Abstract
Infants admitted to neonatal intensive care units for suspicion of bacterial sepsis receive at least two broad-spectrum antibiotics for a minimum of 48 to 72 hours to cover both gram-positive and gram-negative organisms while awaiting blood culture results. On average, bacterial growth becomes detectable within 12 to 24 hours, with an additional 24 to 48 hours required for identification. We have previously described using a 16S rRNA PCR assay for screening neonatal blood for bacterial DNA. Combining PCR with DNA sequencing could prove a faster means of detecting bacteria than culture-based identification. If successful, antibiotic therapy could be appropriately tailored sooner, thus sparing infants the administration of unnecessary antibiotics. Our goal was to assess the potential of pyrosequencing to differentiate between bacteria commonly associated with neonatal sepsis. To begin, full-length sequencing of the 380-bp 16S rRNA amplicons from representative bacteria was conducted (ABI 3100) and several databases queried. These included Staphylococcus sp., Streptococcus sp., Listeria sp., and numerous gram-negative rods. The sequences from clinical isolates were identical to those present in the published databases for the same bacteria. As a result, an informative 15 bases within the 380-bp amplicon was targeted for pyrosequencing following enrichment culture and PCR amplification. A total of 643 bacterial isolates commonly associated with neonatal sepsis, and 15 PCR-positive, culture-positive neonatal whole blood samples were analyzed by pyrosequencing. Results of DNA sequencing and culture identification were compared. In summary, we were successful at using PCR and pyrosequencing together to accurately differentiate between highly diverse bacterial groups.
Diagnosing neonatal sepsis is difficult as signs and symptoms in infants are subtle, and often mimic other medical conditions such as hypothermia, delayed transition, or transient tachypnea.1,2,3,4 If the neonate is infected, then antibiotic therapy is warranted, as sepsis is a life-threatening medical emergency.3,4,5
Nowadays, most blood culturing techniques use automated instrumentation, followed by phenotypic identification of the purified isolate.6 In general, this approach requires 2 to 3 days to complete. Although automated blood culturing is considered the gold standard, even this approach can suffer from low sensitivity due to intermittent seeding of low numbers of bacteria within the blood stream, collection of small blood volumes, or the increasingly common practice of providing intra-partum antibiotics to mothers of high-risk deliveries.7,8,9
As a result, many infants receive antibiotics even though few bacterial infections are documented.2,10,11 In fact, between 5% and 10% of all infants in the U.S. receive systemic antibiotic therapy annually.12 Generally during the initial sepsis evaluation, when the etiological agent is unknown, the infant will receive two broad-spectrum antibiotics. This practice provides coverage for both gram-positive and gram-negative organisms. Unfortunately, this practice can lead to destruction of the infant’s normal gastrointestinal flora and the risk of becoming colonized with drug-resistant microorganisms.13
Designing an approach that could more rapidly differentiate between the blood-borne pathogens commonly associated with neonatal sepsis could result in neonates receiving either fewer doses of unnecessary antibiotics or a more tailored regimen of antibiotics sooner. Molecular techniques such as PCR have been used successfully to diagnose a wide range of infectious agents including bacteria, fungi, virus, and protozoa.14,15
Improving on this approach, beyond generating a positive or negative result, to include sequence information to differentiate between bacterial groups could prove invaluable. Targeting a 16S rRNA gene sequence for this purpose has many advantages; it is present as multiple copies per cell, and contains both highly conserved and variable regions.16,17,18
Pyrosequencing (Biotage, Uppsala, Sweden), a relatively new technology, provides rapid, short-read sequencing of 30 bases in approximately 30 minutes. Other investigators have used this approach to classify, identify, and subtype a variety of bacterial 16S rDNA fragments.19,20,21
The goal here was to assess the potential use of pyrosequencing to distinguish between the different bacterial isolates commonly associated with neonatal sepsis compared to conventional phenotypic identification. Our results indicated that a 15 base sequence generated by pyrosequencing after 16S rRNA PCR analysis consistently differentiated between diverse groups of bacteria commonly associated with neonatal sepsis. The identity by pyrosequencing compared favorably to those determined by conventional phenotypic identification methods. If implemented, combining 16S rRNA PCR screening with pyrosequencing could result in the infant’s antibiotic regimen being tailored sooner and thus help reduce the risks of altering the neonate’s normal intestinal flora and the appearance of emerging bacterial resistance due to the use of unnecessary antibiotics.
Materials and Methods
Bacterial Isolates
The purified bacterial isolates included in this study had been isolated from clinical blood cultures, and were being stored as glycerol stocks at −70°C. These bacteria were not limited to Magee Women’s Hospital (MWH), but includes those from another regional hospital, as well as one from another state. All isolates had been identified using conventional phenotypic analysis.
Patient Population
The patients in this study included neonates admitted to the Magee Women’s Hospital (MWH) NICU for bacterial sepsis evaluations. Each neonatal sepsis evaluation included a blood draw for culture and a complete blood count (CBC). The discarded portions of whole blood remaining after CBC analysis was used for 16S rRNA PCR analysis, and, if PCR-positive, for pyrosequencing. The MWH Investigational Review Board approved this study.
Blood Culturing and Phenotypic Identification
The clinical laboratory uses the BACTEC 9240 (Becton Dickinson, Sparks, MD), for blood culturing. This is an automated blood culture system that uses a fluorescent sensor for detecting microorganisms. The CO2 being produced by actively metabolizing microorganisms is continuously monitored. Whole blood (0.5 to 1.0 ml) was collected from infants in the NICU and inoculated into pediatric-sample-sized, resin-containing blood culture bottles (Peds Plus, Becton Dickinson). The bottles were incubated immediately on receipt in the microbiology laboratory in accordance with the manufacturer’s recommendation.
Fluid from a blood culture bottle that was flagged by the BACTEC 9240 instrument for detectable bacterial growth was gram-stained, and subcultured on the appropriate agar-based culture plates. Purified colonies were analyzed either by an automated identification system (MicroScan Dade, West Sacramento, CA) or using the appropriate individual test reagent necessary for phenotypic identification.
Whole Blood Specimen Collection, Pre-Enrichment, and Preparation for PCR Analysis
Specimen collection and processing were described previously.17 Briefly, the discarded portion of whole blood collected in a pediatric-sized purple top tube (EDTA) for CBC analysis was used. The average blood volume was 250 μl, but ranged between 100 μl to 600 μl. These samples are routinely collected in either one of two ways, direct from an arterial line, or from a heel stick. The method used to collect the individual neonatal blood samples for this study was not noted.
After the sample volume was noted, the contents of the tube was added directly to 4 ml of tryptic soy broth (TSB; Invitrogen, Carlsbad, CA). The inoculated TSB was incubated at 37°C with continuous shaking for between 5 hours and 10 hours. The cellular material was pelleted at 4°C for 5 minutes at 13,000 × g. Red blood cells were lysed using 1 ml 4°C buffer consisting of 0.32 mol/L sucrose, 10 mmol/L Tris-HCl, pH 7.5, 5 mmol/L MgCl2, and 1% Triton X-100. Intact cells were treated for 30 minutes at 37°C with 100 μl phosphate-buffered saline (PBS) containing 50 units mutanolysin (Sigma-Aldrich, St. Louis, MO) before digestion with 10 μl 10 mg/ml proteinase K (PK) (Sigma) for 30 minutes at 70°C. The PK was inactivated in the specimen by heating to 100°C for 10 minutes.
Bacterial DNA Preparation from Purified Colonies for PCR Analysis
A sterile loop was touched to an individual bacterial colony present on an agarose plate. The bacteria on the loop were emulsified in 85 μl PBS and 5 μl (50 units) mutanolysin (Sigma) before incubation at 37°C for 30 minutes. To this mixture, 5 μl of 10 mg/ml PK (Sigma) and 1 μl 10% sodium dodecyl sulfate (SDS) (Sigma) were added before incubation for 30 minutes at 37°C, followed by heating at 95°C for 10 minutes. A 1-μl volume of this prepared bacterial DNA was added to 99 μl of master mix for amplification of the 380-bp 16S rRNA target.
16S rRNA PCR Assay
The 16S rRNA PCR assay, which consistently detects 10 to 50 colony forming units/ml was previously described.17 Briefly, the well-characterized primers, RW01 and DG-74,22 were used to amplify a 380-bp (bp) fragment that resides within a conserved region of the bacterial 16S rRNA gene that is flanked by variable regions V8 and V9. The master mix consisted of 10 mmol/L Tris-HCl, pH 8.3, 50 mmol/L MgCl2, 200 μmol/L each dATP, dCTP, dUTP, and dGTP, 1 unit (U) of UNG (Applied Biosystems, Foster City, CA), 25 μmol/L of each primer and 2.5 U Taq polymerase (Promega, Madison, WI). Ten μl of the prepared specimen were added to 90 μl of master mix. After 30 cycles of amplification 20 μl of material were analyzed by agarose gel electrophoresis. The ethidium-stained gel was evaluated for the presence of the 380-bp DNA fragment compared to a 100-bp ladder molecular weight standard (BioWhitaker, Rockland, ME).
DNA Dot Blot Hybridization
DNA dot blot hybridization analysis was carried out on all PCR-positive specimens to determine the nature of the amplified bacteria compared to the gram stain results (Table 1). Individually labeled oligonucleotide probes, RW03 (gram positive), DL04 (gram negative) and RDR245 (universal probe), all previously described,22 were used to hybridize to the denatured 380-bp PCR product as previously described.17
Table 1.
DNA Sequences of the Various Primers or Probes Used for 16S rRNA PCR Amplification, Pyrosequencing, or Dot Blot Hybridization Reactions
| Oligo-nucleotide name | Oligonucleotide function | DNA sequence (5′−3′) | Base pair location22 |
|---|---|---|---|
| DG74 | PCR primer | AGG AGG TGA TCC AAC CGC A | 1522–1540 |
| RW01 | PCR primer | AAC TGG AGG AAG GTG GGG AT | 1170–1189 |
| RDR245 | Sequencing primer and Dot Blot probe* | GTAC AAG GCC CGG GAA CGT ATT CAC CG | 1369–1395 |
| RW03 | Dot Blot probe† | GAC GTC AAA TCA TCA TGC CCC TTA TGT C | 1190–1217 |
| DL04 | Dot Blot probe‡ | GAC GTA AGG GCC ATG ATG ACT TGA CGTC | 1190–1217 |
, serves as a universal probe recognizing both gram-positive and gram-negative organisms;
, serves as a gram-positive-specific probe;
, serves as a gram-negative-specific probe.
ABI 3100 Full-Length Sequencing of the 380-bp 16S rRNA Amplicon
The 380-bp amplicons from several isolates of each group of bacteria listed in Table 2 were fully sequenced using the ABI 3100 instrument (Applied Biosystems) PCR amplification efficiency of each sample was assessed by agarose gel electrophoresis before sequencing. Once adequate amplification was documented, the 380-bp amplicon was purified using a Centricon centrifugal filter unit (Millipore, Billerica, MA). The purified DNA (10 ng/μl) was sequenced according to the manufacturer’s recommendation. The RW01 and DG74 primers were used as sequencing primers in separate reactions to provide full-length sequences of both strands. Sequence alignment was carried out using Sequencher software (Gene Codes Inc., Ann Arbor, MI).
Table 2.
Sequence Patterns of the First 15 Bases of DNA Generated by Pyrosequencing Using the RDR 245 Sequencing Primer
| Bacterial classification | n | Pyrosequence 5′−3′ fingerprint |
|---|---|---|
| Streptococcus sp.* | 181 | CGG CGT GCT GAT CCG |
| Staphylococcus sp.† | 205 | TAG CAT GCT GAT CTA |
| Listeria sp.‡ | 32 | TGG CAT GCT GAT CCA |
| Pseudomonas sp.§ | 19 | TGA CAT TCT GAT TCA |
| Enteric gram-negative rod I¶ | 99 | TGG CAT TCT GAT CCA |
| Enteric gram-negative rod II|| | 68 | TAG CAT TCT GAT CTA |
| Haemophilus sp.** | 39 | CGA CAT TCT GAT TCG |
n, number of isolates tested within a group.
The following represents the breakdown of the types of bacteria within a single group; (n) represents the number of a specific bacterial type included.
Streptococcus sp.; GBS (119), GAS (10), Enterococcus sp. (49), S. pneumoniae (3).
Staphylococcus sp.; S. aureus (70), Coagulase-negative Staphylococcus sp. (135).
Listeria sp. consisted of Listeria monocytogenes (32).
Pseudomonas sp. consisted of Pseudomonas aeruginosa (19).
Enteric gram negative rod I; E. coli (69), Klebsiella oxytoca (22), Enterobacter agglomerans (1), Enterobacter asburiae (5), Enterobacter gergovia (2).
Enteric gram negative rod II; Klebsiella pneumoniae (34), Proteus mirabilis (11), Serratia marcescens (14), Enterobacter cloacae (5), Enterobacter aerogenes (3), Proteus vulgaris (1).
Haemophilus sp. consisted of Haemophilus influenzae (39).
Pyrosequencing Reaction
Pyrosequencing requires that the DNA to be sequenced contain a biotin label. To this end, 10 μl of pre-amplified, unlabeled 16S rRNA PCR master mix were re-amplified to incorporate a biotinylated RW01 primer. A 40-μl volume of each labeled 380-bp PCR product was purified by immobilization onto streptaviden-coated sepharose beads (3 μl) (Amersham, Piscataway, NJ) in the annealing buffer provided by Biotage (Uppsala, Sweden). The unlabeled strand was dissociated and discarded using alkaline denaturation according to the manufacturer’s instructions. The pyrosequencing vacuum prep tool was used to perform the washing (10 mmol/L Tris acetate, pH 7.6) and bead transferring steps. The bead bound biotinylated strand was added to a 96-well microtiter plate along with annealing buffer and the complementary sequencing primer (RDR 245).22 The plate was heated to 95°C for 2 minutes, and allowed to cool on the bench top, at which time the primer could anneal with its target.
Pyrosequencing analysis was performed using a PSQ96 MA and the SQA reagent kit (Biotage) and a cyclic dispensation program of nucleotides according to the manufacturer.20 The plate containing the annealed product was placed into the PSQ 96MA System (Biotage) where the direct sequencing reaction occurred. The pyrosequencing reaction uses a 4-enzyme cascade system to produce a visible light that is read by a CCD camera. The light detected by the camera is displayed as peaks, with the height of the peak being proportional to the number of bases of a specific dNTP that were incorporated during the reaction. All four bases are added individually, one dNTP base at a time with the unincorporated nucleotide being destroyed enzymatically before the next dNTP is added (dATP, dCTP, dTTP, and finally dGTP). The computer provides the reader with both a figure, referred to as a pyrogram, and a text-based sequence.
The resulting bases of sequence generated from pyrosequencing were compared with the same stretch of sequence within the various bacterial 16S rRNA gene sequences generated by us using the ABI 3100 instrument and with that found in GenBank, EMBL Nucleotide Sequence Database, and the DNA Data Bank of Japan23,24 using the BLAST tool25 at the National Center for Biotechnology Information (NCBI). Sequences were aligned using Sequencher software (Gene Codes Inc.).
Results
Full-Length Sequence of the 380-bp Amplicon Generated from Clinical Isolates Was Virtually Identical to Those Same Bacterial Sequence Found in the Queried Databases
Two to three bacteria from each group listed in Table 2 were analyzed using the ABI 3100 to obtain their full-length 16S rRNA 380-bp sequence. This included the following: GBS, Enterococcus sp., S. pneumoniae, S. aureus, coagulase-negative Staphylococcus, E. coli, Klebsiella oxytoca, Enterobacter agglomerans, E. asburiae, E. gergovia, K. pneumoniae, Proteus mirabilis, Serratia marcescens, E. cloacae, E. aerogenes, P. vulgaris, Pseudomonas aeruginosa, Haemophilus influenzae and Listeria monocytogenes. The purpose of this exercise was to compare the 380-bp DNA sequence obtained on clinical isolates to those present in the various databases queried on the same organism. The 380-bp DNA sequences present in the three databases queried were virtually identical to those generated using clinical isolates (data not shown). The rare non-identical base was always outside of those regions representing primers, probes and, most importantly, the informative 15 bases that had been identified. This 15 base region showed complete sequence identity within a bacterial group regardless of whether they were from clinical isolates or those that were sequenced for database submissions. This 15 base region resides within a constant region of the 16S rRNA gene that is flanked by the variable regions V8 and V9. This important finding allowed this region to be targeted for a more extensive comparison using larger numbers of clinical isolates.
DNA Sequence Generated by Pyrosequencing Using RDR 245 Primer Was Useful in Differentiating between Diverse Groups of Bacteria
After finding complete sequence identity between a small number of clinical isolates and those bacterial sequences present within databases, we went on to examine a much larger number of clinical isolates. A total of 643 clinical isolates were analyzed by pyrosequencing for the 15 base sequences immediately downstream of the RDR 245 primer. For the sake of adequate diversity, the bacteria analyzed here came not only from our hospital, but from another hospital in the region as well as from one in another state within the US. The identity of these purified bacterial isolates was determined phenotypically by conventional culture-based methods.
The RDR 245 oligonucleotide served as the sequencing primer in the pyrosequencing reaction. RDR 245 was chosen based on our previous experience with it as a universal probe for detecting a broad diversity of 380-bp 16S rRNA amplicons in Southern blot assays.17,22 Table 2 illustrates the informative 15 base sequence patterns generated from 643 different clinical isolates that among related isolates was identical. This finding allowed us to differentiate between broadly diverse bacterial groups commonly associated with neonatal bacterial sepsis. This included the ability to distinguish between Streptococcus sp., Staphylococcus sp., Listeria sp, Pseudomonas sp., enteric gram-negative rods, and Haemophilus sp.
Pyrosequencing Provided Useful Information on the Identity of the Amplicon Generated by PCR from Neonatal Whole Blood Samples
16S rRNA PCR is routinely performed on neonatal whole blood that has been pre-enriched in TSB for 5 to 10 hours. Over time, we have detected bacterial DNA in 15 clinical samples. These were subsequently analyzed by pyrosequencing, the results of which were compared to the bacterial identification obtained by culture-based methods and by DNA dot blot hybridization results (380-bp amplicon). Table 3 illustrates this comparison. The pyrosequencing classifications of the bacteria were consistent with the culture identification results and with the results obtained using dot blot hybiridzation for all 15 PCR-positive, culture-positive neonatal blood samples analyzed.
Table 3.
Comparison of Results Generated by Pyrosequencing, Culture, and Dot Blot Hybridization for Bacterial Identification from 15 Neonatal Blood Samples
| Study no. | Culture identification* | Dot Blot classification of the 380-bp 16S rRNA† | Pyrosequencing classification of the 15 bases of 16S rRNA‡ |
|---|---|---|---|
| 1612 | GBS | Gram-positive | Streptococcus sp. |
| 2645 | GBS | Gram-positive | Streptococcus sp. |
| 2810 | GBS | Gram-positive | Streptococcus sp. |
| 2346 | Staphylococcus aureus | Gram-positive | Staphylococcus sp. |
| 1877 | CoNS | Gram-positive | Staphylococcus sp. |
| 2213 | CoNS | Gram-positive | Staphylococcus sp. |
| 2226 | CoNS | Gram-positive | Staphylococcus sp. |
| 2356 | CoNS | Gram-positive | Staphylococcus sp. |
| 2520 | CoNS | Gram-positive | Staphylococcus sp. |
| 2524 | CoNS | Gram-positive | Staphylococcus sp. |
| 2526 | CoNS | Gram-positive | Staphylococcus sp. |
| 2564 | CoNS | Gram-positive | Staphylococcus sp. |
| 1905 | E. coli | Gram-negative | Enteric gram-negative rod I |
| 2498 | Enterobacter asburiae | Gram-negative | Enteric gram-negative rod I |
| 2251 | Klebsiella pneumoniae | Gram-negative | Enteric gram-negative rod II |
Microscan or individual biochemical reactions;
, hybridization reactivity with RW03 (gram-positive), DL04 (gram-negative) and RDR 245 (universal) probes;
, first 15 bases of sequence generated by pyrosequencing using the RDR 245 probe as the sequencing primer; GBS, Group B Streptooccus; CoNS, coagulase-negative Staphylococcus sp.
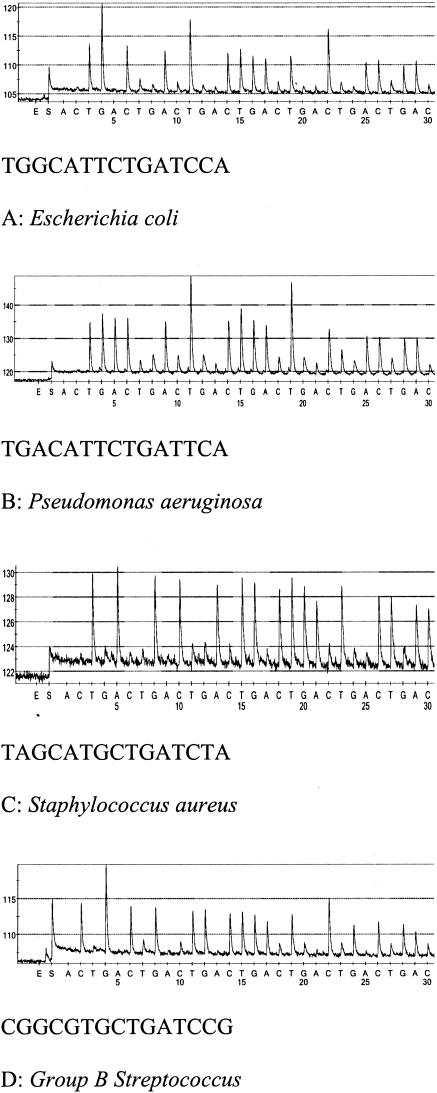
Figure 1, A to D illustrates representative pyrograms and the associated sequences generated in our laboratory on four broadly diverse groups of bacteria that are commonly isolated from neonatal blood cultures. These sequences are identical to those found in Table 2 and illustrated the usefulness of this 15 base fingerprint generated by pyrosequencing for differentiating between diverse bacterial groups.
Figure 1.
A–D: Representative pyrograms and the informative 15 base molecular fingerprint generated from representative bacteria associated with neonatal sepsis.
Discussion
Detecting neonatal sepsis quickly is critical as it is a life-threatening condition. Using an approach that could be more rapid than standard culture and identification techniques for detecting neonatal sepsis would be highly desirable. A molecular-based approach for detecting a highly conserved target like the 16S rRNA gene, which is present among all bacteria associated with septicemia would be useful. Extending the approach beyond providing a “positive-or-negative” result to include a level of discrimination among bacteria would provide invaluable information for physicians making therapeutic management decisions for the neonate.
Applying this approach to the neonatal population is more straightforward than in an adult patient population. There are a rather limited number of bacteria associated with neonatal sepsis, compared to adults. This provides us with a more limited number of sequence comparisons that we need to be concerned with. One must also realize that there is more than one level of specificity built into this approach. The first level is based on the recognition of the RW01 and DG74 primers to generate a 380-bp product. The second level of specificity requires recognition of the RDR 245 primer. It is within this context that the 15 bases of DNA sequence need to be considered. Based on the data presented here, we successfully demonstrated our ability to combine 16S rRNA PCR assay and pyrosequencing to differentiate between broadly diverse bacteria commonly associated with neonatal sepsis compared to the culture-based identification.
This information, if provided to the physician in a timely manner, would allow him/her to appropriately tailor the antibiotic therapy being given to the infected infant sooner than is currently feasible using conventional culture and identification strategies, which require 48 hours to 72 hours to complete.
In laboratory-confirmed cases of neonatal sepsis (true sepsis) at Magee Women’s Hospital the mean time for detecting bacterial growth in blood culture bottles was 18 hours (range, 11 to 28 hours), while the mean time for identifying the purified isolate was 49 hours (range, 23 to 73 hours). Currently physicians may not alter antibiotic therapies being given to neonates suspected of being septic until after the bacterial identification is finalized. For those with clinically suspected cases of sepsis, the no-growth blood cultures are finalized after 5 days of incubation.
Although this combined approach of using PCR and pyrosequencing for detecting bacterial sepsis does not provide definitive bacterial speciation or antibiotic sensitivity testing, it did demonstrate greater discriminating power than the initial gram stain result on the fluid from a positive blood bottle, or a Southern blot result of the 16S rRNA PCR amplicon. The approach of performing rapid, short-read sequencing on a PCR amplicon could provide the physician with valuable information about the etiology of the microorganism sooner than culture, thus enabling them to make more informed decisions sooner about the type(s) of antibiotics to give the infant. Eliminating the need to give the neonate an ineffective or unnecessary broad-spectrum antibiotic sooner would help reduce the risk of destroying the infant’s normal intestinal flora and/or promoting antibiotic resistance. We recognize the importance of trying to exclude the 5 to 10 hours TSB enrichment step from our current protocol if possible, and we are pursuing this goal.
The initial results presented here show promise as a means of rapidly detecting and differentiating among the bacteria commonly associated with neonatal sepsis. However, many more PCR-positive whole blood samples will need to be analyzed before this can be proven.
Footnotes
Supported in full by a grant from the National Institutes of Health National Institute for Child and Health Development R01 HD38559–04 (J.A.J.), and from the generous support of the Jennie Scaife Ovarian Cancer Center within the Scaife Family Foundation for the purchase of the Pyrosequencing PSQ 96 MA Instrument.
References
- Cerase PA. Neonatal sepsis. J Perinatal Neonatal Nursing. 1989;3:48–57. doi: 10.1097/00005237-198910000-00007. [DOI] [PubMed] [Google Scholar]
- Gerdes JS. Clinicopathologic approach to the diagnosis of neonatal sepsis. Clin Perinatol. 1991;18:361–381. [PubMed] [Google Scholar]
- Klein JO, Marcy SM. Bacterial sepsis and meningitis. Infectious Diseases of the Fetus and Newborn. Remington J, Klein J, editors. Philadelphia: W.B. Saunders; 1990:p 601. [Google Scholar]
- Witek-Janusek L, Cusack C. Neonatal sepsis: confronting the challenge. Crit Care Nurs Clin North Am. 1994;6:405–419. [PubMed] [Google Scholar]
- Freedman RM, Ingram DL, Gross I, Ehrenkranz RA, Warshaw JB, Baltimore RS. A half century of neonatal sepsis at Yale: 1928 to 1978. Am J Dis Child. 1981;135:140–144. doi: 10.1001/archpedi.1981.02130260032010. [DOI] [PubMed] [Google Scholar]
- Miller JM, O’Hara CM. Manual and automated systems for microbial identification. Murray PR, Baron EJ, Pfaller MA, Pfaller M, Tenover F, Yolken R, editors. Washington, DC: American Society for Microbiology; Manual of Clinical Microbiology, (ed 7) 1999:193–201. [Google Scholar]
- Brown DR, Kutler D, Rai B, Chan T, Cohen M. Perinatal/neonatal clinical presentation: bacterial concentration and blood volume required for a positive blood culture. J Perinatol. 1995;15:157–159. [PubMed] [Google Scholar]
- Dietzman DE, Fischer GW, Schoenknecht FD. Neonatal Escherichia coli septicemia: bacterial counts in blood. J Pediatr. 1974;85:128–130. doi: 10.1016/s0022-3476(74)80308-2. [DOI] [PubMed] [Google Scholar]
- Washington JA, II, Ilstrup DM. Blood cultures: issues and controversies. Rev Infect Dis. 1986;8:792–802. doi: 10.1093/clinids/8.5.792. [DOI] [PubMed] [Google Scholar]
- Cavaliere TA. Pharmacologic treatment of neonatal sepsis: antimicrobial agents and immunotherapy. JOGNN. 1995;24:647–658. doi: 10.1111/j.1552-6909.1995.tb02547.x. [DOI] [PubMed] [Google Scholar]
- Schuchat A, Whitney C, Zangwill K. Prevention of perinatal group B streptococcal disease: a public health perspective. MMWR Recomm Rep. 1996;45:1–24. [PubMed] [Google Scholar]
- Wegman ME. Annual summary of vital statistics: 1992. Pediatrics. 1993;92:743–754. [PubMed] [Google Scholar]
- Kotloff KL, Blackmon LR, Tenney JH, Rennels MB, Morris JG., Jr Nosocomial sepsis in the neonatal intensive care unit. South Med J. 1989;82:699–704. doi: 10.1097/00007611-198906000-00007. [DOI] [PubMed] [Google Scholar]
- Ehrlich GD, Greenberg SJ. PCR-Based Diagnostics in Infectious Disease. Boston: Blackwell Scientific Publications,; 1994:633–664. [Google Scholar]
- Reischl U. Rapid cycle real-time PCR: methods and applications. Microbiology and Food Analysis. Rischl U, Wittwer C, Cockerill F, editors. Berlin: Springer-Verlag; 2002:1–258. [Google Scholar]
- Bottger EC. Rapid determination of bacterial ribosomal RNA sequences by direct sequencing of enzymatically amplified DNA. FEMS Microbiol Lett. 1989;53:171–176. doi: 10.1016/0378-1097(89)90386-8. [DOI] [PubMed] [Google Scholar]
- Jordan JA, Durso MB. Comparison of 16S rRNA gene PCR and BACTEC 9240 for detection of neonatal bacteremia. J Clin Microbiol. 2000;38:2574–2578. doi: 10.1128/jcm.38.7.2574-2578.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KH, Blitchington RB, Greene RC. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 1990;28:1942–1946. doi: 10.1128/jcm.28.9.1942-1946.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahn N, Olofsson M, Ellnebo-Svedlund K, Monstein HJ, Jonasson J. Identification of mixed bacterial DNA contamination in broad-range PCR amplification of 16S rDNA V1 and V3 variable regions by pyrosequencing of cloned amplicons. FEMS Microbiol Lett. 2003;219:87–91. doi: 10.1016/S0378-1097(02)01190-4. [DOI] [PubMed] [Google Scholar]
- Jonasson J, Olofsson M, Monstein HJ. Classification, identification, and subtyping of bacteria based on pyrosequencing and signature matching of 16S rDNA fragments. APMIS. 2002;110:263–272. doi: 10.1034/j.1600-0463.2002.100309.x. [DOI] [PubMed] [Google Scholar]
- Tarnberg M, Jakobsson T, Jonasson J, Forsum U. Identification of randomly selected colonies of lactobacilli from normal vaginal fluid by pyrosequencing of the 16S rDNA variable V1 and V3 regions. APMIS. 2002;110:802–810. doi: 10.1034/j.1600-0463.2002.1101106.x. [DOI] [PubMed] [Google Scholar]
- Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. GenBank. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. The Molecular Biology Database Collection: 2004 update. Nucleic Acids Res. 2004;32:D3–D22. doi: 10.1093/nar/gkh143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]