Abstract
Meningiomas are cytogenetically heterogeneous tumors in which chromosome gains and losses frequently occur. Based on the intertumoral cytogenetic heterogeneity of meningiomas, hypothetical models of clonal evolution have been proposed in these tumors which have never been confirmed at the intratumoral cell level. The aim of this study was to establish the intratumoral patterns of clonal evolution associated with chromosomal instability in individual patients as a way to establish tumor progression pathways in meningiomas and their relationship with tumor histopathology and behavior. A total of 125 meningioma patients were analyzed at diagnosis. In all cases, multicolor interphase fluorescence in situ hybridization (iFISH) studies were performed on fresh tumor samples for the detection of quantitative abnormalities for 11 different chromosomes. In addition, overall tumor cell DNA content was measured in parallel by flow cytometry. iFISH studies were also performed in parallel on tissue sections in a subset of 30 patients. FISH studies showed that 56 (45%) of the 125 cases analyzed had a single tumor cell clone, all these cases corresponding to histologically benign grade I tumors. In the remaining cases (55%) more than one tumor cell clone was identified: two in 45 cases (36%), three in 19 (15%), and four or more clones in five cases (4%). Overall, flow cytometric analysis of cell DNA contents showed the presence of DNA aneuploidy in 44 of these cases (35%), 30% corresponding to DNA hyperdiploid and 5% to hypodiploid cases; from the DNA aneuploid cases, 35 (28%) showed two clones and 9 (7%) had three or more clones. A high degree of correlation (r ≥ 0.89; P < 0.001) was found between FISH and flow cytometry as regards the overall quantitative DNA changes detected with both techniques, the former being more sensitive. Among the cases with chromosome abnormalities, the earliest tumor cell clone observed was frequently characterized by the loss of one or more chromosomes (64% of all meningiomas); loss of either a single chromosome 22 or, less frequently, of a sex chromosome (X or Y) and del (1p) was commonly found as the single initial cytogenetic aberration (30%, 5%, and 5% of the cases, respectively). Interestingly, an isolated loss of chromosome 22 was only found as the initial abnormality in one out of 14 atypical/anaplastic meningiomas, while the same cytogenetic pattern was present in the ancestral tumor cell clone of 32% of the benign tumors. Cytogenetic patterns based on chromosome gains were found in the ancestral tumor cell clone in 4% of the patients, 2% corresponding to tetraploid tumors. Overall, cytogenetic evolution of the earliest tumor cell clones was frequently associated with tetraploidization (31%). Our results show that meningiomas are genetically heterogeneous tumors that display different patterns of numerical chromosome changes, with the presence of more than one tumor cell clone detected in almost half of the cases including all atypical/anaplastic cases. Interestingly, the pathways of intratumoral clonal evolution observed in the benign tumors were different from those observed in atypical/anaplastic meningiomas, suggesting that the latter tumors might not always represent a more advanced stage of histologically benign meningiomas.
In recent years, an increasing number of studies have been reported which show that chromosome gains and losses are a frequent finding in meningioma tumors;1,2,3,4,5,6 at the same time these studies have provided information on the specific cytogenetic abnormalities accumulated.1,2,3,4,5,6 Of these, monosomy 22/22q−, and to a lesser extent 14q−, 1p− and 10q− abnormalities, together with loss of a sex chromosome (Y in males and X in females) and tetraploid karyotypes, are by far the most commonly observed aberrations. The analysis of large series of patients, using fluorescence in situ hybridization (FISH) techniques on interphase (iFISH) nuclei has provided a further accurate estimation of the incidence of these chromosomal abnormalities and their potential clinical significance.6,7,8,9,10,11
For many years it has been well-established that the development of various human tumors including colorectal carcinomas,12,13 gliomas,14,15 renal cell tumors,16 prostate cancer,17 and head and neck squamous cell carcinomas18 follows a multi-step pathway where an increasing number of genetic aberrations are accumulated due to genetic and/or chromosome instability. Typically, specific patterns of genetic evolution have been associated with both a more advanced stage and a more aggressive course of the disease.19,20,21,22,23 In a similar way, some models of clonal evolution have been proposed in meningiomas based on the intertumoral cytogenetic heterogeneity found, through the analysis of large series of patients by conventional karyotyping.4 These models provide hypothetical evolution pathways for what occurs during the sequential transition from normal meningeal cells to grade I, and grade II/grade III tumors.1,3,11 However, so far such models have not been confirmed at the intratumoral level. This is probably related to the fact that conventional karyotyping techniques have several major limitations for the assessment of intratumoral clonal evolution in meningiomas: 1) they allow the analysis of only a small fraction of all tumor cells; 2) the use of cultured samples may induce selective growth of specific tumor cell clones; and 3) the low number of metaphases analyzed makes it difficult to accurately identify the different tumor cell clones present in a sample.
In recent years, alternative cytogenetic techniques have been developed which facilitate the analysis of chromosome abnormalities in both interphase cells and metaphase chromosomes. Among others, these include iFISH and flow cytometry assessment of the DNA ploidy status of tumor cells. Although neither technique provides specific information on every chromosomal abnormality present in a tumor, they do allow a sensitive, rapid, and precise assessment of intratumoral heterogeneity. Multicolor iFISH is particularly suited to assessing intratumoral genetic heterogeneity, provided that adequate combinations of probes are used.24
In the present study, we have applied multicolor iFISH analysis of 12 different probes specific for DNA sequences of 11 chromosomes in combination with flow cytometry DNA cell content measurements in a series of 125 consecutive patients, to explore the intratumoral cytogenetic heterogeneity of meningiomas. Our goal is to establish the intratumoral pathways of clonal evolution associated with chromosomal instability in individual meningioma patients as a way to identify more precisely the cytogenetic stage of each individual neoplasia and its relationship with the histopathology and behavior of the tumor.
Materials and Methods
Patients
A total of 125 consecutive patients, 44 males and 81 females, diagnosed with meningioma at the Neurosurgical Unit of the University Hospital of Salamanca were analyzed. Mean age at diagnosis was 58 ± 15 years (range, 16 to 82 years). All cases were diagnosed with either intracranial (n = 118; 94%) or spinal (n = 7; 6%) meningiomas based on conventional histological criteria.25,26 Patient distribution according to the World Health Organization classification25,26 was as follows: 89% of the cases were grade I tumors; 10%, grade II; and the remaining 1%, grade III meningiomas. Tumor specimens were obtained by conventional surgical procedures and were then cut into several parts. Part of the tumor showing both macroscopic and microscopic infiltration was used to prepare single cell suspensions as described below. The remaining tumor was fixed in formalin (Paureac Quimicasa, Barcelona, Spain) and embedded in paraffin (Merck, Darmstadt, Germany). From these latter samples, sections were cut from three different areas representative of the tumoral tissue and placed over poly L-lysine coated slides (BioGenex, San Ramon, CA). All tissues were evaluated after hematoxylin-eosin (Merck) staining to confirm the presence and quantity of tumor cells infiltrating the material to be studied. Infiltration by tumor cells in tissue areas in the vicinity of those used for iFISH analysis of single cell suspensions was always 65%.
Flow Cytometry Analysis of Cell DNA Contents
Tumor cell DNA content studies were performed in all cases using fresh tumor samples obtained at diagnostic surgery. Single tumor cell suspensions were obtained by mechanical disaggregation and the cells’ DNA was stained with propidium iodide (PI) (Sigma, St. Louis, MO) using techniques which have been previously described in detail.27,28 Once sample preparation was completed and within a maximum period of 1 hour, DNA cell content measurements were performed in a FACSCalibur flow cytometer (Becton/Dickinson Biosciences (BDB), San Jose, CA). Information on a minimum of 104 tumor cells per sample was acquired using the CellQUEST software program (BDB). A second tube containing a 1/1 mixture of cells from the tumor sample and peripheral blood mononuclear cells from a gender-matched healthy volunteer was prepared and measured in parallel. DNA aneuploidy was defined as the presence of two distinct G0/G1 populations of cells with different DNA cell contents. The presence of more than one tumor cell clone by flow cytometry was defined by the observation of two or more G0/G1 populations of tumor cells. DNA index was calculated for each tumor cell clone as the ratio between the modal intensity of the PI-associated fluorescence of the G0/G1 tumor cells and that obtained for the normal G0/G1 diploid cells present in the same tube. The mean coefficient of variation for the G0/G1 peak of tumor cells was 3.3 ± 1%.
In a subset of 22 meningiomas in which the diploid cells by iFISH could have derived from contamination by leukocytes, simultaneous staining with an anti-pan leukocyte antigen CD45 antibody conjugated with fluorescein isothiocyanate (FITC) (BDB) and the DRAQ5 DNA-dye (Biostatus, Cambridge, UK) was performed as previously described.29
Fluorescence in Situ Hybridization (FISH) Studies
Multicolor interphase FISH (iFISH) studies were performed in all cases on an aliquot of the same single cell suspension prepared from the tumor sample and used for the flow cytometric measurement of cell DNA contents, after fixation in 3/1 methanol/acetic (v/v) (Merck). For the investigation of numerical chromosome abnormalities by iFISH, the following probes (Vysis Inc; Downers Grove, IL) specific for those chromosomes and chromosome regions more frequently gained or deleted in meningiomas6 were systematically used in double-stainings: 1) for chromosomes 9 and 22: LSI BCR/ABL dual-color probe; 2) for chromosomes 15 and 17: LSI PML/RAR-α dual-color probe; 3) for chromosomes 14 and 18: LSI IgH/BCL2 dual-color probe; 4) for chromosomes X and Y: CEP X DNA probe, conjugated with Spectrum Orange (SO) and CEP Y DNA probe, conjugated with Spectrum Green (SG); 5) for chromosomes 7 and 10: CEP 7 DNA probe, conjugated with SO and CEP 10 DNA probe, conjugated with SG; and 6) for chromosome 1: CEP 1 DNA probe, conjugated with SO. In addition, the Midisatellite 1p36 probe directly labeled with FITC (D1, Q-BIOgene, Amsterdam, The Netherlands) was also used. To accurately define the exact abnormalities carried by a tumor cell clone within a tumor sample, further appropriate two-color stainings were performed, whenever necessary.
Fixed cells were dropped into ethanol/ether (1/1, v/v) cleaned slides according to conventional cytogenetic protocols. The slides were then sequentially incubated with a solution containing 0.1 mg/ml pepsin (Sigma) (10 minutes at 37°C), fixed in 1% acid-free formaldehyde (Merck) (10 minutes at room temperature (RT)) and dehydrated in decreasing concentrations of ethanol (Merck) in water (100%, 95%, 70%). Once dried, the slides containing both the cells’ DNA and the probes’ DNA were denatured at 75°C (1 minute) and subsequently hybridized overnight at 37°C in a Hybrite thermocycler (Vysis, Inc.). After this incubation, slides were sequentially washed for 5 minutes at 46°C in 50% formamide (Merck) in 2X SSC (Merck) and then in 2X SSC. Afterward, cells were counter-stained with 35 μl of a mounting medium containing 75 ng/ml of DAPI (Sigma); Vectashield (Vector Laboratories Inc, Burlingame, CA) was used as anti-fading agent.
In addition, FISH analysis was also performed on tissues sections from 30 tumors. Briefly, unstained 5-μm paraffin sections on electrostatically charged slides (BioGenex; San Ramon, CA) were baked overnight (12 hours to 24 hours) at 65°C and immersed in xylene (Merck) (3 × 15 minutes). Afterward, they were dehydrated in ethanol and washed with distilled water. The slides were pre-treated in a pressure cooker with 10 mmol/L citric acid-trisodium salt (Sigma) buffer (pH 6.0) for 4 minutes and washed with 2X SSC. For enzymatic digestion, incubation with proteinase K (Sigma) was performed for 15 minutes at 37°C. After washing with 2X SSC, the slides were fixed with 1% formaldehyde, washed with 2X SSC, and dehydrated with 70%, 90%, and 100% ethanol. The slides were hybridized in the Hybrite thermocycler using 10-μl probe/slide. Denaturation was performed at 90°C for 6 minutes and hybridized overnight at 38°C. Post-hybridization washes were carried out at 46°C with 50% formamide and counterstaining with DAPI was used, as described above.
A BX60 fluorescence microscope (Olympus, Hamburg, Germany) equipped with a 100X oil objective was used for the enumeration of hybridization spots per nuclei. At least 200 nuclei were counted per slide. Only those spots with a similar size, intensity, and shape were counted in areas with <1% unhybridized cells; doublet signals were considered as single spots. A tumor was considered to carry a numerical abnormality for a given chromosome when the proportion of cells displaying an abnormal number of hybridization spots for the corresponding chromosome probe was at a percentage higher than the mean value + 2 SD of the percentage obtained with the same probe in control samples. The mean percentage of cells ± 1 SD showing one chromosome loss/gain in control samples was: chromosome 1p, 1.47 ± 1.22/0.14 ± 0.55; chromosome 1q, 0.47 ± 0.71/1.23 ± 1.30; chromosome 7, 0.33 ± 0.79/0.24 ± 0.46; chromosome 9, 1.47 ± 1.11/0.64 ± 0.7; chromosome 10, 1.04 ± 1.04/0.27 ± 0.50; chromosome 14, 1.53 ± 0.88/0.69 ± 1.11, chromosome 15, 1.09 ± 1.01/0.42 ± 0.68, chromosome 17, 1.84 ± 0.96/0.5 ± 0.85; chromosome 18, 0.38 ± 0.65/0.36 ± 0.99; chromosome 22, 1.73 ± 1.07/0.54 ± 0.85; chromosome X in females, 0.98 ± 0.88/0.13 ± 0.31; chromosome X in males, 0.1 ± 0/1 ± 0.87; and chromosome Y, 0.60 ± 1/1.08 ± 0.8. A tumor cell clone carrying a chromosomal abnormality was defined as being present when cells carrying identical numbers of hybridization spots for all of the probes analyzed, including the altered ones, were found at frequencies higher than 4% of the total cells analyzed in the sample.
The expected amount of total mean DNA content per cell according to the relative amount of DNA abnormally gained or lost corresponding to each altered chromosome by FISH was calculated according to two different mathematical formulas: 1) chromosome index (CI): 1+ [(Ch1p × 0.022) − (2 × 0.022)] + [(Ch1q × 0.022) − (2 × 0.022)] + [(Ch7 × 0.027) − (2 × 0.027)] + [(Ch9 × 0.024) − (2 × 0.024)] + [(Ch10 × 0.023) − (2 × 0.023)] + [(Ch11 × 0.024) − (2 × 0.024)] + [(Ch14 × 0.018) − (2 × 0.018)] + [(Ch15 × 0.017) − (2 × 0.017)] + [(Ch17 × 0.015) − (2 × 0.015)] + [(Ch18 × 0.014) − (2 × 0.014)] + [(Ch22 × 0.009) − (2 × 0.009)] + [(Chx × 0.024) − (x × 0.024)] + [(Chy × 0.009) − (y × 0.009)] and 2) corrected chromosome index (CCI) = [(Ch1p × 0.022) + (Ch1q × 0.022) + (Ch7 × 0.027) + (Ch9 × 0.024) + (Ch10 × 0.023) + (Ch11 × 0.024) + (Ch14 × 0.018) + (Ch15 × 0.017) + (Ch17 × 0.015) + (Ch18 × 0.014)+ (Ch22 × 0.009) + (Chx × 0.024) + (Chy × 0.009)] ÷ [(2 × 0.043) + (2 × 0.027) + (2 × 0.024) + (2 × 0.023) + (2 × 0.024) + (2 × 0.018) + (2 × 0.017) + (2 × 0.015) + (2 × 0.014)+ (2 × 0.009) + (x × 0.027) + (y × 0.009)], where Chn is the number of spots per cell found for each probe; ch x was 2 in women and 1 in men and ch y was 0 in women and 1 in men. Tetraploid tumor cell clones were defined as those showing a corrected chromosome index by iFISH of 2.00 ± 0.10. In turn, tetraploidization was defined by the presence of a tetraploid tumor cell clone or by the coexistence of two different tumor cell clones one showing one or more chromosome losses and the other showing a duplication of the chromosome copy number of the former clone. To define intratumoral clonal evolution in those tumors with multiple subclones, we assumed that karyotypic abnormalities shared by all subclones represented the earliest changes, whereas the latter cytogenetic changes would only be present in some of the tumor cell clones.
Statistical Methods
For all continuous variables included in the present study, their mean value, SD, and range were calculated using the SPSS software package (SPSS 11.0 Inc., Chicago, IL); for dichotomic variables, frequencies were reported. A multivariate step-wise regression analysis (regression, SPSS) was performed to examine the correlation between the DNA index obtained by flow cytometry and both the chromosome and the corrected chromosome indices obtained by iFISH. Statistical significance was considered to be present when P values lower than 0.05 were found.
Results
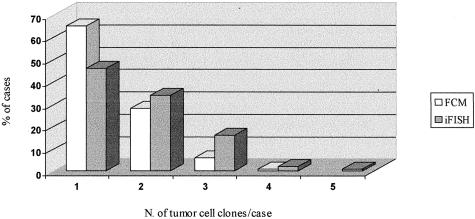
FISH studies showed that 56 (45%) of the 125 cases analyzed had a single tumor cell clone. In 35 of these 56 cases the tumor cell clone constantly displayed no abnormalities for the 12 chromosome-probes analyzed, whereas in the other 21 patients at least one chromosome was altered. In the remaining cases (55%), more than one tumor cell clone was identified by FISH: 2 in 45 cases (36%), 3 in 19 tumors (15%), and 4 or more clones in 5 patients (4%) (Figure 1). FISH analyses of tissue sections from other areas of the tumor performed in a group of 30 cases with two or more tumor cell clones, confirmed the presence of all of the clones identified in single cell suspensions, at the same time no additional clones could be identified in these cases in the areas of the tumor tissue analyzed, as illustrated in Figure 2. Flow cytometric analysis of the overall tumor cell DNA contents showed the presence of DNA aneuploidy in 44 (35%) from which 30% corresponded to DNA hyperdiploid and 5% to DNA hypodiploid cases. Of the DNA aneuploid cases, 35 (28%) showed two clones while 9 (7%) showed three or more clones by flow cytometry (Figure 1).
Figure 1.
Number of tumor cell clones present in meningioma tumors (n = 125) as detected by multicolor interphase FISH (iFISH) and flow cytometry analysis of cell DNA contents (FCM).
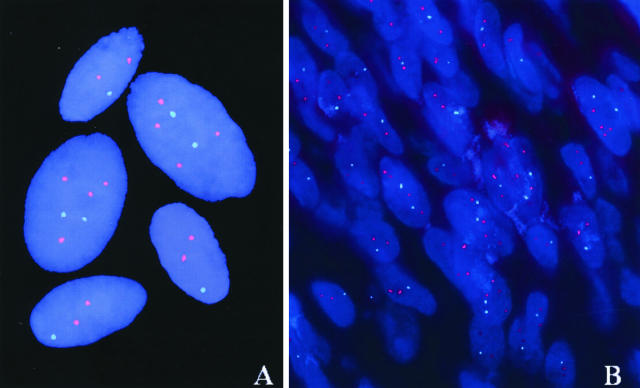
Figure 2.
Representative pictures of meningioma tumoral nuclei from a patient carrying two different tumor cell clones as defined by simultaneous hybridization for chromosomes 22 (green spots) and 9 (red spots): one clone shows monosomy 22 while the second clone displays two copies for chromosome 22 and four copies for chromosome 9. A corresponds to hybridizations performed on single cell suspensions while B shows hybridized nuclei from a tissue section from another area of the same tumor.
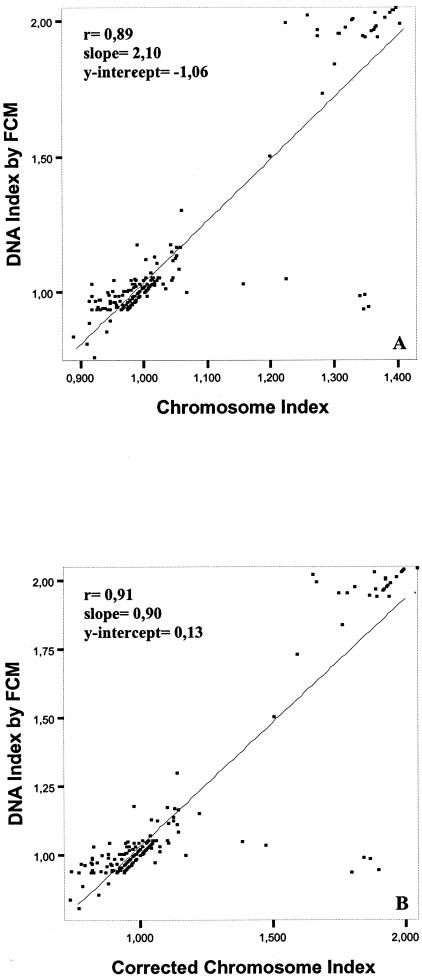
To explore the relationship between the quantitative DNA abnormalities found for the 11 chromosomes analyzed with the overall mean amount of DNA per cell measured by flow cytometry, we compared the chromosome indices obtained for each individual tumor cell clone with the DNA index obtained by flow cytometry. Interestingly, our results showed a significant correlation between the DNA contents per cell measured by flow cytometry and those derived from the iFISH studies assessed by both the chromosome (r = 0.89; P < 0.001) and the corrected chromosome (r = 0.91; P < 0.001) indices (Figure 3). Despite this, it should be noted that in 47 of 81 cases with a homogeneous DNA diploid tumor cell population by flow cytometry, hypodiploid clones were detected by iFISH. These frequently corresponded to near-diploid clones carrying losses of a single chromosome 22 (n = 19; 23%) or of a sex chromosome (n = 2; 3%), del (1p) (n = 4; 5%), a chromosome 22 and a sex chromosome (n = 2; 3%), a chromosome 22 and a chromosome 14 (n = 1; 1%), a chromosome 22 and del (1p) (n = 2; 3%), a chromosome 22, a chromosome 14, and a del (1p) (n = 2; 3%), a chromosome 22, a del (1p), and a chromosome Y (n = 2; 3%), a chromosome 22, a chromosome 7, and del (1p) (n = 1; 1%) and losses of a single chromosome 22 together with a sex chromosome and del (1p) (n = 2; 3%); DNA diploid tumors by flow cytometry displaying complex karyotypes with more than three chromosomal abnormalities by FISH were detected less frequently (n = 10; 12%). According to the World Health Organization classification, all cases showing only one clone (n = 56) by iFISH were diagnosed as being histologically benign (grade I) while all grade II and grade III tumors (n = 14; 100%) displayed two or more tumor cell clones (P < 0.001) (Table 1).
Figure 3.
Meningioma tumors (n = 125): correlation between the DNA index obtained by flow cytometry (FCM) and both the uncorrected (A) and corrected (B) chromosome indices obtained for each tumor cell clone according to the numerical chromosome abnormalities detected by interphase FISH analysis of 12 different chromosome probes.
Table 1.
Relationship between the Number of Tumor Cell Clones Found in Each Sample by Interphase Fluorescence in Situ Hybridization (iFISH) and Tumor Grade According to the WHO Classification
| No. of clones | Tumor grade
|
p-value | |
|---|---|---|---|
| I/(benign) n = 111 | II/III/ (atypical/anaplastic) n = 14 | ||
| 1 | 56 (51%) | 0 (0%) | <0.001 |
| 2 | 39 (35%) | 6 (43%) | |
| 3 | 15 (13%) | 4 (28.5%) | |
| ≥4 | 1 (1%) | 4 (28.5%) | |
Results are expressed as number of cases and percentage in brackets. WHO, World Health Organization.
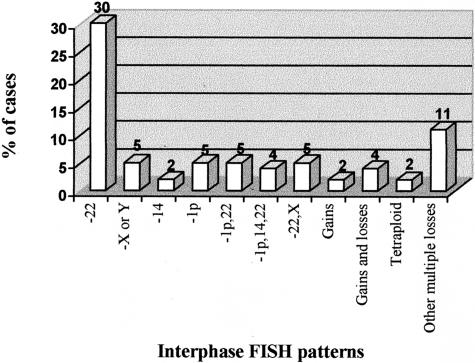
In all those cases in which two or more cell clones were present by iFISH, an ancestral tumoral cell clone could be identified. This earliest tumor cell clone observed (Figure 4) was frequently characterized by the loss of one or more chromosomes as the initial abnormality (64% of the cases). Loss of either a single chromosome 22 or a sex chromosome (X or Y) were commonly found as the earliest cytogenetic aberration (30% and 5% of the cases, respectively); other single chromosome losses detected in the earliest tumor cell clone observed, included del (1p) and monosomy 14/14q− detected in 5% and 2% of the cases, respectively. In the other cases characterized by chromosome losses in the ancestral tumor cell clone, between two and eight of the 11 chromosomes studied were simultaneously involved and corresponded to combined losses of chromosomes 22 and del (1p) (5%), del (1p), monosomy 14 and 22 or del (1p) (4%) two loss of chromosome 22 and an X chromosome (2%); in some cases (11%) these cytogenetic patterns were associated with additional chromosomes losses. Cytogenetic patterns based on chromosome gains were found in the ancestral tumor cell clone from five patients (4%), two of whom showed a tetraploid karyotype; in the other three cases (2%) trisomy 7 (n = 1), trisomy 9 and 17 (n = 1) and simultaneous gains of chromosomes 9, 10, and 14 (n = 1) were observed. Cytogenetic patterns based on the coexistence of chromosome gains and losses were also found in the earliest tumor cell clone, in five patients (4%). The cytogenetic patterns observed in these cases corresponded to loss of a chromosome 22 and trisomy 10 (n = 1), loss of a chromosome 22 and simultaneous gain of chromosomes 10, 17, and 18 (n = 1), loss of a chromosome 22 and trisomy 7, 9, 15, and 17 (n = 1), monosomy 22 and monosomy X with trisomy 7, 10, and 18 (n = 1) and loss of chromosomes 14, 22, Y, and del (1p) in association with trisomy of chromosomes 15 and 18 (n = 1).
Figure 4.
Meningioma tumors (n = 125): cytogenetic patterns found in the ancestral tumor cell clone identified, based on the analysis of 12 different chromosome-probes by multicolor interphase FISH.
As shown in Table 2 a significant association was found between tumor grade and the iFISH pattern observed for the ancestral tumor cell clone. Accordingly, most (64%) histopathologically benign tumors showed either no detectable chromosome abnormalities or monosomy 22/22q− in the earliest tumor cell clone identified with the selected markers, whereas these cytogenetic patterns were only found in one of 14 (7%) atypical/anaplastic meningiomas (P < 0.001). Among these latter cases, the earliest tumor cell lesion observed more frequently corresponded to isolated (six cases; 43%) or combined (two cases; 14%) losses of other chromosomes or tetraploidy (two cases; 14%).
Table 2.
Relationship between Tumor Grade According to the WHO Classification and the Interphase Fluorescence in Situ Hybridization (iFISH) Pattern Representative of the Earliest Tumor Cell Lesion Identified
| iFISH pattern | I/(benign) n = 111 | Tumor grade II/III/(atypical/anaplastic) n = 14 | p-value |
|---|---|---|---|
| Diploid | 35 (32%) | 0 (0%) | |
| −22 | 36 (32%) | 1 (7%) | |
| Loss of one chromosome other than 22 | 9 (8%) | 5 (36%) | |
| −22 associated with other chromosome losses | 21 (19%) | 3 (21%) | |
| Other multiple chromosome losses | 3 (3%) | 2 (14%) | <0.001 |
| Chromosome gains | 3 (3%) | 0 (0%) | |
| Chromosome gains and losses | 4 (4%) | 1 (7%) | |
| Tetraploid | 0 (0%) | 2 (14%) |
Results are expressed as number of cases and percentage in brackets. WHO, World Health Organization.
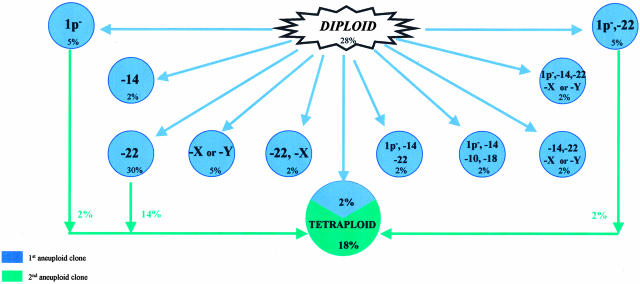
A detailed analysis of the different tumor cell clones present in each meningioma analyzed allowed us to hypothesize about the patterns of intratumoral clonal evolution in these neoplasias. Figure 5 summarizes the hypothetical models for the most frequent intratumoral aneuploidization pathways found for the chromosomes studied. As shown in the figure, 35 of the 125 meningioma patients studied (28%), had a normal chromosome set for the 12 chromosome probes tested. Among those 90 cases showing chromosomal abnormalities, the presence of a tumor cell clone with monosomy 22/22q− as the single earliest detected numerical chromosome abnormality was the most frequently detected aberration (n = 37; 30%). Whereas monosomy 22/22q− was the only cytogenetic aberration found in 17 of these tumors, in the other 20 cases, tumor cell clones with additional chromosome changes were found. These added abnormalities consisted of tetraploidization (n = 17; 14%), gains of individual chromosomes (n = 2) and monosomy 1/1p− (n = 1). Interestingly, monosomy 22q/22q− associated with loss of one chromosome X was the earliest cytogenetic pattern found in the ancestral tumor cell clone detected in another three cases (2%), one of them showing evolution toward tetraploidization. Monosomy 22q/22q− associated with loss of chromosome 1/1p− was also found as the first abnormality in six patients (5%); in a latter stage, one of them displayed monosomy 14, another showed loss of chromosome Y, while the other three patients evolved with tetraploidization. In another eight patients (6%), the ancestral tumor cell clone observed showed monosomy 22/22q− and del (1p) in association with loss of one or more of the other chromosomes studied, in the absence or presence (3%) of further detectable clonal evolution: monosomy Y (n = 1), monosomy 14/14q− (n = 1), monosomy 10/10q− and monosomy X (n = 1), monosomy 14/14q− and monosomy 17/17q− (n = 1), monosomy 10 and 18/18q− (n = 1), monosomy Y and 14/14q− and gains (trisomy) of chromosomes 15 and 17 (n = 1), losses of chromosomes 14/14q−, Y, 10, and 18 (n = 1) and monosomy 7 (n = 1). Loss of a sex chromosome (X or Y) as the unique chromosomal abnormality in the earliest tumor cell clone detected was observed in six cases (5%); four of these cases showed further evolution consisting of loss of chromosome 22 and del (1p) (n = 1), trisomy 10 (n = 1), 1q+ (n = 1) and trisomy 22, 15, 17, 18, and 7 (n = 1). All these cases, except the one showing trisomy 10, displayed further evolution into a tetraploid tumor cell clone. Monosomy 1/1p− as the sole abnormality detected in the ancestral tumor cell clone was found in six patients (5%); two of these cases showed further evolution into tetraploid tumor cell clones, one case gained an extra copy of chromosome 9 and the other showed trisomy 15. Two cases (2%) showed monosomy 14/14q− as the sole chromosome change present in the ancestral tumor cell clone; from these cases, one evolved with additional losses involving chromosome 22 and del (1p) while the other case sequentially showed gains (trisomy) of chromosomes 22 and X and later on, in a third clone, of chromosomes 1 and 9. Interestingly, in 14 additional cases (11%), monosomy 14/14q− was also present in the earliest tumor cell clone identified, in the context of a more complex karyotype, being associated with other chromosome losses; such losses involved chromosome 22 alone (n = 1; 1%) or along with a sex chromosome (n = 4; 3%) associated (n = 2) or not (n = 2) with additional del (1p); in the latter two patients, further clonal evolution was associated with trisomy 1 in one case and tetraploidization in the other patient. In another case, monosomy 14/14q− was associated with del (1p) and loss of chromosomes 22, Y, 10, and 18 in the ancestral tumor cell clone without further clonal evolution. In the other three cases (3%) displaying monosomy 14/14q− in the earliest tumor cell clone detected, additional chromosome changes consisted of del (1p) associated to monosomy 10 (n = 1) or to monosomy 10 and 18 (n = 2). Interestingly, a high proportion of these patients having monosomy 14/14q− associated with complex karyotypes in the ancestral tumor cell clone (6 of 14 cases) showed further evolution into tetraploid tumor cell clones either directly (n = 4) or after losing other chromosomes (n = 2). Another 11 different patterns of chromosome changes were observed in individual meningioma tumors.
Figure 5.
Genetic heterogeneity of meningiomas: hypothetical intratumoral aneuploidization pathways for the 11 chromosomes (12 probes) analyzed which were recurrently observed (≥2% of the cases). Percentages show the incidence of cases with a tumor cell clone displaying a specific cytogenetic pattern.
Discussion
Previous studies have shown that meningiomas are cytogenetically heterogeneous tumors which frequently display complex karyotypes (more than three numerical or structural aberrations). Interestingly, quantitative changes in the tumor cell genome are the most frequently detected, and, as such, the most common cytogenetic abnormality in these tumors is monosomy 22/22q−.1,30,31,32,33 In addition, a variety of other chromosomal changes which are mainly associated with losses of an entire chromosome or a major part of it, have also been described in meningiomas. These involve chromosomes 1, 10, 14, and 18 together with the sex chromosomes.2,4,34,35,36,37,38 Based on these findings, genetic changes associated with chromosome instability are believed to play an important role in both tumorgenesis and tumor progression in meningiomas. In recent years, a number of studies have attempted to describe the genetic pathways of tumor progression through the comparison of the karyotypes of meningioma tumors from different individuals. However, to the best of our knowledge, no study has been reported so far in which these genetic pathways have been analyzed at the intratumoral cell level. The potential existence of intratumoral genetic heterogeneity has long been suspected in meningiomas.9,33,39,40,41 Nevertheless, it has only recently been suggested that it could represent a common finding.6 A prerequisite to the study of intratumoral genetic heterogeneity is the availability of techniques that, through the simultaneous identification of two or more chromosome abnormalities at the single cell level, allow evaluation of the entire tumor cell populations in the absence of clonal selection. Accordingly, multicolor iFISH analysis of the most common chromosome aberrations was used in the present study to assess intratumoral genetic heterogeneity in meningiomas. Our results showed that in the great majority of the meningioma tumors analyzed two or more tumor cell clones carrying different chromosomal abnormalities were present. Since only part of each meningioma specimen was studied and FISH analyses only addressed part of the whole genome, we cannot rule out the possibility that additional tumor cell clones could exist. However, it should be noted that further analyses of tissue sections confirmed the results obtained in single cell suspensions for a subset of 30 tumors with two or more tumor cell clones. Interestingly, in all cases analyzed, a relationship could be clearly established between all of the genetically abnormal cell clones found within a tumor, and hypothetical models of clonal evolution pathways could be developed. Although several patterns of chromosome changes observed at the intratumoral cell level were common to two or more tumor samples, the exact incidence of each of the patterns detected was largely variable.
Development of a tumor is thought to start with the clonal expansion of a single cell carrying a mutation that leads to a growth/survival advantage;42 subsequently, any cell of this original clone may acquire additional genetic alterations giving rise to more rapidly growing subclones. Therefore, tumors develop in a multi-step process through the accumulation of genetic changes. In line with this, models have been proposed describing the genetic pathways of tumor progression in various human tumor types including colorectal carcinomas,12,13 gliomas,14,15 renal cell tumors,16 prostate cancer,17 and head and neck squamous cell carcinomas.18 To identify the exact sequence of accumulation of chromosomal abnormalities in a given tumor carrying multiple subclones, we assumed that karyotypic abnormalities shared by all subclones should represent relatively early changes; in contrast, latter cytogenetic changes would only be present in some of the tumor cell subclones. According to this concept of clonal evolution, complete or partial loss of either chromosome 22, a sex chromosome (Y in males and X in females), del (1p) or, less frequently, of chromosome 14/14q− alone or in combination with other chromosomes, eg, monosomy 10/10q−, 18/18q−, would frequently represent the earliest detectable cytogenetic event in meningioma tumor cells. These results would be in line with those observed by Zang4 who found similar karyotypes for the earliest tumor cell clones: diploid, loss of a sex chromosome, loss of a single chromosome 22 or del (22q), losses of a chromosome 22 and one sex chromosome, del (1p), or losses of a chromosome 22 and del (1p). Interestingly, tetraploidization, which has been associated with a worse clinical outcome,7 despite occurring as an early event in a low proportion of tumors, would usually correspond to a late stage during clonal evolution of meningiomas. The relatively high incidence of tumor cell clones that underwent tetraploidization found in the present study, contrasts with that reported by Zang4 using conventional karyotyping techniques. Such a discrepancy could be explained, at least in part, by the fact that tumor cell clones that underwent tetraploidization usually represented only a minor fraction of all tumor cells (between 5% and 10%) and therefore could either be missed or underestimated if only a small proportion/number of tumor cells/metaphases are analyzed as is the case when conventional cytogenetic techniques are used; alternatively, tetraploid tumor cells could undergo a negative clonal selection during cell culture.
To the best of our knowledge, this is the first time that intratumoral patterns of clonal evolution are reported in meningiomas. Until now, only Zang4 has proposed a model of clonal evolution in meningiomas based on conventional cytogenetic studies. However, in this model only those cases with monosomy 22/22q− were selected; such cases were further divided according to the coexistence or not of del (1p) in the majority of the metaphases analyzed, with the relative frequency found for this latter abnormality, similar to that observed in the present study. According to the model proposed, further genetic changes occurring in cases with monosomy 22/22q− would exclusively consist of the acquisition of other chromosome losses, in contrast to what we have observed at the intratumoral cell level, as discussed above. Other reports in which cytogenetic models of tumor progression are proposed are based either on the same patients reported by Zang3,4 or on the cytogenetic patterns observed in the different histopathological subtypes of meningiomas.2,36,38 These latter models assume that atypical and anaplastic meningiomas are sequentially related to histologically benign tumors and represent more advanced stages of the disease. In line with this, in the present study a clear association was observed between a more advanced tumor grade and a higher number of tumor cell clones and complex karyotypes. However, our results also show that the cytogenetic patterns present in histologically benign tumors are different from those in atypical/anaplastic tumors. While monosomy 22/22q− was commonly present in the earliest tumor cell clone observed in grade I meningiomas, it was only detected in a minor proportion of all atypical/anaplastic tumors. In turn, isolated losses of a sex chromosome, del (1p) and monosomy 14 in the ancestral tumor cell clone were characteristic of atypical/anaplastic tumors. Overall, these results may indicate that histologically benign tumors and atypical/anaplastic meningiomas may progress along different genetic pathways.
To explore whether or not the abnormal FISH probe-spot counts really reflected extensive quantitative changes in the chromosome material and were not restricted to the DNA sequences detected by the probes used, FISH results were correlated with the DNA index obtained by flow cytometry for the same patients. For that purpose, two different mathematical formulas were used in which the relative DNA content weight of each numerical chromosome abnormality detected was taken into account. Interestingly, a high degree of correlation was obtained between both types of information, clearly indicating that the chromosomal abnormalities obtained by iFISH largely reflected the presence of extensive gains or losses of chromosome material usually involving either the whole chromosome or a major part of it, in line with previous observations.43 Despite this, it should be noted that in approximately half of the meningioma tumors displaying a diploid tumor cell DNA content by flow cytometry, numerical chromosome changes existed. Interestingly, these alterations consisted of balanced chromosome gains and losses or, more frequently, of near-diploid clones with single chromosome changes, (eg, monosomy 22/22q−) which remained undetectable by flow cytometry.
In summary, our results support the notion that, as in other tumors, progression of meningiomas is a multi-step process in which neoplastic cells develop genome-wide instability and variants are selected, leading to the emergence of clonal populations with multiple chromosome abnormalities which potentially confer them selective proliferative advantages. Interestingly, benign tumors displayed different intratumoral clonal evolution pathways from atypical/anaplastic meningiomas, suggesting that these latter tumors might not always represent a more advanced stage of histologically benign meningiomas.
Footnotes
Supported by grants from the Fondo de Investigaciones Sanitarias (FIS 01/1564 and FIS PI 020010, Madrid, Spain), the Fundación Memoria de Don Samuel Solórzano Barruso (Salamanca, Spain), the Ayuda Biomedicina of the Consejería de Sanidad y Bienestar Social, Junta de Castilla y León (Exp SA 02/02, Valladolid, Spain), grants 02/9103 and 020010 from the Ministerio de Sanidad y Consumo, (Madrid, Spain) (J.M.S. and A.E.), and by a grant from the Ministerio de Ciencia y Tecnología (Programa Ramón y Cajal; Madrid, Spain) (M.D.T.).
J.M.S. and M.D.T contributed equally to this study and are joint first authors.
References
- Zattara-Cannoni H, Gambarelli D, Dufour H, Figarella D, Vollot F, Grisoli F, Vagner-Capodano AM. Contribution of cytogenetics and FISH in the diagnosis of meningiomas: a study of 189 tumors. Ann Genet. 1998;41:164–175. [PubMed] [Google Scholar]
- Weber RG, Bostrom J, Wolter M, Baudis M, Collins VP, Reifenberger G, Lichter P. Analysis of genomic alterations in benign, atypical, and anaplastic meningiomas: toward a genetic model of meningioma progression. Proc Natl Acad Sci USA. 1997;94:14719–14724. doi: 10.1073/pnas.94.26.14719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketter R, Henn W, Niedermayer I, Steilen-Gimbel H, Konig J, Zang KD, Steudel WI. Predictive value of progression-associated chromosomal aberrations for the prognosis of meningiomas: a retrospective study of 198 cases. J Neurosurg. 2001;95:601–607. doi: 10.3171/jns.2001.95.4.0601. [DOI] [PubMed] [Google Scholar]
- Zang KD. Meningioma: a cytogenetic model of a complex benign human tumor, including data on 394 karyotyped cases. Cytogenet Cell Genet. 2001;93:207–220. doi: 10.1159/000056986. [DOI] [PubMed] [Google Scholar]
- Cai DX, Banerjee R, Scheithauer BW, Lohse CM, Kleinschmidt-Demasters BK, Perry A. Chromosome 1p and 14q FISH analysis in clinicopathologic subsets of meningioma: diagnostic and prognostic implications. J Neuropathol Exp Neurol. 2001;60:628–636. doi: 10.1093/jnen/60.6.628. [DOI] [PubMed] [Google Scholar]
- Sayagués JM, Tabernero MD, Maillo A, Diaz P, Rasillo A, Bortoluci A, Gomez-Moreta J, Santos-Briz A, Morales F, Orfao A. Incidence of numerical chromosome aberrations in meningioma tumors as revealed by fluorescence in situ hybridization (FISH) using 10 chromosome-specific probes. Cytometry. 2002;50:153–159. doi: 10.1002/cyto.10075. [DOI] [PubMed] [Google Scholar]
- Maillo A, Diaz P, Sayagues JM, Blanco A, Tabernero MD, Ciudad J, Lopez A, Goncalves JM, Orfao A. Gains of chromosome 22 by fluorescence in situ hybridization in the context of an hyperdiploid karyotype are associated with aggressive clinical features in meningioma patients. Cancer. 2001;92:377–385. doi: 10.1002/1097-0142(20010715)92:2<377::aid-cncr1333>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Maillo A, Orfao A, Sayagues JM, Diaz P, Caballero M, Morales F, Tabernero D. A new classification scheme for the prognostic stratification of meningioma based on chromosome 14 abnormalities, patient’s age, and tumor histopathology. J Clin Oncol. 2003;21:3285–3295. doi: 10.1200/JCO.2003.07.156. [DOI] [PubMed] [Google Scholar]
- Scholz M, Gottschalk J, Striepecke E, Firsching R, Harders A, Fuzesi L. Intratumorous heterogeneity of chromosome 10 and 17 in meningiomas using non-radioactive in situ hybridization. J Neurosurg Sci. 1996;40:17–23. [PubMed] [Google Scholar]
- Kasai H, Kawamoto K. Cytogenical analysis of brain tumors by FISH (fluorescence in situ hybridization) and FCM (flow cytometry). Noshuyo Byori. 1995;12:75–82. [PubMed] [Google Scholar]
- Lekanne Deprez RH, Riegman PH, van Drunen E, Warringa UL, Groen NA, Stefanko SZ, Koper JW, Avezaat CJ, Mulder PG, Zwarthoff EC. Cytogenetic, molecular genetic, and pathological analyses in 126 meningiomas. J Neuropathol Exp Neurol. 1995;54:224–235. doi: 10.1097/00005072-199503000-00009. [DOI] [PubMed] [Google Scholar]
- Giaretti W. A model of DNA aneuploidization and evolution in colorectal cancer. Lab Invest. 1994;71:904–910. [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Collins VP, James CD. Gene and chromosomal alterations associated with the development of human gliomas. FASEB J. 1993;7:926–930. doi: 10.1096/fasebj.7.10.8344489. [DOI] [PubMed] [Google Scholar]
- Louis DN, Gusella JF. A tiger behind many doors: multiple genetic pathways to malignant glioma. Trends Genet. 1995;11:412–415. doi: 10.1016/s0168-9525(00)89125-8. [DOI] [PubMed] [Google Scholar]
- Kovacs G. Molecular cytogenetics of renal cell tumors. Adv Cancer Res. 1993;62:89–124. doi: 10.1016/s0065-230x(08)60316-4. [DOI] [PubMed] [Google Scholar]
- Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinanen R, Palmberg C, Palotie A, Tammela T, Isola J, Kallioniemi OP. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet. 1995;9:401–406. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- Califano J, van der Riet P, Westra W, Nawroz H, Clayman G, Piantadosi S, Corio R, Lee D, Greenberg B, Koch W, Sidransky D. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res. 1996;56:2488–2492. [PubMed] [Google Scholar]
- Jin C, Jin Y, Wennerberg J, Akervall J, Dictor M, Mertens F. Karyotypic heterogeneity and clonal evolution in squamous cell carcinomas of the head and neck. Cancer Genet Cytogenet. 2002;132:85–96. doi: 10.1016/s0165-4608(01)00535-0. [DOI] [PubMed] [Google Scholar]
- Garcia SB, Novelli M, Wright NA. The clonal origin and clonal evolution of epithelial tumours. Int J Exp Pathol. 2000;81:89–116. doi: 10.1046/j.1365-2613.2000.00142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulci G, Ishii N, Maurici D, Gernert KM, Hainaut P, Kaur B, Van Meir EG. Initiation of human astrocytoma by clonal evolution of cells with progressive loss of p53 functions in a patient with a 283H TP53 germ-line mutation: evidence for a precursor lesion. Cancer Res. 2002;62:2897–2905. [PubMed] [Google Scholar]
- Kattar MM, Kupsky WJ, Shimoyama RK, Vo TD, Olson MW, Bargar GR, Sarkar FH. Clonal analysis of gliomas. Hum Pathol. 1997;28:1166–1179. doi: 10.1016/s0046-8177(97)90255-0. [DOI] [PubMed] [Google Scholar]
- Coons SW, Johnson PC, Shapiro JR. Cytogenetic and flow cytometry DNA analysis of regional heterogeneity in a low grade human glioma. Cancer Res. 1995;55:1569–1577. [PubMed] [Google Scholar]
- Rasillo A, Tabernero D, Sánchez ML, Pérez de Andrés M, Martín M, Hernández J, Moro MJ, Fernández-Calvo J, Sayagues JM, Bortoluci A, San Miguel J, Orfao A. Fluorescence in situ hybridization analysis of aneuploidization patterns in monoclonal gammopathy of undetermined significance versus multiple myeloma and plasma cell leukemia. Cancer. 2003;97:601–609. doi: 10.1002/cncr.11100. [DOI] [PubMed] [Google Scholar]
- Louis DN, Scheithauer BW, Budka H, von Deimling A, Kepes JJ. Meningeal tumours. Kleihues P, Cavenee WK, editors. Lyon; World Heath Organization Classification of TumoursPathology and Genetics of Tumours of the Nervous System. 2000:176–184. [Google Scholar]
- Kleihues P, Burger PC, Scheithauer BW. The new WHO classification of brain tumours. Brain Pathol. 1993;3:255–268. doi: 10.1111/j.1750-3639.1993.tb00752.x. [DOI] [PubMed] [Google Scholar]
- Vindelov LL, Christensen IJ, Nissen NI. A detergent-trypsin method for the preparation of nuclei for flow cytometric DNA analysis. Cytometry. 1983;3:323–327. doi: 10.1002/cyto.990030503. [DOI] [PubMed] [Google Scholar]
- Maillo A, Diaz P, Blanco A, Lopez A, Ciudad J, Hernandez J, Morales F, Perez-Simon JA, Orfao A. Proportion of S-phase tumor cells measured by flow cytometry is an independent prognostic factor in meningioma tumors. Cytometry. 1999;38:118–123. doi: 10.1002/(sici)1097-0320(19990615)38:3<118::aid-cyto5>3.3.co;2-a. [DOI] [PubMed] [Google Scholar]
- Smith PJ, Wiltshire M, Davies S, Patterson LH, Hoy T. A novel cell permeant and far red-fluorescing DNA probe, DRAQ5, for blood cell discrimination by flow cytometry. J Immunol Methods. 1999;229:131–139. doi: 10.1016/s0022-1759(99)00116-7. [DOI] [PubMed] [Google Scholar]
- Al Saadi A, Latimer F, Madercic M, Robbins T. Cytogenetic studies of human brain tumors and their clinical significance: II. meningioma. Cancer Genet Cytogenet. 1987;26:127–141. doi: 10.1016/0165-4608(87)90140-3. [DOI] [PubMed] [Google Scholar]
- Collins VP, Nordenskjold M, Dumanski JP. The molecular genetics of meningiomas. Brain Pathol. 1990;1:19–24. doi: 10.1111/j.1750-3639.1990.tb00634.x. [DOI] [PubMed] [Google Scholar]
- Katsuyama J, Papenhausen PR, Herz F, Gazivoda P, Hirano A, Koss LG. Chromosome abnormalities in meningiomas. Cancer Genet Cytogenet. 1986;22:63–68. doi: 10.1016/0165-4608(86)90138-x. [DOI] [PubMed] [Google Scholar]
- Perry A, Jenkins RB, Dahl RJ, Moertel CA, Scheithauer BW. Cytogenetic analysis of aggressive meningiomas: possible diagnostic and prognostic implications. Cancer. 1996;77:2567–2573. doi: 10.1002/(SICI)1097-0142(19960615)77:12<2567::AID-CNCR21>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Bello MJ, de Campos JM, Kusak ME, Vaquero J, Sarasa JL, Pestana A, Rey JA. Allelic loss at 1p is associated with tumor progression of meningiomas. Genes Chromosomes Cancer. 1994;9:296–298. doi: 10.1002/gcc.2870090411. [DOI] [PubMed] [Google Scholar]
- Menon AG, Rutter JL, von Sattel JP, Synder H, Murdoch C, Blumenfeld A, Martuza RL, von Deimling A, Gusella JF, Houseal TW. Frequent loss of chromosome 14 in atypical and malignant meningioma: identification of a putative ‘tumor progression’ locus. Oncogene. 1997;14:611–616. doi: 10.1038/sj.onc.1200853. [DOI] [PubMed] [Google Scholar]
- Lamszus K, Kluwe L, Matschke J, Meissner H, Laas R, Westphal M. Allelic losses at 1p, 9q, 10q, 14q, and 22q in the progression of aggressive meningiomas and undifferentiated meningeal sarcomas. Cancer Genet Cytogenet. 1999;110:103–110. doi: 10.1016/s0165-4608(98)00209-x. [DOI] [PubMed] [Google Scholar]
- Logan JA, Seizinger BR, Atkins L, Martuza RL. Loss of the Y chromosome in meningiomas: a molecular genetic approach. Cancer Genet Cytogenet. 1990;45:41–47. doi: 10.1016/0165-4608(90)90064-h. [DOI] [PubMed] [Google Scholar]
- Simon M, von Deimling A, Larson JJ, Wellenreuther R, Kaskel P, Waha A, Warnick RE, Tew JM, Jr, Menon AG. Allelic losses on chromosomes 14, 10, and 1 in atypical and malignant meningiomas: a genetic model of meningioma progression. Cancer Res. 1995;55:4696–4701. [PubMed] [Google Scholar]
- Janowska K, Kordek R, Biernat W, Liberski PP, Krul W, Debiec-Rychter M. Biologic heterogeneity of angiomatous meningiomas. Pol J Pathol. 1998;49:279–284. [PubMed] [Google Scholar]
- Sawyer JR, Husain M, Pravdenkova S, Krisht A, Al Mefty O. A role for telomeric and centromeric instability in the progression of chromosome aberrations in meningioma patients. Cancer. 2000;88:440–453. [PubMed] [Google Scholar]
- Simon M, Kokkino AJ, Warnick RE, Tew JM, Jr, von Deimling A, Menon AG. Role of genomic instability in meningioma progression. Genes Chromosomes Cancer. 1996;16:265–269. doi: 10.1002/(SICI)1098-2264(199608)16:4<265::AID-GCC7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- Tabernero D, San Miguel JF, Garcia-Sanz M, Najera L, Garcia-Isidoro M, Perez-Simon JA, Gonzalez M, Wiegant J, Raap AK, Orfao A. Incidence of chromosome numerical changes in multiple myeloma: fluorescence in situ hybridization analysis using 15 chromosome-specific probes. Am J Pathol. 1996;149:153–161. [PMC free article] [PubMed] [Google Scholar]