Abstract
The methylation status of genes in hydatidiform mole and choriocarcinoma and its significance is relatively unexplored. We investigated the methylation status of the promoter regions of six genes, p16, HIC-1, TIMP3, GSTP1, death-associated protein kinase (DAPK), and E-cadherin in 54 hydatidiform moles, five choriocarcinomas, and 10 first trimester placenta by methylation-specific polymerase chain reaction (PCR). Immunohistochemical expression of p16, TIMP3, and E-cadherin, and quantitative real-time RT-PCR of p16 was also performed. Among the six genes examined, the promoter region of four genes (E-cadherin, HIC-1, p16, TIMP3) in choriocarcinoma and three genes (E-cadherin, HIC-1, p16) in hydatidiform mole exhibited aberrant methylation whereas none was hypermethylated in normal placenta. There was a significant correlation between methylation and reduced expression of p16, E-cadherin, and TIMP3 (P < 0.001). Fifteen of the 54 patients with hydatidiform mole developed gestational trophoblastic neoplasia requiring chemotherapy. Promoter hypermethylation of p16 alone, or combined with E-cadherin, was significantly correlated to such development (P = 0.001, 0.0005, respectively). Hypermethylation of multiple genes, especially p16, might be related to the subsequent development of gestational trophoblastic neoplasia.
Hydatidiform mole and choriocarcinoma are two major subtypes of gestational trophoblastic disease (GTD) in Orientals. The reported incidence of GTD is approximately 1 in 500 to 1 in 1000 pregnancies in Asia, two to ten times higher than that in United States and Europe.1,2 Choriocarcinoma is a highly malignant epithelial tumor arising from the trophoblast of any type of gestational event, most often a hydatidiform mole. Hydatidiform mole represents abnormally formed placenta characterized by swelling of chorionic villi and excessive proliferation of trophoblast. Although not a frank malignancy, hydatidiform mole may behave in an aggressive way with a predisposition for malignant transformation. About 8 to 30% of patients with hydatidiform mole will develop gestational trophoblastic neoplasia and require chemotherapy.1,2 Most choriocarcinomas are related to previous hydatidiform moles. Similar to other human cancers, neoplastic transformation of trophoblast is likely to be a multi-step process and involves multiple genetic alterations including activation of oncogenes and inactivation of tumor suppressor genes.3,4 Potential biological markers related to malignant transformation of trophoblast include c-erbB-2,5 cyclin E,6 DOC-2/hDab2,7 Ras GTPase activating protein,8 telomerase activity,9 apoptotic index,10,11 and matrix metalloproteinases,12 but no molecular parameter can currently replace serial serum human chorionic gonadotrophin (hCG) titers as the mainstay for predicting gestational trophoblastic neoplasia.
Promoter hypermethylation has recently been found to be an important epigenetic mechanism causing gene inactivation. Tumor suppressor genes, involving cell cycle regulation (p16), DNA repair and protection (BRCA1 and GSTP1), apoptosis (DAPK), cell adherence and metastasis process (E-cadherin, TIMP3), may be silenced by promoter CpG island methylation in many human tumors, thus contribute to carcinogenesis. Previously, we have shown that hypermethylation of E-cadherin may be related to its reduced expression in hydatidiform mole using a relatively small number of cases.13 While hypermethylation of the H19 and TFPI-2 genes has been reported in choriocarcinoma,14,15,16 little is currently known about the methylation status of tumor suppressor genes in GTD.
In this study, we examined the frequency of methylation of six genes, p16, E-cadherin, TIMP3, DAPK, GSTP1, and HIC-1, which are known to undergo epigenetic inactivation in other human cancers, in 54 hydatidiform moles, five human gestational choriocarcinomas, and 10 normal first trimester placentas by methylation-specific polymerase chain reaction (MSP). Expressions of p16, TIMP3, and E-cadherin in these samples were investigated by immunohistochemistry and quantitative real-time RT-PCR. In addition, the relationship between the methylation status and subsequent development of gestational trophoblastic neoplasia was analyzed.
Materials and Methods
Patients and Samples
All specimens of trophoblastic tissue were collected at the Department of Pathology, University of Hong Kong, Queen Mary Hospital. Clinically and ultrasonographically suspected cases of hydatidiform moles were suction evacuated. First trimester samples were obtained after induced abortion by suction evacuation. Fresh chorionic villi, molar vesicles, and decidua were dissected, selected and snap-frozen in liquid nitrogen, and stored at −70°C. The rest of tissue was fixed in formalin. Frozen tissue blocks of 10 normal first trimester placentas (gestational age 9.0 ± 2.2 weeks) and 54 hydatidiform moles with follow-up data were selected for this study. The patients’ age ranged from 16 to 51 (mean, 32.7) while the gestational age of the hydatidiform mole ranged from 6 weeks to 34 weeks (mean, 13.6 weeks). Archival formalin-fixed, paraffin-embedded tissue blocks from five choriocarcinomas before chemotherapy were also retrieved. The histological features of these cases were assessed using generally agreed and accepted diagnostic criteria.1,2 Some of the hydatidiform moles had also been examined previously, by chromosome in situ hybridization and genotyping using DNA extracted from laser-capture-microdissected tissues.17
Thirty-nine of the 54 hydatidiform moles were complete moles while 15 were partial moles. Gestational trophoblastic neoplasia was diagnosed if there was a plateau in hCG level for 4 weeks or if there was a further increase in hCG for 3 consecutive weeks when pregnancy was excluded.18 According to these criteria, 39 patients had spontaneous regression of the hydatidiform mole while the other 15 cases developed gestational trophoblastic neoplasia and received chemotherapy.
MSP
Genomic DNA extracted from the frozen tissues was modified by sodium bisulfite as previously described with minor modification.19,20 Briefly, one μg genomic DNA was denatured by adding freshly prepared sodium hydroxide (final concentration of 0.3 mol/L) at 37°C for 10 minutes, and 95°C for 2 minutes. After cooling down to room temperature, hydroquinone (30 mmol/L) and sodium bisulfite (3.6 mol/L, pH 5.0) were added. The mixture was then incubated at 55°C for 15 to 16 hours in the dark. Wizard DNA Cleanup System (Promega, Madison, WI) was applied to purify the sodium bisulfite-modified DNA according to manufacturer’s recommendations. The modified DNA was then incubated in 0.3 mol/L sodium hydroxide at 37°C for 15 minutes, then neutralized by adding sodium acetate (pH 4.0). The DNA was ethanol-precipitated and re-suspended in 30 μl of 10 mmol/L Tris water (pH 8.0). The primers and reaction conditions of MSP for the investigated genes were listed in Table 1. Five μl of each PCR products were separated on 2.5% agarose gel, which was stained with ethidium bromide, and directly visualized under UV illumination. DNA from normal placenta treated with SssI methylase (New England Biolabs, Beverly, MA) was used as a methylation-positive control. Genomic DNAs without bisulfite modification were used as negative control in both methylated and unmethylated reactions. Samples demonstrated to have methylation in either gene were repeated for confirmation.
Table 1.
Primer Sequences and PCR Conditions for MSP
| Gene* | Forward primers (5′→ 3′) | Reverse primer (5′→ 3′) | Annealing temperature (°C) | Product size (bp) | |
|---|---|---|---|---|---|
| p16 | M | TTATTAGAGGGTGGGGCGGATCGC | GACCCCGAACCGCGACCGTAA | 65 | 150 |
| U | TTATTAGAGGGTGGGGTGGATTGT | CAACCCCAAACCACAACCATAA | 60 | 151 | |
| E-cadherin | M | TTAGGTTAGAGGGTTATCGCGT | TAACTAAAAATTCACCTACCGAC | 57 | 115 |
| U | TAATTTTAGGTTAGAGGGTTATTGT | CACAACCAATCAACAACACA | 53 | 97 | |
| TIMP3 | M | CGTTTCGTTATTTTTTGTTTTCGGTTTTC | CCGAAAACCCCGCCTCG | 59 | 116 |
| U | TTTTGTTTTGTTATTTTTTGTTTTTGGTTTT | CCCCCAAAAACCCCACCTCA | 59 | 122 | |
| DAPK | M | GGATAGTCGGATCGAGTTAACGTC | CCCTCCCAAACGCCGA | 60 | 98 |
| U | GGAGGATAGTTGGATTGAGTTAATGTT | CAAATCCCTCCCAAACACCAA | 60 | 106 | |
| GSTP1 | M | TTCGGGGTGTAGCGGTCGTC | GCCCCAATACTAAATCACGACG | 59 | 91 |
| U | GATGTTTGGGGTGTAGTGGTTGTT | CCACCCCAATACTAAATCACAACA | 59 | 97 | |
| HIC-1 | M | TCGGTTTTCGCGTTTTGTTCGT | AACCGAAAACTATCAACCCTCG | 64 | 95 |
| U | TTGGGTTTGGTTTTTGTGTTTTG | CACCCTAACACCACCCTAAC | 62 | 118 |
m, methylated sequence; u, unmethylated sequence.
Immunohistochemistry
Sections 5-μm thick were cut from representative paraffin blocks of each case and mounted on 2% aminopropyltriethoxysilane-coated glass slides. To avoid antigen loss after preparation, immunostaining was carried out within 48 hours. After microwave pretreatment for antigen retrieval, primary antibodies for E-cadherin (monoclonal, at a dilution 1:150; Transduction Laboratories, Lexington, KY), p16 (monoclonal, at a dilution 1:100; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), TIMP3 (monoclonal, at a dilution 1:100; Oncogene Research Products, Boston, MA) were added and incubated overnight at 4°C. Biotin-labeled anti-mouse immunoglobulin (Ig) G (Dako, Glostrup, Denmark) was used as secondary antibody. Diaminobenzidine-hydrogen peroxide was used as chromogen. A light Mayer’s hematoxylin counter-stain was used. Negative controls were prepared by replacing the primary antibody with Tris-buffered saline. According to a previous report, we defined the expression status as normal or reduced if ≥80%, or <80% of trophoblast cells displayed positive staining, respectively.21
Quantitative Real-Time RT-PCR
Twelve hydatidiform moles, six subsequently regressed, and six developed gestational trophoblastic neoplasia, were selected for quantitative RT-PCR analysis according to previously published procedures.22 First-strand cDNA was synthesized from DNase-treated total RNA with oligo-dT primer and SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA), followed by PCR amplification using the corresponding gene-specific primers. Primer sequences for p16 are as follows: 5′- TAC GGT CGG AGG CCG ATC CAG GTC-3′ (sense) and 5′- GGG GAT GTC TGA GGG ACC TTC CGC -3′ (antisense). GAPDH (housekeeping gene used as control in RT-PCR study): 5′-CTC AGA CAC CAT GGG GAA-3′ (sense) and 5′-ATG ATC TTG AGG CTG TTG-3′ (antisense). Quantitative real-time RT-PCR was performed in a 25-μl reaction, which included 1 μl of cDNA template, 50 mmol/L KCl, 3 mmol/L MgCl2, 10 mmol/L Tris-HCl, pH 9.0, 0.05 mmol/L of each dNTP, 0.2 mmol/L of each forward and reverse oligo primer, 2.5 U of TaqDNA polymerase (Amersham Biosciences, Piscataway, NJ), and 0.1X SYBR Green I (Molecular Probes, Eugene, OR), using an iCycler iQ Multi Color Real Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA). Each PCR reaction was optimized to ensure that a single PCR product was amplified and no product corresponding to primer-dimer pairs was present. PCR reactions of each template were performed in duplicate in one 96-well plate. The PCR cycling conditions were as follows: 95°C for 3 minutes followed by 50 cycles of three steps at 95°C for 30 seconds, 68°C for 30 seconds, and then 72°C for 30 seconds. The relative fold change method was used to determine the relative quantitative gene expression for p16 compared with the GAPDH.
Statistical Analysis
Statistical analysis was performed using the Statistical Package for Social Science 10.1 for Windows (SPSS Inc., Chicago, IL). Differences between groups were analyzed by the χ2 test or Fisher’s exact test where applicable. Quantitative real time RT-PCR data were analyzed by Wilcoxon test. Regression analysis was performed to test the correlation between the methylation status and the clinical parameter. P values less than 0.05 was considered statistically significant. All statistical tests were calculated in two-sided.
Results
MSP
We have analyzed the methylation status of the promoter region of six genes (p16, E-cadherin, TIMP3, DAPK, GSTP1, and HIC-1) in choriocarcinoma, hydatidiform mole, and normal placenta. In normal placenta, only unmethylated alleles could be detected for all of the six investigated genes.
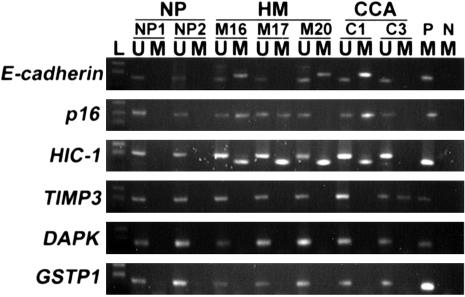
The results of MSP in choriocarcinoma and hydatidiform mole were shown in Table 2. Representative examples of the MSP analysis were shown in Figure 1. At least one gene was methylated in 64% (38 of 59) of the GTD samples including 61% (33 of 54) of hydatidiform mole and 100% (5 of 5) of choriocarcinoma. Four genes showed aberrant methylation in choriocarcinoma at the frequencies of 40% (2 of 5) for E-cadherin, p16, TIMP3, and 60% (3 of 5) for HIC-1. Three genes showed aberrant methylation in hydatidiform mole at frequencies of 13.0% (7 of 54) for E-cadherin, 20.4% (11 of 54) for p16 and 40.7% (22 of 54) for HIC-1. No significant correlation was found between methylation status of E-cadherin, p16, and HIC-1 (P > 0.05).
Table 2.
Methylation Profiles of the Investigated Genes in Hydatidiform Moles and Choriocarcinomas
| Code | Diagnosis | Patient’s age (years) | Gestational age (weeks) | E-cadherin | HIC-1 | p16 | TIMP3 | DAPK | GSTP1 |
|---|---|---|---|---|---|---|---|---|---|
| M3 | CM, GTN | 31 | 22 | ▪ | |||||
| M4 | CM | 23 | 9 | ||||||
| M5 | CM | 28 | 11 | ||||||
| M6 | PM | 44 | 6 | ||||||
| M12 | PM, GTN | 38 | 28 | ||||||
| M14 | CM, GTN | 24 | 16 | ▪ | |||||
| M16 | PM | 45 | 14 | ▪ | ▪ | ▪ | |||
| M17 | CM | 25 | 10 | ▪ | ▪ | ||||
| M20 | CM | 31 | 9 | ▪ | ▪ | ||||
| M24 | CM | 35 | 11 | ||||||
| M25 | PM | 32 | NA | ||||||
| M26 | CM | 32 | 10 | ▪ | |||||
| M27 | CM | 34 | 12 | ||||||
| M28 | CM, GTN | 35 | 12 | ||||||
| M30 | PM, GTN | 30 | NA | ▪ | |||||
| M31 | CM, GTN | 27 | 11 | ▪ | |||||
| M32 | CM, GTN | 45 | 16 | ▪ | ▪ | ||||
| M33 | CM | 51 | 16 | ||||||
| M34 | CM | 24 | NA | ||||||
| M35 | CM | 21 | NA | ||||||
| M42 | CM | 45 | NA | ▪ | |||||
| M48 | CM | 28 | NA | ||||||
| M49 | PM | 31 | 15 | ||||||
| M50 | CM, GTN | 35 | 18 | ▪ | ▪ | ||||
| M51 | CM | 40 | NA | ▪ | |||||
| M52 | CM | 16 | 7 | ||||||
| M54 | CM | 48 | NA | ||||||
| M55 | CM | 24 | 17 | ||||||
| M56 | PM | 29 | NA | ▪ | |||||
| M57 | CM, GTN | 30 | NA | ▪ | ▪ | ||||
| M61 | PM | 37 | 14 | ▪ | |||||
| M62 | CM | 33 | NA | ||||||
| M63 | CM | 25 | 10 | ▪ | |||||
| M64 | CM, GTN | 44 | NA | ▪ | ▪ | ||||
| M66 | CM, GTN | 43 | NA | ▪ | ▪ | ||||
| M67 | CM, GTN | 37 | 11 | ▪ | |||||
| M70 | CM | 34 | NA | ||||||
| M71 | CM | 25 | 18 | ||||||
| M72 | CM | 19 | 13 | ▪ | |||||
| M73 | CM | 18 | NA | ||||||
| M74 | CM | 29 | NA | ▪ | |||||
| M77 | CM, GTN | 38 | 12 | ▪ | |||||
| M78 | PM | 39 | 34 | ▪ | |||||
| M79 | PM | 32 | 7 | ▪ | |||||
| M80 | PM | 29 | 9 | ▪ | |||||
| M81 | CM | 23 | 19 | ||||||
| M82 | CM | 30 | NA | ▪ | |||||
| M89 | PM | 42 | 6 | ▪ | |||||
| M91 | CM, GTN | 24 | 9 | ▪ | |||||
| M92 | CM | 33 | 13 | ▪ | |||||
| M95 | PM | 50 | 12 | ▪ | |||||
| M99 | PM | 35 | NA | ||||||
| M100 | CM | 31 | NA | ▪ | |||||
| M102 | PM, GTN | 35 | 19 | ||||||
| C1 | CCA | 33 | NA | ▪ | ▪ | ▪ | |||
| C2 | CCA | 42 | NA | ▪ | ▪ | ||||
| C3 | CCA | 39 | NA | ▪ | |||||
| C4 | CCA | 28 | NA | ▪ | |||||
| C5 | CCA | 37 | NA | ▪ | ▪ |
Filled boxes, methylated results; open boxes, unmethylated results; CM, complete mole; PM, partial mole; GTN, subsequent development of gestational trophoblastic neoplasia; NA, not available.
Figure 1.
Representative examples of MSP findings in normal placenta (NP), hydatidiform mole (HM), and choriocarcinoma (CCA). The PCR products in the lanes marked U showed the presence of unmethylated alleles of each gene, whereas the products in the lanes marked M indicate the presence of methylated alleles. L, size marker (pBSK/MSP I DNA marker); P, positive control; N, negative control.
The findings regarding correlation between methylation status of these genes, in isolation or combination, with subsequent development of gestational trophoblastic neoplasia was tabulated in Table 3. The adjusted P value for multiple testing was estimated by the Bonferroni procedure α’ = 1 − (1 − 0.05)1/7 = 0.007.
Table 3.
Correlation of Methylation Status in Hydatidiform Moles with the Development of Gestational Trophoblastic Neoplasia (GTN)
| Gene | Status | Regressive moles
|
GTN
|
P-value (Fisher or *Chi-square) | OR, 95% CI | ||
|---|---|---|---|---|---|---|---|
| Freq | (%) | Freq | (%) | ||||
| E–cad HIC-1 p16 | U | 20 | 51.3% | 3 | 20.0% | ||
| M
|
19
|
48.7%
|
12
|
80.0%
|
0.037* | 4.211, 1.026 to 17.288 | |
| Total | 39 | 15 | |||||
| E–cad p16 | U | 34 | 87.2% | 4 | 26.7% | ||
| M
|
5
|
12.8%
|
11
|
73.3%
|
<0.0005 | 18.7, 4.256 to 82.161 | |
| Total | 39 | 15 | |||||
| E–cad HIC-1 | U | 21 | 53.8% | 8 | 53.3% | ||
| M
|
18
|
46.2%
|
7
|
46.7%
|
0.973* | ||
| Total | 39 | 15 | |||||
| HIC1 p16 | U | 21 | 53.8% | 4 | 26.7% | ||
| M
|
18
|
46.2%
|
11
|
73.3%
|
0.073 | 3.208, 0.869 to 11.845 | |
| Total | 39 | 15 | |||||
| E–cad | U | 36 | 92.3% | 11 | 73.3% | ||
| M
|
3
|
7.7%
|
4
|
26.7%
|
0.084 | ||
| Total | 39 | 15 | |||||
| HIC-1 | U | 22 | 56.4% | 10 | 66.7% | ||
| M
|
17
|
43.6%
|
5
|
33.3%
|
0.492* | ||
| Total | 39 | 15 | |||||
| p16 | U | 36 | 92.3% | 7 | 46.7% | ||
| M
|
3
|
7.7%
|
8
|
53.3%
|
0.001 | 13.714, 2.898 to 62.899 | |
| Total | 39 | 15 | |||||
Those hydatidiform moles, which subsequently developed gestational trophoblastic neoplasia, were found to have significantly more frequent p16 promoter hypermethylation comparing to those that subsequently regressed (P = 0.001, Fisher’s exact test, OR = 13.714, 95% CI = 2.898 to 62.899). The sensitivity and specificity of p16 hypermethylation in predicting subsequent development of gestational trophoblastic neoplasia were 53.5% (8 of 15) and 92.3% (36 of 39), respectively. Methylation status of E-cadherin and HIC-1 by themselves showed no such correlation (P = 0.084 and 0.492, respectively). However, when the E-cadherin and p16 methylation status were analyzed together, statistically significant correlation between promoter hypermethylation of either or both genes and development of gestational trophoblastic neoplasia was demonstrated (P < 0.0005, Fisher’s exact test, OR = 18.7, 95% CI = 4.256 to 82.161). Moreover, the sensitivity and specificity in predicting subsequent development of gestational trophoblastic neoplasia became 73.5% (11 of 15) and 87.2% (34 of 39), respectively. On the other hand, combined analysis on the E-cadherin, p16, and HIC-1 methylation status showed no significant correlation to the development of gestational trophoblastic neoplasia (P = 0.037, more than adjusted P–value 0.013, Fisher’s exact test) with sensitivity and specificity of 80% (12 of 15) and 51.3% (20 of 39), respectively.
Regression analysis showed that E-cadherin, HIC-1, and p16 promoter methylation neither correlated with patients’age (P = 0.12, 0.24, and 0.46, the adjusted R2 = 0.028, 0.0080, and 0, respectively) nor with the gestational age of hydatidiform moles (P = 0.92, 0.97, and 0.39, respectively, the adjusted R2 = 0) in our studied samples.
Methylated alleles of DAPK and GSTP1 cannot be detected in either choriocarcinoma or hydatidiform mole. Aberrant methylation of TIMP3 was detectable in choriocarcinoma but not in hydatidiform mole.
Immunohistochemical Analysis
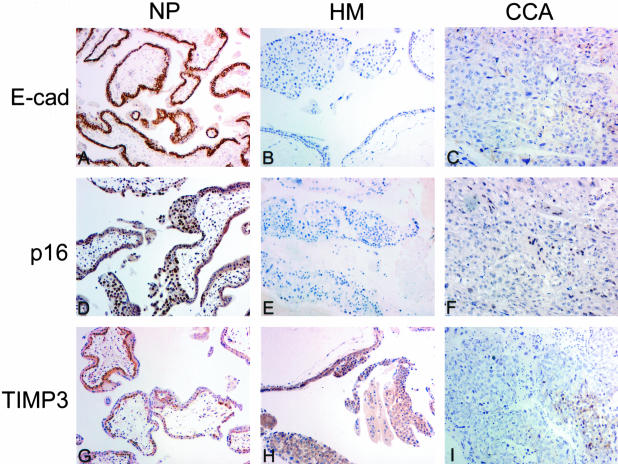
To determine whether the observed aberrant methylation was associated with gene silencing, we examined the expression of p16, E-cadherin, and TIMP3 in 54 hydatidiform moles and five choriocarcinomas using immunohistochemistry. P16 immunoreactivity was mainly demonstrated in the nuclei of the trophoblasts while TIMP3 and E-cadherin expression was predominantly found at the cytoplasm and cell membrane. Reduced expression was defined if less than 80% trophoblastic cells showed positive staining.21 Table 4 and Figure 2 showed the findings for the immunohistochemical staining of these three proteins and their relationship with the MSP results. A close correlation was noted between hypermethylation of p16, E-cadherin, and TIMP3 and reduced expression of the corresponding protein (P < 0.001).
Table 4.
Correlation of MSP Results with Protein Expression in Hydatidiform Moles and Choriocarcinomas
| Protein | Expression status | MSP results
|
P–value | |
|---|---|---|---|---|
| M | U | |||
| E-cadherin | Normal | 0 | 44 | <0.001 |
| Reduced | 9 | 6 | ||
| p16 | Normal | 2 | 42 | <0.001 |
| Reduced | 11 | 4 | ||
| TIMP3 | Normal | 0 | 57 | <0.001 |
| Reduced | 2 | 0 | ||
Figure 2.
Immunostaining of E-cadherin, p16, and TIMP3 in normal placenta (NP), hydatidiform mole (HM), and choriocarcinoma (CCA). All three proteins were strongly expressed in normal placentas, but significantly reduced in choriocarcinomas showing hypermethylation. In hydatidiform moles showing hypermethylation of E-cadherin (M50) or p16 (M31), expression of corresponding proteins was reduced. Expression of TIMP3 was strong in hydatidiform mole.
Quantitative Real-Time RT-PCR
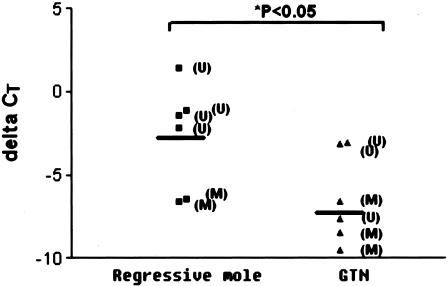
Six cases of hydatidiform mole which had subsequently developed gestational trophoblastic neoplasia, and six cases which had subsequently regressed, were further analyzed by quantitative real-time RT-PCR to explore the RNA expression of p16. The expression levels of p16 in these 12 cases were shown in Figure 3 and concomitant to the immunostaining results. The mRNA level of p16 in methylated cases was significantly less than in those without methylation (P < 0.01). Moreover, in hydatidiform mole cases that subsequently developed gestational trophoblastic neoplasia, the expression of p16 was significantly lower than in hydatidiform moles with spontaneous remission (P = 0.026).
Figure 3.
Quantitative real-time RT-PCR analysis of p16 expression in regressive mole and those developed gestational trophoblastic neoplasia (GTN) as illustrated by the ratio of signals from p16 and GAPDH housekeeping gene, measured by threshold cycle value (ΔCT). Expression of p16 was significantly lower in GTN than in regressive mole.
Discussion
Accumulating evidence suggests that changes in gene expression through epigenetic mechanism play important roles for tumor progression. Hydatidiform mole is associated with an increased risk of subsequent development of gestational trophoblastic neoplasia and choriocarcinoma but the mechanism by which such aggressive progression is mediated remains unknown. In this study, a relatively large collection of GTD samples and normal placentas was analyzed for methylation status of six genes which have been found to be frequently methylated in other human cancers.21,23 At least one gene was hypermethylated in two-thirds of the hydatidiform mole samples and up to 100% of choriocarcinoma samples. In contrast, no aberrant methylation was detected in 10 normal first trimester placentas at all of the tested genes. This indicated that aberrant CpG island methylation is a frequent and probably disease-restricted event in GTD, including both hydatidiform mole and choriocarcinoma, though more common in choriocarcinoma.
In particular, our data suggested that inactivation of the p16 gene through methylation was more likely to be associated with the malignant transformation of trophoblast. As observed in this study, hypermethylation of p16 occurred more frequently among patients with gestational trophoblastic neoplasia or choriocarcinoma than those with regressed disease.
We also demonstrated that p16 promoter hypermethylation by itself was relatively specific in predicting gestational trophoblastic neoplasia development while combined analysis of p16 and E-cadherin methylation status was more sensitive in such prediction. While methylation status of E-cadherin alone showed no statistically significant correlation with clinical outcome, it improves the sensitivity of predicting gestational trophoblastic neoplasia when analyzed together with p16 methylation status. The methylation of E-cadherin may thus still play a role in the progress of hydatidiform mole. Further studies on may be able to elucidate the mechanisms. On the other hand, combined analysis on the E-cadherin, p16, and HIC-1 methylation status showed no significant correlation to the development of gestational trophoblastic neoplasia. HIC-1 methylation status may not play a role in such disease progress.
p16 encodes a protein belonging to the cyclin-dependent-kinase-inhibit family, inhibits Cdk4/6 and cyclin D association, thus prevents the cell entering the S phase.24 Aberrant methylation of p16 associated with a loss of expression in tumor cells was first reported by Herman et al20 This phenomenon may be analogous to homozygous deletion, leading to reduced p16 expression and a selective growth advantage for tumor cells. Methylation silencing of p16 was found to be frequent in some types of tumors, including colon, prostate, esophageal, and pancreatic carcinomas.25,26,27,28 In normal placenta, p16 remains unmethylated, as confirmed in the present report.29 In GTD, our findings corroborated previous immunohistochemical study in which reduction of p16 expression was observed in malignant trophoblastic tumors.30 However, this is the first report evaluating the methylation status of p16 and its clinical significance. In this study, we documented a statistically significant correlation between methylation status and expression level of p16 in GTD.
Little is known about the molecular events involved in the pathogenesis of choriocarcinoma. Genes involved in the control of metastatic potential in human cancers, such as nm23, Kiss-1, and E-cadherin have been reported to be down-regulated in choriocarcinoma.5,31,32,13 Recently, reduced expression of TFPI-2 due to promoter hypermethylation was observed in choriocarcinoma cells.15 Hypermethylation of TIMP3, which was detected in choriocarcinoma but not in hydatidiform mole in our current study, might be a late event involved in the development and progression of choriocarcinoma. Interestingly, both TFPI-2 and TIMP3 effectively decreased the activation of metalloproteinases, thus inhibiting degradation of the extracellular matrix and reducing the invasive potential of tumor cells.15,33 In addition, TIMP3 also has some unique properties in human cancer development, such as growth suppression,34 inhibition of angiogenesis,35,36 and induction of apoptosis.37,38 Transcriptional inactivation of TIMP3 by hypermethylation has been found to associated with a more malignant and invasive phenotype in some human cancer cells.21,39,40 Therefore, down-regulation of TIMP3 in choriocarcinoma might be involved in inactivation of several tumor suppressive mechanisms. The accurate roles of TIMP3 in malignant trophoblast remained to be uncovered by further study.
The HIC-1 gene, located on chromosome 17p13.3, is unusual in that the entire gene is contained in a CpG-rich region. Previous studies showed that HIC-1 was commonly hypermethylated and transcriptionally inactivated in leukemia, medulloblastoma, and cancers of breast, prostate, and uterine cervix.23,41,42,43,44 In the present study, HIC-1 was the most frequently methylated gene among the panel of genes studied in both hydatidiform moles as well as in choriocarcinoma. This indicated that methylation of HIC-1 might be an early event involved in the pathogenesis of GTD.
Regarding DAPK and GSTP1, we could not detect hypermethylation in either gene in the GTD samples examined. Although these genes were frequently methylated in other tumor types, they were apparently not the targets for methylation in GTD. Similar organ-specific methylation patterns have been reported.21,45,46
Recent improvements in the quality of sonography have resulted in the earlier detection of abnormal pregnancy and earlier evacuation, thus increasing the chances of encountering difficulties in histological distinction between hydatidiform mole and a non-molar abortion exhibiting hydropic change and trophoblast hyperplasia.4 Given the relative high rate of methylation of HIC-1 and to a lesser extent, p16 and E-cadherin in hydatidiform mole, detection of aberrant methylation of these genes by MSP might be an adjunct tool in differentiating molar and non-molar pregnancy.
The careful follow-up of patients with hydatidiform mole has undoubtedly contributed to the dramatic reduction in mortality from choriocarcinoma. However, such follow-up involves frequent serial clinic visits and β-hCG assay, creating burden on both the patients and the community resources. In reality, less than 30% of hydatidiform mole patients need such an intensive care. In addition, much debate exists on the timing regarding initiation of chemotherapy for women at risk for gestational trophoblastic neoplasia. If the patients who will develop gestational trophoblastic neoplasia could be identified at the time of the original diagnosis, there would be a significant benefit in reduction of unnecessary follow-up. The outcome of hydatidiform mole can be evaluated using MSP and immunohistochemical studies on candidate genes immediately after evacuation and the selection of high-risk patients and initiation of prophylactic chemotherapy can thus be guided. Our data suggested that p16 may be a candidate deserving further study.
In conclusion, using candidate genes approach, we found that hypermethylation of tumor suppressor genes frequently occurred in choriocarcinoma and hydatidiform mole. This report provides preliminary evidence that hypermethylation of p16 in relation to its decreased expression might serve as a potential marker predicting subsequent development of gestational trophoblastic neoplasia. However, more conclusive interpretation of our observation needs to be confirmed in further studies involving larger sample size and preferably quantitative methylation study.
Footnotes
Supported by a grant from the Research Grants Council of the Hong Kong Special Administrative Region, China (RGC Project No. HKU 7281/00M) and the Conference and Research Council Grant from the University of Hong Kong.
W.-C.X. and K.Y.K.C. contributed equally to the study.
References
- Paradinas FJ, Elston CW. Gestational trophoblastic diseases. Fox H, Wells M, editors. Edinburgh: Churchill Livingstone; Haines & Taylor Obstetrical and Gynaecological Pathology. 2003:1359–1430. [Google Scholar]
- Shih IM, Mazur MT, Kurman RJ. Gestational trophoblastic disease and related lesion. Kurman RJ, editor. New York: Springer Verlag; Blaustein’s Pathology of the Female Genital Tract. 2002:1193–1247. [Google Scholar]
- Li HW, Tsao SW, Cheung AN. Current understandings of the molecular genetics of gestational trophoblastic diseases. Placenta. 2002;23:20–31. doi: 10.1053/plac.2001.0744. [DOI] [PubMed] [Google Scholar]
- Cheung AN. Pathology of gestational trophoblastic diseases. Best Pract Res Clin Obstet Gynaecol. 2003;17:849–868. doi: 10.1016/s1521-6934(03)00094-4. [DOI] [PubMed] [Google Scholar]
- Yang X, Zhang Z, Jia C, Li J, Yin L, Jiang S. The relationship between expression of c-ras, c-erbB-2, nm23, and p53 gene products and development of trophoblastic tumor and their predictive significance for the malignant transformation of complete hydatidiform mole. Gynecol Oncol. 2002;85:438–444. doi: 10.1006/gyno.2002.6652. [DOI] [PubMed] [Google Scholar]
- Kim YT, Cho NH, Ko JH, Yang WI, Kim JW, Choi EK, Lee SH. Expression of cyclin E in placentas with hydropic change and gestational trophoblastic diseases: implications for the malignant transformation of trophoblasts. Cancer. 2000;89:673–679. doi: 10.1002/1097-0142(20000801)89:3<673::aid-cncr24>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Fulop V, Colitti CV, Genest D, Berkowitz RS, Yiu GK, Ng SW, Szepesi J, Mok SC. DOC-2/hDab2, a candidate tumor suppressor gene involved in the development of gestational trophoblastic diseases. Oncogene. 1998;17:419–424. doi: 10.1038/sj.onc.1201955. [DOI] [PubMed] [Google Scholar]
- Stahle-Backdhal M, Inoue M, Zedenius J, Sandstedt B, DeMarco L, Flam F, Silfversward C, Andrade J, Friedman E. Decreased expression of Ras GTPase activating protein in human trophoblastic tumors. Am J Pathol. 1995;146:1073–1078. [PMC free article] [PubMed] [Google Scholar]
- Cheung AN, Zhang DK, Liu Y, Ngan HY, Shen DH, Tsao SW. Telomerase activity in gestational trophoblastic disease. J Clin Pathol. 1999;52:588–592. doi: 10.1136/jcp.52.8.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu PM, Ngan YS, Khoo US, Cheung AN. Apoptotic activity in gestational trophoblastic disease correlates with clinical outcome: assessment by the caspase-related M30 CytoDeath antibody. Histopathology. 2001;38:243–249. doi: 10.1046/j.1365-2559.2001.01065.x. [DOI] [PubMed] [Google Scholar]
- Wong SY, Ngan HY, Chan CC, Cheung AN. Apoptosis in gestational trophoblastic disease is correlated with clinical outcome and Bcl-2 expression but not Bax expression. Mod Pathol. 1999;12:1025–1033. [PubMed] [Google Scholar]
- Vegh GL, Selcuk TZ, Fulop V, Genest DR, Mok SC, Berkowitz RS. Matrix metalloproteinases and their inhibitors in gestational trophoblastic diseases and normal placenta. Gynecol Oncol. 1999;75:248–253. doi: 10.1006/gyno.1999.5564. [DOI] [PubMed] [Google Scholar]
- Xue WC, Feng HC, Tsao SW, Chan KY, Ngan HY, Chiu PM, MacCalman CD, Cheung ANY. Methylation status and expression of E-cadherin and cadherin-11 in gestational trophoblastic diseases. Int J Gynecol Cancer. 2003;13:879–888. doi: 10.1111/j.1525-1438.2003.13400.x. [DOI] [PubMed] [Google Scholar]
- Arima T, Matsuda T, Takagi N, Wake N. Association of IGF2 and H19 imprinting with choriocarcinoma development. Cancer Genet Cytogenet. 1997;93:39–47. doi: 10.1016/s0165-4608(96)00221-x. [DOI] [PubMed] [Google Scholar]
- Hube F, Reverdiau P, Iochmann S, Rollin J, Cherpi-Antar C, Gruel Y. Transcriptional silencing of the TFPI-2 gene by promoter hypermethylation in choriocarcinoma cells. Biol Chem. 2003;384:1029–1034. doi: 10.1515/BC.2003.115. [DOI] [PubMed] [Google Scholar]
- Kanduri C, Kanduri M, Liu L, Thakur N, Pfeifer S, Ohlsson R. The kinetics of deregulation of expression by de novo methylation of the h19 imprinting control region in cancer cells. Cancer Res. 2002;62:4545–4548. [PubMed] [Google Scholar]
- Lai CYL, Chan KY, Khoo US, Ngan HY, Xue WC, Chiu PM, Tsao SW, Cheung ANY. Analysis of gestational trophoblastic disease by genotyping and chromosome in situ hybridization. Mod Pathol. 2004;17:40–48. doi: 10.1038/modpathol.3800010. [DOI] [PubMed] [Google Scholar]
- Wong LC, Ngan HY, Cheng DK, Ng TY. Methotrexate infusion in low-risk gestational trophoblastic disease. Am J Obstet Gynecol. 2000;183:1579–1582. doi: 10.1067/mob.2000.108077. [DOI] [PubMed] [Google Scholar]
- Chan KY, Ozcelik H, Cheung AN, Ngan HY, Khoo US. Epigenetic factors controlling the BRCA1 and BRCA2 genes in sporadic ovarian cancer. Cancer Res. 2002;62:4151–4156. [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kim WH, Jung HY, Yang MH, Kang GH. Aberrant CpG island methylation of multiple genes in intrahepatic cholangiocarcinoma. Am J Pathol. 2002;161:1015–1022. doi: 10.1016/S0002-9440(10)64262-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo AK, Liu Y, Wang XH, Huang DP, Yuen PW, Wong YC, Tsao GS. Alterations of biologic properties and gene expression in nasopharyngeal epithelial cells by the Epstein-Barr virus-encoded latent membrane protein 1. Lab Invest. 2003;83:697–709. doi: 10.1097/01.lab.0000067480.44925.10. [DOI] [PubMed] [Google Scholar]
- Dong SM, Kim HS, Rha SH, Sidransky D. Promoter hypermethylation of multiple genes in carcinoma of the uterine cervix. Clin Cancer Res. 2001;7:1982–1986. [PubMed] [Google Scholar]
- Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific-inhibition of cyclin-D/Cdk4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- Esteller M, Tortola S, Toyota M, Capella G, Peinado MA, Baylin SB, Herman JG. Hypermethylation-associated inactivation of p14(ARF) is independent of p16(INK4a) methylation and p53 mutational status. Cancer Res. 2000;60:129–133. [PubMed] [Google Scholar]
- Konishi N, Nakamura M, Kishi M, Nishimine M, Ishida E, Shimada K. Heterogeneous methylation and deletion patterns of the INK4a/ARF locus within prostate carcinomas. Am J Pathol. 2002;160:1207–1214. doi: 10.1016/S0002-9440(10)62547-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing EP, Nie Y, Song Y, Yang GY, Cai YC, Wang LD, Yang CS. Mechanisms of inactivation of p14ARF, p15INK4b, and p16INK4a genes in human esophageal squamous cell carcinoma. Clin Cancer Res. 1999;5:2704–2713. [PubMed] [Google Scholar]
- Fukushima N, Sato N, Ueki T, Rosty C, Walter KM, Wilentz RE, Yeo CJ, Hruban RH, Goggins M. Aberrant methylation of preproenkephalin and p16 genes in pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Am J Pathol. 2002;160:1573–1581. doi: 10.1016/S0002-9440(10)61104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian YS, Yan P, Osterheld MC, Fontolliet C, Benhattar J. Promoter methylation analysis on microdissected paraffin-embedded tissues using bisulfite treatment and PCR-SSCP. Biotechniques. 2001;30:66–72. doi: 10.2144/01301st02. [DOI] [PubMed] [Google Scholar]
- Fu C, Zhang Q, Lu F, Zhang Z, Hu Y, Lin L. The expressions of p16, CDK4 and PCNA proteins in trophoblastic tumors. Hunan Yi Ke Da Xue Xue Bao. 1998;23:17–20. [PubMed] [Google Scholar]
- Janneau JL, Maldonado-Estrada J, Tachdjian G, Miran I, Motte N, Saulnier P, Sabourin JC, Cote JF, Simon B, Frydman R, Chaouat G, Bellet D. Transcriptional expression of genes involved in cell invasion and migration by normal and tumoral trophoblast cells. J Clin Endocrinol Metab. 2002;87:5336–5339. doi: 10.1210/jc.2002-021093. [DOI] [PubMed] [Google Scholar]
- Li HW, Cheung AN, Tsao SW, Cheung AL, O WS. Expression of e-cadherin and beta-catenin in trophoblastic tissue in normal and pathological pregnancies. Int J Gynecol Pathol. 2003;22:63–70. doi: 10.1097/00004347-200301000-00013. [DOI] [PubMed] [Google Scholar]
- Bass KE, Li H, Hawkes SP, Howard E, Bullen E, Vu TK, McMaster M, Janatpour M, Fisher SJ. Tissue inhibitor of metalloproteinase-3 expression is up-regulated during human cytotrophoblast invasion in vitro. Dev Genet. 1997;21:61–67. doi: 10.1002/(SICI)1520-6408(1997)21:1<61::AID-DVG7>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Bian J, Wang Y, Smith MR, Kim H, Jacobs C, Jackman J, Kung HF, Colburn NH, Sun Y. Suppression of in vivo tumor growth and induction of suspension cell death by tissue inhibitor of metalloproteinases (TIMP)-3. Carcinogenesis. 1996;17:1805–1811. doi: 10.1093/carcin/17.9.1805. [DOI] [PubMed] [Google Scholar]
- Langton KP, Barker MD, McKie N. Localization of the functional domains of human tissue inhibitor of metalloproteinases-3 and the effects of a Sorsby’s fundus dystrophy mutation. J Biol Chem. 1998;273:16778–16781. doi: 10.1074/jbc.273.27.16778. [DOI] [PubMed] [Google Scholar]
- Qi JH, Ebrahem Q, Moore N, Murphy G, Claesson-Welsh L, Bond M, Baker A, Anand-Apte B. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat Med. 2003;9:407–415. doi: 10.1038/nm846. [DOI] [PubMed] [Google Scholar]
- Ahonen M, Baker AH, Kahari VM. Adenovirus-mediated gene delivery of tissue inhibitor of metalloproteinases-3 inhibits invasion and induces apoptosis in melanoma cells. Cancer Res. 1998;58:2310–2315. [PubMed] [Google Scholar]
- Bond M, Murphy G, Bennett MR, Newby AC, Baker AH. Tissue inhibitor of metalloproteinase-3 induces a Fas-associated death domain-dependent type II apoptotic pathway. J Biol Chem. 2002;277:13787–13795. doi: 10.1074/jbc.M111507200. [DOI] [PubMed] [Google Scholar]
- Kang SH, Choi HH, Kim SG, Jong HS, Kim NK, Kim SJ, Bang YJ. Transcriptional inactivation of the tissue inhibitor of metalloproteinase-3 gene by DNA hypermethylation of the 5′-CpG island in human gastric cancer cell lines. Int J Cancer. 2000;86:632–635. doi: 10.1002/(sici)1097-0215(20000601)86:5<632::aid-ijc5>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Zochbauer-Muller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61:249–255. [PubMed] [Google Scholar]
- Fujii H, Biel MA, Zhou W, Weitzman SA, Baylin SB, Gabrielson E. Methylation of the HIC-1 candidate tumor suppressor gene in human breast cancer. Oncogene. 1998;16:2159–2164. doi: 10.1038/sj.onc.1201976. [DOI] [PubMed] [Google Scholar]
- Melki JR, Vincent PC, Clark SJ. Cancer-specific region of hypermethylation identified within the HIC1 putative tumour suppressor gene in acute myeloid leukaemia. Leukemia. 1999;13:877–883. doi: 10.1038/sj.leu.2401401. [DOI] [PubMed] [Google Scholar]
- Rood BR, Zhang H, Weitman DM, Cogen PH. Hypermethylation of HIC-1 and 17p allelic loss in medulloblastoma. Cancer Res. 2002;62:3794–3797. [PubMed] [Google Scholar]
- Yamanaka M, Watanabe M, Yamada Y, Takagi A, Murata T, Takahashi H, Suzuki H, Ito H, Tsukino H, Katoh T, Sugimura Y, Shiraishi T. Altered methylation of multiple genes in carcinogenesis of the prostate. Int J Cancer. 2003;106:382–387. doi: 10.1002/ijc.11227. [DOI] [PubMed] [Google Scholar]
- Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- Ogi K, Toyota M, Ohe-Toyota M, Tanaka N, Noguchi M, Sonoda T, Kohama G, Tokino T. Aberrant methylation of multiple genes and clinicopathological features in oral squamous cell carcinoma. Clin Cancer Res. 2002;8:3164–3171. [PubMed] [Google Scholar]