Abstract
Hereditary non-polyposis colorectal cancer (HNPCC) accounts for approximately 2 to 4% of the total colorectal cancer burden. For economic reasons a diagnostic “stepladder” is recommended. After evaluation of the family history, diagnostic microsatellite instability (MSI) analysis has found its place as a valuable screening tool for HNPCC. Immunohistochemical analysis can help to pinpoint the affected gene. The detection of a mutation in one of the responsible mismatch repair gene confirmed the diagnosis HNPCC. Here we demonstrate our experience of some important pitfalls that will be discussed in this study. In MSI testing, one potential source for false-negative results is intralesional heterogeneity. We demonstrate examples of a flat adenoma and a carcinoma, which required laser microdissection to correctly determine the microsatellite status. In these lesions manual microdissection, the most frequently applied method, was not sufficient. However, the number of cells obtained by using laser microdisssection can fall below a necessary minimum, which can also cause false-negative results of MSI analysis, as shown here in a mucinous carcinoma. In addition, evaluation of immunohistochemically stained tissue slides requires experience to avoid false-positive or false-negative interpretation. A case with two synchronous colorectal cancers revealed loss of MSH2 expression in one carcinoma, whereas the second carcinoma stained positively leading to a false-negative interpretation. In some cases, false-positive results can be obtained, if a perinuclear-staining pattern is interpreted as positive. In summary, there are several potential pitfalls in the molecular screening for HNPCC. Therefore the importance of correct interpretation of clinical data, immunohistochemistry, and microsatellite analysis in combination, performed by a pathologist with experience in molecular genetics is essential. In addition, laser microdissection of tumor areas that have been chosen by a pathologist is highly recommended in cases that cannot be resolved with manual microdissection.
Hereditary non-polyposis colorectal cancer (HNPCC) is one of the most common hereditary cancer predisposition syndromes accounting for approximately 2 to 4% of the total colorectal cancer burden.1,2,3,4,5 The high predisposition to cancer is due to a germ-line mutation of one allele of a mismatch repair (MMR) gene. To date, pathogenic mutations have been found at four mismatch repair genes (MSH2, MHL1, PMS2 and MSH6), but so far most HNPCC cases caused by mutations in either MLH1 or MSH2.6,7
As soon as a “second hit” inactivates the remaining intact allele in a cell, genomic instability results from the loss of DNA repair function. High microsatellite instability (MSI-H) is a hallmark for deficient MMR.8,9,10,11,12,13,14,15 In more than 90% of HNPCC patients fulfilling the Amsterdam criteria, microsatellite instability is found in the tumor tissue.14,15 Therefore microsatellite analysis is commonly used as the first diagnostic screening test for HNPCC. Immunohistochemical staining for the MMR proteins MSH2, MLH1, and MSH6 in tumor tissue is recommended in addition to identify the gene that is most likely to be mutated.16,17,18,19,20,21 Since many pathogenic mutations of MMR genes cause protein truncation, a negative or less intense nuclear staining is observed in such cases with MMR gene mutations. In these cases a sequence analysis of the presumed mutated gene can be performed after appropriate genetic counseling. Alternatively, a loss of MLH1 expression may also caused by methylation of the 5′CpG island in the promotor region.22 In these cases, the age of the patients is mostly over 60 years. Clinical screening of mutation carriers can help to prevent the cancer and therefore it is essential to identify all HNPCC-associated carcinomas as well as adenomas. False-positive results of the molecular diagnostic may lead to over-treatment and unnecessary psychological stress for the individual and the family. False-negative results may lead to the exclusion of a HNPCC patient from the necessary surveillance program and subsequent failure to detect secondary cancer development in an early stage. In addition, if a mutation is detected in a family, unaffected family members can be tested for the presence of the mutation and appropriate measures for prevention and early detection of cancer in mutation carriers can be taken.
Therefore, screening for MMR deficiencies in adenomas as well as in carcinomas should be as specific and sensitive as possible. Although a lot of work has been done over the last decade, since the first MMR gene was described,23,24 identification of HNPCC patients and mutation carriers is far from being satisfactory. There are still some diagnostic pitfalls in the detection of HNPCC patients, which will be discussed in this study.
Materials and Methods
Four cases were taken as examples from a prospectively studied series of more than 800 colorectal cancer cases that were investigated at the Institutes of Pathology, Klinikum Kassel and University Hospital Bonn, Germany, after obtaining informed consent. Our study was done as a part of the multi-centric German HNPCC-consortium which currently serves for over 1550 registered families almost meeting the Bethesda criteria.
Patient 1
A 46-year-old patient was diagnosed during a routine colonoscopy with a flat adenomatous lesion in the cecum, measuring 4 cm in diameter. Histologically, biopsy specimens showed foci of poorly differentiated, intramucosal carcinoma arising in an adenoma with high-grade dysplasia. The patient was treated by right hemicolectomy and the diagnosis of a large flat intramucosal carcinoma was confirmed with two (<1 mm) small foci of moderately differentiated adenocarcinoma invading the submucosa. Histologically, intra- and peritumoral lymphocytes were noted, which was most particularly prominent in the early invasive carcinomatous tumor part. Lymph node metastases were not present (pT1 (sm1), pN0, M0). Screening for HNPCC was initiated because of the conspicuous histological findings, although the patient did not fulfill the Bethesda (I)25 criteria.
Patient 2
Paraffin-embedded tissue blocks from a 54-year-old woman fulfilling the Bethesda criteria diagnosed with a carcinoma of the right colon were sent for HNPCC screening. The TNM classification of the carcinoma was documented as pT3, pN0, M0. Histopathological examination revealed a completely mucinous carcinoma without intratumoral but with some lymphocytic peritumoral infiltration corresponding to Crohn’s-like lesions.
Patient 3
A 52-year-old women was diagnosed with two synchronous colorectal carcinomas and was treated by right hemicolectomy. Histopathological analysis showed a 6-cm polypoid carcinoma with medullary growth pattern in the ascending colon (pT3, pN0, M0) and a 9-cm ulcerated carcinoma in the transverse colon (pT3, pN0, M0) showing a mixture of glandular and mucinous growth patterns. Both tumors exhibited a dense intra- and peritumoral lymphocytic infiltrate. HNPCC screening was initiated, prompted by the findings of synchronous colonic cancers with histological features suspicious of HNPCC.
Patient 4
Paraffin-embedded tissue of a cecal carcinoma obtained from a 64-year-old male index patient of a family fulfilling the Amsterdam I criteria was analyzed.26 The tissue showed a mixed histological pattern (mucinous and solid differentiation) and an elevated count of intratumoral lymphocytes. TNM-classification was pT3, pN1, pM0.
Microdissection and DNA Extraction
Paraffin-embedded tumor, adenoma, and corresponding normal tissue sections were mounted on glass slides and stained with hematoxylin and eosin (H&E). Adenomas were additionally stained with periodic acid Schiff (PAS) reagent. The dominant degree of dysplasia was determined and graded as mild (D1), moderate (D2), or severe (D3) with D1 and D2 corresponding to low-grade and D3 to high-grade intraepithelial neoplasia according to the updated World Health Organization classification of tumors.27
Both manual microdissection and laser microdissection were performed in parallel on all samples (by G.G.). Manual microdissection was performed on paraffin sections stained briefly with 0.1% methylene blue. Areas of interest measuring ∼1 cm2 were selected and marked by a pathologist, scraped off the slide with a scalpel under microscopic control, and subjected to proteinase K digestion at a final concentration of 2 μg/ml (Qiagen, Hilden, Germany). DNA was extracted with the QIAamp DNA Mini Kit (Qiagen) according to the manufacturer’s recommendations.
For laser microdissection, sections were mounted onto a polyethylene membrane and attached to a glass slide with rubber cement. After staining with hematoxylin, laser microdissection was carried out using the Leica AS LMD system (Leica Microsystems Wetzlar GmbH, Germany). About 100 to 300 epithelial cells were harvested into microtubes from each sample. Cell lysis was performed by dispensing 20 μl lysis buffer into each lid. The buffer consisted of 10% proteinase K (4 mg/ml) (Roche, Mannheim, Germany), 0.5% Tween 20 (Merck, Darmstadt, Germany) and 1X TaqPCR buffer (Roche). After digestion for 18 hours at 50°C, proteinase K was inactivated by incubation at 94°C for 10 minutes.
Microsatellite Analysis
In accordance to the recommendations by the NCI and the ICG-HNPCC, five microsatellite loci were used to detect microsatellite instability (MSI): two loci with mononucleotide runs (BAT 25, BAT 26) and three loci with CA dinucleotide repeats (D2S123, D5S346, D17S250).16 Primers were 5′-labeled with HEX, FAM, or TET (Perkin Elmer, Foster City, CA). DNA was amplified in a standard reaction mix with cycling conditions using Ampli Taq Gold (Perkin Elmer). The PCR products were run on an ABI 310 automated sequencer using fragment analysis software (Gene Scan, Perkin Elmer). Additional peaks in tumor tissue DNA in comparison to normal tissue DNA indicated MSI. If only one marker was classified as unstable, a second panel with five additional markers (BAT40, D10S197, D13S153, MYCL1, D18S58) was run. Instability in 30% or more of the markers was reported as high instability (MSI-H), less than 30% was classified as low instability (MSI-L) and no shifts or additional peaks as microsatellite stable (MSS).
Immunostaining for MSH2, MLH1, MSH6, and PMS2
To investigate for loss of MMR protein expression, the samples were subjected to immunohistochemical analysis. Briefly, slides were deparaffinized and hydrated by graded alcohols. Antigen retrieval was achieved by microwave treatment (800 Watt, 20 minutes) and incubation with 3% H2O2. Non-specific binding was blocked by incubation with goat serum for 30 minutes. The slides were incubated overnight at 4°C with primary antibody against MSH2 (0.5 μg/ml; Oncogene Sciences, Cambridge, MA), MLH1 (1 μg/ml Clone G168–728, PharMingen, San Diego, CA, dilution 1:200), MSH6 (mAB Clone 44, Becton Dickinson Transduction Laboratories, San Diego, CA, dilution 1:600) or PMS2 (mAB Clone sc-25315, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, dilution 1:25). The sections were washed with phosphate-buffered saline (PBS) and incubated with secondary biotinylated antibody (Vector Laboratories, Burlingame, CA). After rinsing with PBS, the sections were incubated with streptavidin-conjugated horseradish peroxidase (Vector Laboratories). For detection, the chromogen 3-amino-9-ethylcarbazole (Sigma, St. Louis, MO) was used according to the manufacturer’s recommendations and finally the slides were counter-stained with hematoxylin.
Results
Patient 1
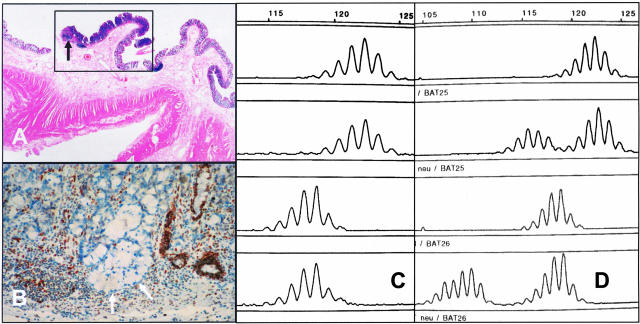
Using DNA obtained by manual microdissection of the complete adenoma, a microsatellite instability was not detected, the adenoma was therefore classified as MSS. However, using DNA obtained by laser microdissection of the small (<1 mm) areas of early invasion the adenoma (Figure 1A) was found to be MSI-H (Figure 1, C and D). Immunohistochemical staining revealed loss of MLH1 expression supporting the diagnosis of MMR deficiency (see arrows in Figure 1B). Finally, mutation analysis later demonstrated a germline missense mutation at codon 292 within exon 10 (exchange from leucine to proline) of the MLH1 gene. The mutation is predicted to destroy an important β-strand and therefore most likely inactivates the MLH1 function (according to personal communication by Rick Fishel, Kimmel Cancer Institute, Philadelphia, PA).
Figure 1.
A–D: Results of MSI analysis in patient 1 diagnosed with a flat adenoma in the cecum. A: An overview of the lesion shows slightly thickened mucosa within the marked area (box) and a minute focus of invasion (arrow). H&E, magnification, ×4. B: Immunohistochemistry showed distinct loss of MLH1 expression in the neoplastic crypts (arrows). Anti-MLH1, magnification, ×10. C: MSI-analysis after crude manual microdissection displayed MSS status. D: MSI-analysis of the mononucleotide markers BAT 25 (upper two lanes) and BAT 26 (lower two lanes) after laser microdissection disclosed the MSI-H nature particularly in the invasive part (normal tissue: lanes 1 and 3; corresponding tumor tissue: lanes 2 and 4).
Patient 2
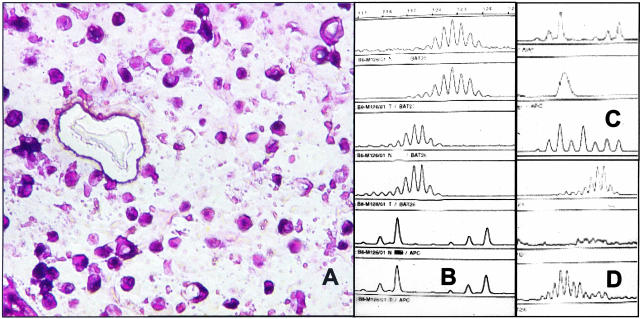
MSI analysis after manual dissection of the mucinous carcinoma shown in Figure 2A initially indicated MSS (Figure 2B). Because of the discrepancy with histological data, including loss of MSH2 protein expression in the immunohistochemical analysis, the MSI analysis was repeated by using laser microdissection. Again the data indicated MSS and a loss of heterozygosity at the APC (D5S346) locus was observed (Figure 2C, lane 2). Although the other four markers displayed no additional peaks in comparison with the normal tissue of the patient, the fragment signals were so low that we suspected that the number of cells for a correct analysis had been fallen below the detection level (Figure 2D, lane 2). Therefore, in the second run of laser microdissection, we counted the collected cells and used over 100 cells for analysis. Indeed, we then observed appropriate amplification of microsatellite loci and the result indicated microsatellite instability in five of five tested markers (Figure 2, C and D, lane 3).
Figure 2.
A–D: MSI analysis in a mucinous carcinoma. A: Histological tumor section after laser microdissection of selected tumor cells. H&E, magnification, ×40. B: Crude manual dissection of the mucinous carcinoma revealed MSS status (shown: BAT25, BAT26, D5S346; normal tissue: lanes 1, 3, and 5; corresponding tumor tissue: lanes 2, 4, and 6). C and D: After laser microdissection MSI analysis revealed a loss of the second peak at the dinucleotide marker (APC, D5S346) in the tumor sample when compared to normal tissues (normal tissue, lane 1; corresponding tumor tissue, lane 2). This result, suggesting LOH, was observed only when less than 50 cells were used for the MSI-analysis, whereas laser microdissection of a larger tumor cell number revealed the correct MSI-H result (C, lane 3). D: Corresponding results for BAT 26 with the PCR artifact resulting from too few analyzed cells in lane 2 and the correct MSI result shown in lane 3.
Patient 3
From this patient we tested two synchronous colon carcinomas. MSI analysis using the NIH consensus panel of microsatellite loci revealed MSI-H-phenotype in both carcinomas. All five loci showed additional peaks, although shifts were less prominent in one of the carcinomas (Figure 3G). After immunohistochemical staining, in the tumor showing less distinct shifts, a moderate to strong nuclear MSH2 protein staining was observed and thus this carcinoma initially was scored as positive for MSH2 expression (Figure 3E). In contrast, the second tumor, located more proximal, showed complete loss of the MSH2 protein expression and was correctly interpreted as negative for MSH2 (Figure 3B).
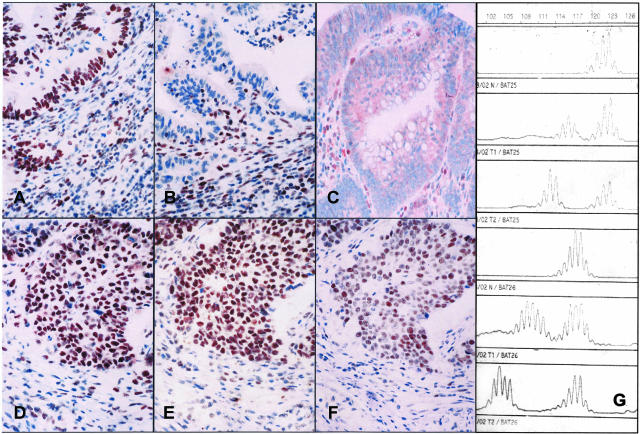
Figure 3.
A-G: Immunohistochemical staining and MSI-analyses of two synchronous colon carcinomas. A–F: The immunohistochemical analysis showed loss of MSH2 protein expression in one adenocarcinoma (B) but clearly detectable MSH2 protein expression in the second, more solid carcinoma (E) (A and D: anti-MLH1, B and E: anti-MSH2, C and F: anti-MSH6 immunostaining, magnification, ×20.). G: Results of MSI-analysis in both tumors (lanes 1 and 4, normal tissue; lanes 2 and 5, second solid tumor; lanes 3 and 6, adenocarcinoma). Less distinct shifts in the MSH2 expressing carcinoma.
Taking the MSI result into account the immunostaining of the first tumor was re-evaluated. We first compared MSH2 expression to adjacent normal mucosa and inflammatory cells which was not unequivocally different. Most interestingly, however, MSH6 expression was markedly reduced in both carcinomas (Figure 3, C and F) which then rendered the correct classification of complete loss in one and reduced MSH2 protein expression in the other tumor.
Patient 4
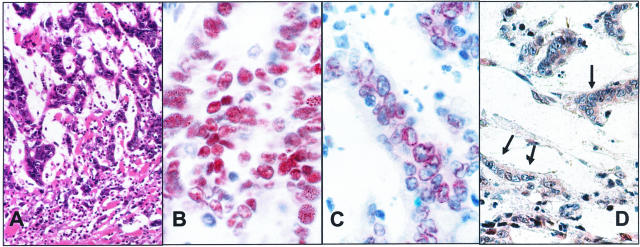
Heterogeneity in tumor differentiation as well as the intratumoral lymphocytes were highly suspicious for a carcinoma of the mutator phenotype. Analysis of the microsatellite status revealed MSI-H with shifts in 4 of 5 markers. Immunohistochemistry showed a strong nuclear staining pattern of the internal control tissue for MLH1 and MSH2. The same was observed for MSH2 expression in the carcinoma. In contrast, MLH1 did not show a nuclear but rather a distinct perinuclear staining (Figure 4). Thus, loss of MLH1 expression could not be diagnosed with certainty but, on the other hand, was not ruled out completely. Mutation analysis of the MLH1 gene then revealed a pathogenic splice mutation of the highly conserved splice donor site of intron 17. MSI data are not shown.
Figure 4.
A–D: Histological and immunohistological findings in patient 4. A: Carcinoma with dense intratumoral lymphocytic infiltrate suggestive of an MSI-H tumor. H&E, magnification, ×10. B: Strong intranuclear staining of MSH2 in the tumor cells. Anti-MSH2, mgnification, ×40. C: MLH1 immunostaining shows only a perinuclear rim-like reaction, but no distinct intranuclear staining. Anti-MLH1, magnification, ×40. D: Complete loss of PMS2 staining in the tumor cell nuclei (arrows).
Discussion
Identification of patients with a predisposition for hereditary cancer is important for the initiation of an intensive clinical cancer screening regimen to detect tumors at an early stage. Clinically, both the clustering of malignant tumors in a single family, as well as an early age of onset of the disease, is highly suggestive of an hereditary cancer predisposition and should prompt appropriate diagnostic procedures.28,29
A high degree of microsatellite instability in carcinomas and its precursor lesions is the hallmark for the underlying MMR deficiency.8,9,10,11,12,13,14,15 Most recently, it has been confirmed that MSI status is also a strong predictor of the benefit of adjuvant chemotherapy with fluorouracil in stage II and stage III colon cancer.30 Therefore, analyzing the MSI-H status in colon tumors is important not only for recognition of HNPCC families but also for the management of the individual patient in terms of further therapy decisions. Results in this study indicate that there are a number of pitfalls that need to be considered to avoid false-positive or false-negative interpretations of microsatellite analysis and immunohistochemical analysis in tumors from patients suspected of HNPCC. Therefore, we will discuss problems in MSI analysis especially in adenomas, the dissection technique, as well as the occasional problematic interpretation of immunohistochemical staining results.
In the literature, the detection rate of MSI-H in adenomas of HNPCC patients varies between 57 to 90%.30,31,32 As demonstrated by our first case, this might be related to intralesional heterogeneity of MSI in these early lesions. To overcome this problem, we strongly recommend applying microdissection and analyzing, in particular, the most advanced tumor areas. False-negative results may arise when manual microdissection is used or if tissue is not microdissected at all, because, in these cases, areas with a predominance of stromal or inflammatory cells may be selected for analysis.31 For MSI analysis at least 70% of the cells examined should be tumor cells. This high degree of cell separation in small precursor lesions can only be achieved by precise microdissection.
Inaccuracy of manual microdissection was demonstrated in our study in one case (patient 1). In our hands, advanced lesions and the highest-grade dysplasia show the highest sensitivity to detect MSI-H. This is especially true for adenomas.8 The correct classification of the case as MMR deficient by laser microdissection was confirmed by immunohistochemistry, revealing loss of MLH1 expression and subsequent detection of a germ-line mutation in the corresponding gene. However, laser microdissection cannot be used without the clinico-pathological context. One technical limitation is the number of cells that is required to correctly amplify the microsatellite loci and to reliably detect instability. In mucinous carcinomas especially, the analysis may be very difficult due to the small number of cancer cells within the mucinous areas and reliable MSI data can only be obtained if enough tumor cells are analyzed.33 Based on our experience, we recommend that at least 100 tumor cells are dissected for microsatellite analysis.
In samples with less than 50 cells pseudo-LOH may be found. This was exemplarily demonstrated in our case where the laser microdissection with less then 100 cells resulted in a false-negative MSI result. Using cell sections (as opposed to uncut cells) from very small cell numbers may pose a problem because parts of the nuclei are usually deleted on routine histological sections. In such cases, the clinico-pathological characteristics, the immunohistological staining results, and the family history should provide additional information to warrant further studies.
Although immunohistochemical analysis complements the interpretation of the microsatellite analysis and helps to identify the deficient protein, tumors with MSI-H may not always entirely lose expression of a mismatch repair protein. This phenomenon is exemplarily demonstrated in one of the two synchronous tumors of our patient 3. A possible explanation for this phenomenon might be that the two tumors differ in their second hit mutation leading to a still expressed, albeit not functional, protein in one (leading to false-positive result) and to a truncated, not detectable protein, in the other tumor. At the same time, loss of MMR protein expression in the absence of a mutation leading to a false-negative result can be caused by over-fixation of the tissue,34 therefore only cases with positively stained internal normal control tissue cells should be evaluated. Analysis of a panel of MMR proteins and other markers might also help to identify false-negatives and/or false-positives.
Loss of MSH2 is often accompanied by loss of MSH6 as a secondary event, because MSH6 contains a microsatellite that might be mutated as a consequence of the microsatellite instability or because the heterodimer MSH2-MSH6 cannot be formed because of the lack of MSH2. Thus, in cases showing some equivocal reduction of MSH2 staining, weak or even loss of MSH6 expression strongly favors a MSH2 mutation.35 This could clearly be confirmed in our case with two synchronous MSI-H carcinomas which differently expressed MSH2 but showed almost complete loss of MSH6 staining in both neoplasms. Similarly, loss of MLH1 is associated with a loss of PMS2.36 Immunohistochemical staining for MMR proteins in locations other than the nucleus is also suspicious for a mutation demonstrated in our patient 4 of this series. We therefore recommend using vigorous internal and external controls for interpretation of immunohistochemical staining results and performing MSI analysis in parallel.
In conclusion, although the molecular testing for HNPCC screening does not require really sophisticated equipment, the interpretation of the data may be difficult, at least in some cases. Therefore the importance of interpreting MSI analysis and immunohistochemical profiles in the context of the clinico-pathological findings and the family history has to be emphasized. The selection of the areas for microdissection and the interpretation of immunohistochemical stainings should be performed by a pathologist familiar with the molecular pathology of MMR.37,38
Appendix
The German HNPCC-Consortium consists of the following centers (in alphabetic order): clinical centers in Bochum (in addition to author: Frank Brasch, Jörg T. Epplen, Stefan Hahn, Erdmute Kunstmann, Christian Pox Jörg Willert), Bonn (in addition to authors: Constanze Pagenstecher, Waltraut Friedl, Holger Lauschke, Andreas Hirner, Christof Lamberti, Peter Propping, Tilman Sauerbruch), Düsseldorf (in addition to author: PD Dr. med. G. Möslein), Dresden (in addition to authors: Daniela E. Aust, Friedrich Balck, Ruth Höhl, Friedmar R. Kreuz, Stefan Krüger, Steffen R. Pistorius, Jens Plaschke), Heidelberg (in addition to author; Prof. M. von Knebel-Döberitz), München-Regensburg (in addition to author: Wolfgang Dietmaier, Gross, Reinhard Kopp, Peter Lohse, Michael Muders, Yvonne Müller-Koch, Holger Vogelsang), and center for documentation and biometry in Leipzig (in addition to author: Jochen Forberg, Marlies Herold, Markus Löffler).
Footnotes
Supported by the German Cancer Aid (Deutsche Krebshilfe) to J.R. and R.B. (Grant 70–303I-Rü 2), by the Deutsche Forschungsgemeinschaft to R.B. (Bu 672/10–1, 10–2) and by the Ministero dell′Istruzione, dell′Universita′ e della Ricerca (MIUR) and the Consiglio Nazionale delle Ricerche (CNR) Legge 449/97 to G.T. (Progetto Diagnostica Molecolare in Oncologia, contribution nr. CU02.00343. ST97).
References
- Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomäki P, Chadwick RB, Kääriäinen H, Eskelinen M, Järvinen H, Mecklin JP, de la Chapelle A. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;21:1481–1487. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- Aarnio M, Sankila R, Pukkala E, Salovaara R, Aaltonen LA, de la Chapelle A, Peltomäki P, Mecklin JP, Jarvin HJ. Cancer risk in mutation carrier of DNA-mismatch-repair genes. Int J Cancer. 1999;81:214–218. doi: 10.1002/(sici)1097-0215(19990412)81:2<214::aid-ijc8>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- Salovaara R, Loukola A, Kristo P, Kääriäinen H, Ahtola H, Eskelinen M, Härkönen N, Julkunen R, Kangas E, Ojala S, Tulikoura J, Valkamo E, Järvinen H, Mecjklin JP, Aaltonen LA, de la Chapelle A. Population-based molecular detection of hereditary nonpolyposis colorectal cancer. J Clin Oncol. 2000;11:2193–2200. doi: 10.1200/JCO.2000.18.11.2193. [DOI] [PubMed] [Google Scholar]
- Vasen HF, Wijnen JT, Menko FH, Kleibeuker JH, Taal BG, Griffioen G, Nagengast FM, Meijers-Heijboer EH, Bertario L, Varesco L, Bisgaard ML, Mohr J, Fodde R, Khan PM. Cancer risk in families with hereditary nonpolyposis colorectal cancer diagnosed by mutation analysis. Gastroenterology. 1996;110:1020–1027. doi: 10.1053/gast.1996.v110.pm8612988. [DOI] [PubMed] [Google Scholar]
- Boland CR, Sato J, Saito K, Carethers JM, Marra G, Laghi L, Chauhan DP. Genetic instability and chromosomal aberrations in colorectal cancer: a review of the current models. Cancer Detect Prev. 1998;22:377–382. doi: 10.1046/j.1525-1500.1998.00050.x. [DOI] [PubMed] [Google Scholar]
- Papadopoulos N, Lindblom A. Molecular basis of HNPCC: mutations of MMR genes. Hum Mutat. 1997;10:89–99. doi: 10.1002/(SICI)1098-1004(1997)10:2<89::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Loukola A, Salovaara R, Kristo P, Moisio AL, Kääriäinen H, Ahtola H, Eskelinen M, Harkonen N, Julkunen R, Kangas E, Ojala S, Tulikoura J, Valkamo E, Jarvinen H, Mecklin JP, de la Chapelle A, Aaltonen LA. Microsatellite instability in adenomas as a marker for hereditary nonpolyposis colorectal cancer. Am J Pathol. 1999;155:1849–1853. doi: 10.1016/S0002-9440(10)65503-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocker T, Diermann J, Friedl W, Gebert J, Holinski-Feder E, Karner-Hanusch J, von Knebel-Doeberitz M, Koelble K, Moeslein G, Schackert HK, Wirtz HC, Fishel R, Ruschoff J. Microsatellite instability analysis: a multi-center study for reliability and quality control. Cancer Res. 1997;57:4739–4743. [PubMed] [Google Scholar]
- Gryfe R, Gallinger S. Microsatellite instability, mismatch repair deficiency, and colorectal cancer. Surgery. 2001;130:17–20. doi: 10.1067/msy.2001.112738. [DOI] [PubMed] [Google Scholar]
- Iino GH, Simms L, Young J, Arnold J, Winship IM, Webb SI, Furlong KL, Leggett B, Jass JR. DNA microsatellite instability and mismatch repair protein loss in adenomas presenting in hereditary non-polyposis colorectal cancer. Gut. 2000;47:37–42. doi: 10.1136/gut.47.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jass JR, Walsh MD, Barker M, Simms LA, Young J, Leggett BA. Distinction between familial and sporadic forms of colorectal cancer showing DNA microsatellite instability. Eur J Cancer. 2002;38:858–866. doi: 10.1016/s0959-8049(02)00041-2. [DOI] [PubMed] [Google Scholar]
- Loukola A, Eklin K, Laiho P, Salovaara R, Kristo P, Jarvinen H, Mecklin JP, Launonen V, Aaltonen LA. Microsatellite marker analysis in screening for hereditary nonpolyposis colorectal cancer (HNPCC). Cancer Res. 2001;61:4545–4549. [PubMed] [Google Scholar]
- Potocnik U, Glavac D, Golouh R, Ravnik-Glavac M. Causes of microsatellite instability in colorectal tumors: implications for hereditary non-polyposis colorectal cancer screening. Cancer Genet Cytogenet. 2001;126:85–96. doi: 10.1016/s0165-4608(00)00399-x. [DOI] [PubMed] [Google Scholar]
- Ruschoff J, Bocker T, Schlegel J, Stumm G, Hofstaedter F. Microsatellite instability: new aspects in the carcinogenesis of colorectal carcinomas. Virchows Arch. 1995;426:215–222. doi: 10.1007/BF00191357. [DOI] [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshlemann JR, Burt RW, Meltzer SJ, Rodriguez-Bias MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- Jass JR. hMLH1 and hMSH2 immunostaining in colorectal cancer. Gut. 2000;47:315–316. doi: 10.1136/gut.47.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraf F, Gilquin M, Longy M, Gilbert B, Gorry P, Petit B, Labrousse F. MLH1 and MSH2 protein immunohistochemistry is useful for detection of hereditary non-polyposis colorectal cancer in young patients. Histopathology. 2002;39:250–258. doi: 10.1046/j.1365-2559.2001.01203.x. [DOI] [PubMed] [Google Scholar]
- Salahshor S, Koelble K, Rubio C, Lindblom A. Microsatellite instability and hMLH1 and hMSH2 expression analysis in familial and sporadic colorectal cancer. Lab Invest. 2001;81:535–41. doi: 10.1038/labinvest.3780262. [DOI] [PubMed] [Google Scholar]
- Plaschke J, Krüger S, Pistorius S, Theissig F, Saeger HD, Schackert HK. Involvement of hMSH6 in the development of hereditary and sporadic colorectal cancer revealed by immunostaining is based on germline mutations, but rarely on somatic inactivation. Int J Cancer. 2002;97:643–648. doi: 10.1002/ijc.10097. [DOI] [PubMed] [Google Scholar]
- Wahlberg SS, Schmeits J, Thomas G, Loda M, Garber J, Syngal S. Evaluation of microsatellite instability and immunohistochemistry for the prediction of germ-line MSH2 and MLH1 mutations in hereditary nonpolyposis colon cancer families. Cancer Res. 2002;62:3485–3492. [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Wilson JKV, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstin B, Kunkel TA, Baylin SB. Incidence and functional consequences of MLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parson R, Petlomaki P, Sistonen P, Aaltonen LA, Trent JM, de la Chapelle A, Kinzler KW, Vogelstein B. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Bigas MA, Boland CR, Hamilton SR, Henson DE, Jass JR, Khan PM, Lynch HT, Perucho M, Smyrk T, Sobin L. A national cancer workshop on hereditary nonpolyposis colorectal cancer syndrome: meeting highlights and Bethesda criteria. J Natl Cancer Inst. 1997;23:1758–1762. doi: 10.1093/jnci/89.23.1758. [DOI] [PubMed] [Google Scholar]
- Vasen HF, Mecklin J-P, Meera KP, Lynch HT. The international collaborative group on hereditary non-polyposis colorectal cancer (ICG-HNPCC). Dis Colon Rectum. 1991;34:424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- Hamilton SR, Aaltonen LA, editors. Lyon, France: IARC Press; Tumours of the digestive System. Pathology and Genetics: World Health Organization Classification of Tumours. 2000 [Google Scholar]
- Aarnio M, Sankila R, Pukkala E, Salovaara R, Aaltonen LA, de la Chapelle A, Peltomäki P, Mecklin JP, Jarvin HJ. Cancer risk in mutation carrier of DNA-mismatch-repair genes. Int J Cancer. 1999;81:214–218. doi: 10.1002/(sici)1097-0215(19990412)81:2<214::aid-ijc8>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Syngal S, Fox EA, Eng C, Kolodner RD, Garber JE. Sensitivity and specificity of clinical criteria for hereditary non-polyposis colorectal cancer associated mutations in MSH2 and MLH1. J Med Genet. 2000;37:641–645. doi: 10.1136/jmg.37.9.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CM, Dent OF, Barker M, Newland RC, Chapuis PH, Bokey EL, Young JP, Leggett BA, Jass JR, Macdonald GA. Prognostic significance of extensive microsatellite instability in sporadic clinicopathological stage C colorectal cancer. Br J Surg. 2000;87:1197–1202. doi: 10.1046/j.1365-2168.2000.01508.x. [DOI] [PubMed] [Google Scholar]
- Lanspa SJ, Lynch HT, Smyrk TC, Strayhorn P, Watson P, Lynch JF, Jenkins JX, Appelman HD. Colorectal adenomas in the Lynch syndromes: results of a colonoscopy screening program. Gastroenterology. 1990;98:1117–1122. doi: 10.1016/0016-5085(90)90323-s. [DOI] [PubMed] [Google Scholar]
- E Heinmoller E, W Dietmaier W, H Zirngibl H, P Heinmoller P, W Scaringe W, KW Jauch KW, F Hofstadter F, J Ruschoff J: Molecular analysis of microdissected tumors and preneoplastic intraductal lesions in pancreatic carcinoma. Am J Pathol 2000, 157:83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüschoff J, Dietmaier W, Lüttges J, Seitz G, Bocker T, Zirngibl Z, Schlegel J, Schackert HK, Jauch KW, Hofstädter F. Poorly differentiated colon adenocarcinoma, medullary type: clinical, phenotypic, and molecular characteristics. Am J Pathol. 1997;150:1815–1825. [PMC free article] [PubMed] [Google Scholar]
- Monzon F, Kovatich A, Barusevicius A, Bocker T, Palazzo J. Effect of fixation time on the immunohistochemical detection of hMLH1 and hMSH2 in paraffin-embedded tissue. Mod Pathol. 1999;12:193A. [Google Scholar]
- Rigau V, Sebbagh N, Olschwang S, Paraf F, Mourra N, Parc Y, Flejou JF. Microsatellite instability in colorectal carcinoma: the comparison of immunohistochemistry and molecular biology suggests a role for hMSH6 [correction of hMLH6] immunostaining. Arch Pathol Lab Med. 2003;127:694–700. doi: 10.5858/2003-127-694-MIICC. [DOI] [PubMed] [Google Scholar]
- Wu X, Platt JL, Cascalho M. Dimerization of MLH1 and PMS2 limits nuclear localization of MutLalpha. Mol Cell Biol. 2003;23:3320–3328. doi: 10.1128/MCB.23.9.3320-3328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüschoff J, Dietmaier W, Bocker T, Wallinger S, Kullmann F, Beham A, Hofstadter F. Molecular cancer disposition diagnosis exemplified by colorectal carcinoma. What is the contribution of pathology? Pathologe. 1998;19:269–278. doi: 10.1007/s002920050283. [DOI] [PubMed] [Google Scholar]
- Alexander J, Watanabe T, Wu TT, Rashid A, Li S, Hamilton SR. Histopathological identification of colon cancer with microsatellite instability. Am J Pathol. 2001;158:527–535. doi: 10.1016/S0002-9440(10)63994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]