53 of the same tumors with duplex PCR,15 we found good concordance. More cases with homozygous deletion were detected with RTQ-PCR, reflecting the higher sensitivity of the assay and the use of objective scoring criteria. One surprising result was that three of the cases in which homozygous deletion had been scored in the previous study were scored as under-represented but not homozygously deleted by RTQ-PCR. The control primer pair used in the previous assays was enolase (ENO), which maps to 12p. One possible reason for this discrepancy in results is that these three tumors may have over-representation/amplification of this 12p gene, leading to the generation of false-positive results for p16 exon 2. Alternatively, it may reflect the relative insensitivity and lack of objective criteria for scoring the previous duplex PCR results.
We were able to test the novel technique MPLA on 32 samples for which DNA was available. This provided independent confirmation of the RTQ-PCR results and had the significant advantage that breakpoints could be mapped with much greater precision. We found the commercially available kit gave reproducible results and performed well in the same normal:homozygously deleted sample titration experiment that had been used to assess RTQ-PCR (Figure 2 and data not shown). A major advantage of this assay is that many controls are included in the kit, thus overcoming the problems discussed above. As illustrated in case 248 (Figure 6e) this led to an assessment of under-representation rather than homozygous deletion as had been scored by RTQ-PCR. The most likely reason for this is that the 21q control used in RTQ-PCR was over-represented in this sample. This sample was one of those with deletion of p16 exon 2 alone and although the level of loss was scored differently by the two techniques, the reduction in copy number of p16 exon 2 alone was clearly defined by MLPA. The ease of use of MLPA and the robust results obtained even in this series of samples with normal DNA contamination makes this a very attractive alternative choice of assay for future studies.
Our results present a clear picture of the patterns of homozygous deletion at 9p21. To date there has not been a study in which all three potential gene targets have been assessed simultaneously. The most common pattern of homozygous deletion in bladder tumors was loss of all three targets, or p14ARF co-deletion with p16 exon 2. p15 deletion was not found alone and the frequency was lower than for the other two genes leading to the suggestion that this target is not critical. p14ARF-specific homozygous deletion was found in 2 of 83 tumors and p14ARF + p15 homozygous deletion in a further two tumors. This has not been reported in TCC though some melanoma cell lines have been identified with exon-1β deletion alone37 and germline deletions have been recorded in cases of familial melanoma.35,55 No p14ARF-specific deletions were identified in TCC cell lines but the number studied was too small to ensure detection of rare events. To our knowledge, such deletions have not been reported in other adult sporadic tumors though there have been relatively few comprehensive homozygous deletion analyses reported to date. For example, using the same technique, no p14ARF-specific homozygous deletions were detected in gliomas40 although 41% of cases showed co-deletion of p14ARFand p16 exon 2. Our results suggest that p14ARF can act as an independent tumor suppressor gene in at least some cases of TCC. The importance of p14ARF is also suggested by our finding of under-representation of p14ARF relative to p15 and p16 exon 2 in five cases. As homozygous deletion of p16 exon 2 also affects p14, it is likely that this gene is at least as important a target as p16. To date, there have been no studies that have examined the p16-specific exon 1α and it will be important to examine this in the future in both bladder and other cancers. It will also be of great interest to examine the expression status of p16 in cases with retention of at least one copy of p16 exon 2 but homozygous deletion of p14ARF as retention of p16 expression in such cases would provide evidence that p14ARF has independent biological significance. This was not possible in the present study since RNA was not available from the tumors in question. In the future, it will also be desirable to confirm deletions by fluorescence in situ hybridization on fresh tissue samples or urine sediments from the same patient and to confirm small homozygous deletions by long-range PCR.
There is some functional evidence that loss of p16 function plays an important role in urothelial carcinogenesis. For example, gene replacement studies have shown a marked effect on TCC tumor cell phenotype.32 Studies of papillary superficial TCC in vitro have shown that many of these tumors retain at least one copy of p16 and that the cells derived from them have limited lifespan in culture with typical up-regulation of p16 protein at senescence. Loss of p16 or Rb expression is required for establishment of such tumor cells as immortal lines in vitro.33 This therefore argues for an important role in immortalization but not necessarily for initial papillary tumor formation in vivo. The role of p14ARF in human tumor development is not well elucidated though several biochemical functions of the protein have been described. In the mouse, the equivalent protein (p19 in mouse) has been shown to play a key role in cell senescence30 but current evidence in human cells suggests that p16 rather than p14 plays a more major role at senescence.56
There has been some debate regarding the relationship between LOH and homozygous deletion of CDKN2A/ARF and tumor phenotype. Several studies indicate that LOH of 9q is more frequent than LOH of 9p in superficial TCC57,58 and there is some evidence that LOH of 9p21 is associated with a more aggressive phenotype.59 Orlow et al50 reported an association of LOH/homozygous deletion with tumor grade but not stage and a significant association with tumor size and recurrence-free interval. Jung et al60 reported a similar association with recurrence but others have not61 and some studies suggest that LOH on 9q rather than 9p21 is associated with tumor recurrence.62,63 In the present study we found no association with tumor grade or stage. In light of the finding that large tumor size is associated with 9p21 alterations,50 it is worth noting that the 53 tumors from our previous series that were re-assessed here are those for which we still had DNA remaining and therefore represented larger tissue samples. These may therefore have a higher frequency of 9p21 alteration than a panel of unselected primary tumors matched for grade and stage. This may explain in part the relatively high frequency of homozygous deletion found (38%), though our comparison with previous results on the same samples shows that increased sensitivity of the assay accounts for many of the new homozygous deletions detected. Previous studies have recorded homozygous deletion frequencies ranging from 11% to 70%.15,50,64 A recent study that also used RTQ-PCR found only 14% of homozygous deletion of p14ARF. However this may be related to the low frequency of 9p21 LOH measured in the tumors studied (22%) which is lower than usually found in superficial TCC.53
In conclusion, we have developed a sensitive RTQ-PCR assay for three potential homozygous deletion targets at 9p21 and have shown that this provides improved detection rates for homozygous deletion over semi-quantitative multiplex PCR. The study has provided evidence that p16 and p14ARF are the major targets of deletion in TCC and that each may be targeted independently in some cases. It will now be important to apply the technique to a large series of microdissected tissue samples to determine the true overall frequency of homozygous deletion of these two genes in TCC.
Acknowledgments
We thank the patients and urology surgeons from the following hospitals in the West Midlands who provided tissues for this study: Burton General Hospital, Dudley Road Hospital, East Birmingham Hospital, Queen Elizabeth Hospital, Reddich Alexandra Hospital, Sandwell District General Hospital, Selly Oak Hospital, Solihull Hospital, Walsgrave Hospital, and Wolverhampton New Cross Hospital.
Footnotes
Supported by Cancer Research UK.
References
- Tsai YC, Nichols PW, Hiti AL, Williams Z, Skinner DG, Jones PA. Allelic losses of chromosomes 9, 11, and 17 in human bladder cancer. Cancer Res. 1990;50:44–47. [PubMed] [Google Scholar]
- Cairns P, Shaw ME, Knowles MA. Initiation of bladder cancer may involve deletion of a tumour-suppressor gene on chromosome 9. Oncogene. 1993;8:1083–1085. [PubMed] [Google Scholar]
- Cairns P, Shaw ME, Knowles MA. Preliminary mapping of the deleted region of chromosome 9 in bladder cancer. Cancer Res. 1993;53:1230–1232. [PubMed] [Google Scholar]
- Miyao N, Tsai YC, Lerner SP, Olumi AF, Spruck CH, III, Gonzalez-Zulueta M, Nichols PW, Skinner DG, Jones PA. Role of chromosome 9 in human bladder cancer. Cancer Res. 1993;53:4066–4070. [PubMed] [Google Scholar]
- Habuchi T, Ogawa O, Kakehi Y, Ogura K, Koshiba M, Hamazaki S, Takahashi R, Sugiyama T, Yoshida O. Accumulated allelic losses in the development of invasive urothelial cancer. Int J Cancer. 1993;53:579–584. doi: 10.1002/ijc.2910530409. [DOI] [PubMed] [Google Scholar]
- Devlin J, Keen AJ, Knowles MA. Homozygous deletion mapping at 9p21 in bladder carcinoma defines a critical region within 2cM of IFNA. Oncogene. 1994;9:2757–2760. [PubMed] [Google Scholar]
- Keen AJ, Knowles MA. Definition of two regions of deletion on chromosome 9 in carcinoma of the bladder. Oncogene. 1994;9:2083–2088. [PubMed] [Google Scholar]
- James MC, Peters G. Alternative product of the p16/CKDN2A locus connects the Rb and p53 tumor suppressors. Prog Cell Cycle Res. 2000;4:71–81. doi: 10.1007/978-1-4615-4253-7_7. [DOI] [PubMed] [Google Scholar]
- Orian A, Eisenman RN. TGF-beta flips the Myc switch. Sci STKE. 2001;2001:PE1. doi: 10.1126/stke.2001.88.pe1. [DOI] [PubMed] [Google Scholar]
- Haber DA. Splicing into senescence: the curious case of p16 and p19ARF. Cell. 1997;91:555–558. doi: 10.1016/s0092-8674(00)80441-9. [DOI] [PubMed] [Google Scholar]
- Lloyd AC. p53: only ARF the story. Nat Cell Biol. 2000;2:E48–E50. doi: 10.1038/35004078. [DOI] [PubMed] [Google Scholar]
- Stone S, Jiang P, Dayananth P, Tavtigian SV, Katcher H, Parry D, Peters G, Kamb A. Complex structure and regulation of the P16 (MTS1) locus. Cancer Res. 1995;55:2988–2994. [PubMed] [Google Scholar]
- Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Mao L, Merlo A, Bedi G, Shapiro GI, Edwards CD, Rollins BJ, Sidransky D. A novel p16INK4A transcript. Cancer Res. 1995;55:2995–2997. [PubMed] [Google Scholar]
- Williamson MP, Elder PA, Shaw ME, Devlin J, Knowles MA. p16 (CDKN2) is a major deletion target at 9p21 in bladder cancer. Hum Mol Genet. 1995;4:1569–1577. doi: 10.1093/hmg/4.9.1569. [DOI] [PubMed] [Google Scholar]
- Cairns P, Tokino K, Eby Y, Sidransky D. Homozygous deletions of 9p21 in primary human bladder tumors detected by comparative multiplex polymerase chain reaction. Cancer Res. 1994;54:1422–1424. [PubMed] [Google Scholar]
- Orlow I, Lacombe L, Hannon GJ, Serrano M, Pellicer I, Dalbagni G, Reuter VE, Zhang Z-F, Beach D, Cordon-Cardo C. Deletion of the p16 and p15 genes in human bladder tumors. J Natl Cancer Inst. 1995;87:1524–1529. doi: 10.1093/jnci/87.20.1524. [DOI] [PubMed] [Google Scholar]
- Fountain JW, Karayiorgou M, Ernstoff MS, Kirkwood JM, Vlock DR, Titus-Ernstoff L, Bouchard B, Vijayasaradhi S, Houghton AN, Lahti J, Kidd VJ, Housman DE, Dracopoli NC. Homozygous deletions within human chromosome band 9p21 in melanoma. Proc Natl Acad Sci USA. 1992;89:10557–10561. doi: 10.1073/pnas.89.21.10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James CD, He J, Collins VP, Allalunis-Turner MJ, Day RS., III Localization of chromosome 9p homozygous deletions in glioma cell lines with markers constituting a continuous linkage group. Cancer Res. 1993;53:3674–3676. [PubMed] [Google Scholar]
- Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, Seymour AB, Weinstein CL, Hruban RH, Yeo CJ, Kern SE. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- Zhou X, Tarmin L, Yin J, Jiang HY, Suzuki H, Rhyu MG, Abraham JM, Meltzer SJ. The MTS1 gene is frequently mutated in primary human esophageal tumors. Oncogene. 1994;9:3737–3741. [PubMed] [Google Scholar]
- Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- Spruck CH, III, Gonzalez-Zulueta M, Shibata A, Simoneau AR, Lin MF, Gonzales F, Tsai YC, Jones PA. p16 gene in uncultured tumours. Nature. 1994;370:183–184. doi: 10.1038/370183a0. [DOI] [PubMed] [Google Scholar]
- Cairns P, Mao L, Merlo A, Lee DJ, Schwab D, Eby Y, Tokino K, van der Riet P, Blaugrund JE, Sidransky D. Rates of p16 (MTS1) mutations in primary tumors with 9p loss. Science. 1994;265:415–417. doi: 10.1126/science.8023167. [DOI] [PubMed] [Google Scholar]
- Florl AR, Franke KH, Niederacher D, Gerharz CD, Seifert HH, Schulz WA. DNA methylation and the mechanisms of CDKN2A inactivation in transitional cell carcinoma of the urinary bladder. Lab Invest. 2000;80:1513–1522. doi: 10.1038/labinvest.3780161. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Serrano M. The INK4a/ARF locus in murine tumorigenesis. Carcinogenesis. 2000;21:865–869. doi: 10.1093/carcin/21.5.865. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ, Wu EA, Horner JW, DePinho RA. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- Carnero A, Hudson JD, Price CM, Beach DH. p16INK4A and p19ARF act in overlapping pathways in cellular immortalization. Nat Cell Biol. 2000;2:148–155. doi: 10.1038/35004020. [DOI] [PubMed] [Google Scholar]
- Arap W, Nishikawa R, Furnari FB, Cavenee WK, Huang HJ. Replacement of the p16/CDKN2 gene suppresses human glioma cell growth. Cancer Res. 1995;55:1351–1354. [PubMed] [Google Scholar]
- Wu Q, Possati L, Montesi M, Gualandi F, Rimessi P, Morelli C, Trabanelli C, Barbanti-Brodano G. Growth arrest and suppression of tumorigenicity of bladder carcinoma cell lines induced by the p16/CDKN2 (p16INK4A, MTS1) gene and other loci on chromosome 9. Int J Cancer. 1996;65:840–846. doi: 10.1002/(SICI)1097-0215(19960315)65:6<840::AID-IJC22>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Yeager TR, DeVries S, Jarrard DF, Kao C, Nakada SY, Moon TD, Bruskewitz R, Stadler WM, Meisner LF, Gilchrist KW, Newton MA, Waldman FM, Reznikoff CA. Overcoming cellular senescence in human cancer pathogenesis. Genes Dev. 1998;12:163–174. doi: 10.1101/gad.12.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, III, Johnson BE, Skolnick MH. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- Randerson-Moor JA, Harland M, Williams S, Cuthbert-Heavens D, Sheridan E, Aveyard J, Sibley K, Whitaker L, Knowles M, Bishop JN, Bishop DT. A germline deletion of p14(ARF) but not CDKN2A in a melanoma-neural system tumour syndrome family. Hum Mol Genet. 2001;10:55–62. doi: 10.1093/hmg/10.1.55. [DOI] [PubMed] [Google Scholar]
- Glendening JM, Flores JF, Walker GJ, Stone S, Albino AP, Fountain JW. Homozygous loss of the p15INK4B gene (and not the p16INK4 gene) during tumor progression in a sporadic melanoma patient. Cancer Res. 1995;55:5531–5535. [PubMed] [Google Scholar]
- Kumar R, Sauroja I, Punnonen K, Jansen C, Hemminki K. Selective deletion of exon 1 beta of the p19ARF gene in metastatic melanoma cell lines. Genes Chromosomes Cancer. 1998;23:273–277. doi: 10.1002/(sici)1098-2264(199811)23:3<273::aid-gcc11>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Heyman M, Rasool O, Brandter LB, Liu Y, Grander D, Einhorn S, Soderhall S. Exclusive p15INK4B gene deletions in acute lymphocytic leukemia include the E1 beta exon of the p16INK4 gene. Blood. 1996;87:1657–1658. [PubMed] [Google Scholar]
- Ginzinger DG, Godfrey TE, Nigro J, Moore DH, II, Suzuki S, Pallavicini MG, Gray JW, Jensen RH. Measurement of DNA copy number at microsatellite loci using quantitative PCR analysis. Cancer Res. 2000;60:5405–5409. [PubMed] [Google Scholar]
- Labuhn M, Jones G, Speel EJ, Maier D, Zweifel C, Gratzl O, Van Meir EG, Hegi ME, Merlo A. Quantitative real-time PCR does not show selective targeting of p14(ARF) but concomitant inactivation of both p16(INK4A) and p14(ARF) in 105 human primary gliomas. Oncogene. 2001;20:1103–1109. doi: 10.1038/sj.onc.1204197. [DOI] [PubMed] [Google Scholar]
- World Health Organization Mosotofi FK, Sobin LH, Tosoni I, editors. Geneva: World Health Organization; Histological typing of urinary bladder tumours. International Histological Classification of Tumours. 1973 [Google Scholar]
- UICC Geneva: Union Internationale Contre le Cancer; TNM classification of malignant tumors, bladder. 1978:113–117. [Google Scholar]
- Proctor AJ, Coombs LM, Cairns JP, Knowles MA. Amplification at chromosome 11q13 in transitional cell tumours of the bladder. Oncogene. 1991;6:789–795. [PubMed] [Google Scholar]
- Higuchi R, Dollinger G, Walsh PS, Griffith R. Simultaneous amplification and detection of specific DNA sequences. Biotechnology (NY) 1992;10:413–417. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (NY) 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Flood SJ, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SV, Sibley KD, Davies AM, Nishiyama H, Hornigold N, Coulter J, Kennedy WJ, Skilleter A, Habuchi T, Knowles MA. Molecular genetic analysis of chromosome 9 candidate tumor-suppressor loci in bladder cancer cell lines. Genes Chromosomes Cancer. 2002;34:86–96. doi: 10.1002/gcc.10050. [DOI] [PubMed] [Google Scholar]
- Orlow I, LaRue H, Osman I, Lacombe L, Moore L, Rabbani F, Meyer F, Fradet Y, Cordon-Cardo C. Deletions of the INK4A gene in superficial bladder tumors: association with recurrence. Am J Pathol. 1999;155:105–113. doi: 10.1016/S0002-9440(10)65105-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packenham JP, Taylor JA, Anna CH, White CM, Devereux TR. Homozygous deletions but no sequence mutations in coding regions of p15 or p16 in human primary bladder tumors. Mol Carcinog. 1995;14:147–151. doi: 10.1002/mc.2940140303. [DOI] [PubMed] [Google Scholar]
- Stadler WM, Sherman J, Bohlander SK, Roulston D, Dreyling M, Rukstalis D, Olopade OI. Homozygous deletions within chromosomal bands 9p21–22 in bladder cancer. Cancer Res. 1994;54:2060–2063. [PubMed] [Google Scholar]
- Berggren P, Kumar R, Sakano S, Hemminki L, Wada T, Steineck G, Adolfsson J, Larsson P, Norming U, Wijkstrom H, Hemminki K. Detecting homozygous deletions in the CDKN2A(p16(INK4a))/ARF(p14(ARF)) gene in urinary bladder cancer using real-time quantitative PCR. Clin Cancer Res. 2003;9:235–242. [PubMed] [Google Scholar]
- Bertin R, Acquaviva C, Mirebeau D, Guidal-Giroux C, Vilmer E, Cave H. CDKN2A, CDKN2B, and MTAP gene dosage permits precise characterization of mono- and bi-allelic 9p21 deletions in childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2003;37:44–57. doi: 10.1002/gcc.10188. [DOI] [PubMed] [Google Scholar]
- Rizos H, Darmanian AP, Holland EA, Mann GJ, Kefford RF. Mutations in the INK4a/ARF melanoma susceptibility locus functionally impair p14ARF. J Biol Chem. 2001;276:41424–41434. doi: 10.1074/jbc.M105299200. [DOI] [PubMed] [Google Scholar]
- Drayton S, Peters G. Immortalisation and transformation revisited. Curr Opin Genet Dev. 2002;12:98–104. doi: 10.1016/s0959-437x(01)00271-4. [DOI] [PubMed] [Google Scholar]
- Simoneau M, Aboulkassim TO, LaRue H, Rousseau F, Fradet Y. Four tumor suppressor loci on chromosome 9q in bladder cancer: evidence for two novel candidate regions at 9q22.3 and 9q31. Oncogene. 1999;18:157–163. doi: 10.1038/sj.onc.1202277. [DOI] [PubMed] [Google Scholar]
- Knowles MA, Elder PA, Williamson M, Cairns JP, Shaw ME, Law MG. Allelotype of human bladder cancer. Cancer Res. 1994;54:531–538. [PubMed] [Google Scholar]
- Orlow I, Lianes P, Lacombe L, Dalbagni G, Reuter VE, Cordon-Cardo C. Chromosome 9 allelic losses and microsatellite alterations in human bladder tumors. Cancer Res. 1994;54:2848–2851. [PubMed] [Google Scholar]
- Jung I, Reeder JE, Cox C, Siddiqui JF, O’Connell MJ, Collins L, Yang Z, Messing EM, Wheeless LL. Chromosome 9 monosomy by fluorescence in situ hybridization of bladder irrigation specimens is predictive of tumor recurrence. J Urol. 1999;162:1900–1903. doi: 10.1016/S0022-5347(05)68064-0. [DOI] [PubMed] [Google Scholar]
- Friedrich MG, Blind C, Milde-Langosch K, Erbersdobler A, Conrad S, Loning T, Hammerer P, Huland H. Frequent p16/MTS1 inactivation in early stages of urothelial carcinoma of the bladder is not associated with tumor recurrence. Eur Urol. 2001;40:518–524. doi: 10.1159/000049829. [DOI] [PubMed] [Google Scholar]
- Bartlett JM, Watters AD, Ballantyne SA, Going JJ, Grigor KM, Cooke TG. Is chromosome 9 loss a marker of disease recurrence in transitional cell carcinoma of the urinary bladder? Br J Cancer. 1998;77:2193–2198. doi: 10.1038/bjc.1998.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J, Duncan P, Going JJ, Watters AD, Grigor KM, Bartlett JM. Identification of loci associated with putative recurrence genes in transitional cell carcinoma of the urinary bladder. J Pathol. 2002;196:380–385. doi: 10.1002/path.1052. [DOI] [PubMed] [Google Scholar]
- Cairns P, Polascik TJ, Eby Y, Tokino K, Califano J, Merlo A, Mao L, Herath J, Jenkins R, Westra W, Rutter JL, Buckler A, Gabrielson E, Tockman M, Cho K, Hedrick L, Bova GS, Isaacs W, Koch W, Schwab D, Sidransky D. Frequency of homozygous deletion at p16/CDKN2 in primary human tumours. Nat Genet. 1995;11:210–212. doi: 10.1038/ng1095-210. [DOI] [PubMed] [Google Scholar]