Abstract
Combined use of microdissection and high-density oligonucleotide arrays is a powerful technique to study in vivo gene expression. Because microdissection generally yields ng quantities of RNA, RNA amplification is necessary but affects array results. We tested the reliability and reproducibility of oligonucleotide array data obtained from small sample amplified RNA isolated from primary tissues via laser capture microdissection, to determine whether gene expression measurements obtained under these now customary conditions are reliable and reproducible enough to detect authentic expression differences between clinical samples. We performed eight U133A Affymetrix GeneChip oligonucleotide array hybridizations using RNA isolated from a single normal human breast specimen: two standard and six small samples prepared using independent microdissections, RNA isolations, and amplifications. We then performed six array hybridizations using RNA obtained similarly from paired normal epithelium and ductal carcinoma in situ from three independent breast specimens. We determined reliability by analysis of hybridization quality metrics, and reproducibility by analysis of the number of more than twofold changed genes, linear regression, and principal components analysis. All amplified RNA generated good quality hybridizations. From the initial specimen, correlations between replicates (r = 0.96 to 0.99) and between small samples (r = 0.94 to 0.98) were high, and between standard and small samples (r = 0.84) were moderate. In contrast, in the three normal cancer pairs, the differences in gene expression were large among the normal samples, the ductal carcinoma in situ samples, and between normal and ductal carcinoma in situ within each pair. These differences were a much larger source of variability than the technical variability introduced by the processes of laser capture microdissection, small sample amplification, and array hybridization. Nanogram quantities of RNA isolated from primary tissue using laser-capture microdissection generates reliable and reproducible gene expression measurements. These measurements do not mirror those obtained using micrograms of RNA. Biological variability in gene expression between independent specimens, and between histologically distinct samples within a specimen, is greater than the technical variability associated with the procedures. Future studies of in vivo gene expression using this approach will identify functionally important differences within or between specimens.
High-density oligonucleotide array technology is a powerful tool to analyze gene expression. However, standard array protocols generally require 5 to 10 μg of total RNA as starting material to generate sufficient hybridization signal for accurate detection and quantitation of relative RNA levels. This limits analysis of most in vivo samples, which rarely generate μg quantities of RNA. Because in vivo samples usually generate only ng quantities of RNA, techniques for RNA amplification have been developed.1 Amplified anti-sense RNA (aRNA) has been used for cDNA microarray experiments2,3,4,5 and is of sufficient quantity and quality for use with high-density oligonucleotide microarrays.6,7,8,9,10,11,12 Because they can be manufactured with feature densities that are currently impossible to achieve with spotted cDNA arrays, oligonucleotide microarrays allow cost-efficient analysis of gene expression on a genome-wide scale. Oligonucleotide arrays are also particularly attractive for gene expression studies because they can readily distinguish expression levels of closely related gene family members and splice variants.
In addition to material from fine needle aspirates and primary cell culture, many in vivo clinical samples are acquired by microdissection, particularly laser capture microdissection (LCM), which is a widely available technique used to harvest homogeneous cell populations within complex tissues.13 Although the use of LCM is now standard, few studies have tested the reliability and reproducibility of amplified LCM-generated RNA hybridizations on oligonucleotide arrays. Ohyama and colleagues7 have shown that LCM-captured cells can generate sufficient quantity of aRNA (after T7-based amplification) for oligonucleotide arrays. Luzzi and colleagues11 demonstrated that LCM-captured cell-amplified RNA could provide interpretable hybridization results, assessed using ∼1800 genes. This group also compared two independent 30-ng samples of LCM-captured cells-amplified RNA and found ∼4.3% variability between them (fold change >2).6 These studies are highly promising. However, limited information is available about several important issues, including: 1) the reproducibility of gene expression measurements made using LCM-captured cells-amplified RNA and oligonucleotide arrays; 2) the comparability of standard and LCM-captured cell gene expression measurements using these arrays; 3) whether the technical variability introduced into gene expression measurements by the processes of LCM, RNA amplification, and array hybridization is large enough to obscure differences in gene expression between biologically distinct samples. This information is very important because most studies of in vivo tissue will use microdissected, small-sample RNAs.
To address these issues, we performed two series of oligonucleotide array hybridizations. The first was designed to examine the reproducibility and reliability of the technique, and used as starting material RNA from a single primary human breast specimen. Microgram quantities of this RNA were examined by microarray analysis as standard samples, and compared with small-sample (100 ng) quantities that were obtained in a series of independent isolations (with and without LCM) and amplifications. The second experiment was designed to determine whether the technical variability associated with LCM and the small sample protocol would interfere with the identification of differences in gene expression because of biological variation. The starting material for this experiment was small samples of microdissected, paired normal epithelium, and ductal carcinoma in situ (DCIS) from three independent breast specimens.
Materials and Methods
Sample Acquisition
After obtaining Institutional Review Board approval, breast tissue not needed for pathological diagnosis was collected from mastectomy specimens from the Department of Pathology at Boston Medical Center and deidentified. To preserve RNA quality, the tissue was obtained within an hour of surgery and multiple pieces were immediately snap-frozen in liquid nitrogen or embedded in O.C.T. medium (Sakura Finetek, Torrance, CA), and stored at −80°C.
Study Design
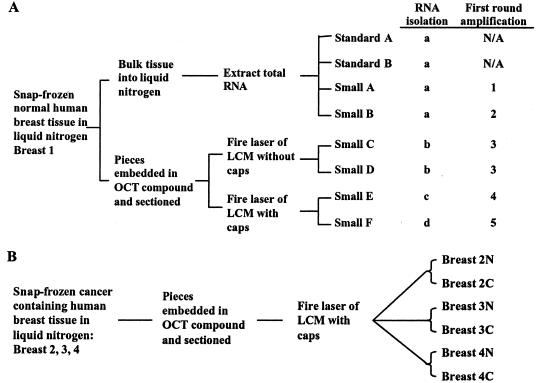
Figure 1 summarizes sample preparation and methods. As shown in Figure 1A, a normal human breast specimen (breast 1), containing a mixture of epithelium, stroma, vasculature, adipocytes, lymphocytes, and possibly other cell types, but no tumor, was snap-frozen. From one piece, total RNA was extracted. Two 5-μg aliquots were processed using the standard Affymetrix protocol of one round of RNA amplification and labeling, and the resulting biotinylated aRNA was hybridized to U133A GeneChip arrays (standard A, standard B). In parallel, two 100-ng aliquots of the same total RNA underwent two rounds of amplification followed by labeling and hybridization (small A, small B). A piece of breast 1 was also embedded in optimal cutting temperature (O.C.T.), sectioned, and microdissected. One set of microdissections used no LCM caps: instead, after laser firing, all tissue elements were scraped from the slides and RNA extracted (small C, small D). The other set of microdissections used caps and RNA was extracted from all tissue elements on the caps (small E, small F). One hundred ng of RNA from each collection method was amplified using the two-round protocol for small samples. For small C and D, a first round amplification was performed on a single RNA aliquot that was then split for two independent second round amplifications. In sum, these samples were analyzed by four independent RNA isolations, five independent RNA amplifications, and eight independent biotin-labelings and hybridizations. As shown in Figure 1B, samples from three different cancer-containing human breast specimens (breast 2, 3, 4) were snap-frozen and embedded in O.C.T. From each specimen, normal epithelium and DCIS were separately microdissected using caps, ie, the protocol used with small E and F. Equal amounts of RNA from each specimen’s normal and cancer pair (breast 2, 43 ng; breast 3, 100 ng; breast 4, 81 ng) were isolated, amplified using the two-round protocol, labeled, and hybridized as described below.
Figure 1.
Schema of tissue acquisition and sample preparation. A: Normal human breast tissue was collected from breast 1 and snap-frozen in liquid nitrogen. In one set of experiments, RNA was directly extracted from pieces of this tissue. Two 5-μg aliquots of RNA (standard A, standard B) were labeled and hybridized according to the standard protocol. Two 100-ng aliquots of RNA (small A, small B) were first amplified and then labeled and hybridized. In the other set of experiments, pieces of snap-frozen tissue were embedded in OCT and sectioned. In small C and small D, the tissue was pulsed with the laser and scraped from the slides (no cap used for collection). In small E and small F, the tissue was acquired as usual, ie, pulsed with the laser through the cap. In small C to small F, RNA was processed in independent amplification, labeling, and hybridization steps. Independent RNA isolations are indicated by a, b, c, and d. Independent first-round amplifications are indicated by 1, 2, 3, 4, and 5. All samples were independently biotin-labeled and hybridized. B: Samples from three different cancer-containing human breast specimens (breast 2, 3, 4) were snap-frozen and embedded in OCT. Paired normal epithelium and DCIS were separately microdissected with caps using the protocol used with small E and small F.
RNA Purification
Total RNA
Snap-frozen pieces of breast 1 were homogenized in a guanidinium isothiocyanate-based buffer. Total RNA was extracted (Micro RNA isolation kit; Stratagene, La Jolla, CA), DNase-treated (Roche Diagnostics Corp., Indianapolis, IN) and quantified (Nanodrop ND-1000 spectrophotometer; NanoDrop Technologies, Inc., Montchanin, DE). RNA quality was assessed by electrophoresing 2 μg on a denaturing 1% agarose gel with SYBR Gold stain (SYBR Gold Nucleic Acid Gel Stain; Molecular Probes, Inc., Eugene, OR).
Microdissected RNA
O.C.T.-embedded tissue was sectioned at 12-μm intervals, mounted on uncoated slides, and immediately returned to −80°C. Subsequently, these sections were fixed in 70% ethanol for 1 minute, lightly stained with dilute Mayer’s hematoxylin and eosin (50% concentration hematoxylin and 10% concentration eosin), and dehydrated using two dips each of 70%, 95%, and 100% ethanol and two 2-minute xylene rinses, followed by air-drying for 2 minutes. In breast 1, all tissue elements on the slides were microdissected (PixCell; Arcturus Engineering, Mountain View, CA). For small E and F, microdissection was performed using the standard technique using caps. For small C and D, microdissection was performed without a cap: the laser was fired onto the tissue, without a cap in place, and all tissue elements were then scraped from the slide. All other specimens (breast 2, 3, 4) were microdissected using caps, as with small E and F. Slides awaiting microdissection were stored briefly under desiccation in a slide box at room temperature. Total RNA was extracted from each sample as described above.
Target Preparation
Standard RNA
Double-stranded cDNA was synthesized from 5 μg of total RNA using a SuperScript double-stranded cDNA synthesis kit (Invitrogen, Carlsbad, CA) and a (dT)24-T7 promoter primer. The products were purified by phenol/chloroform extraction using PLG Heavy (Brinkmann Instruments, Westbury, NY). Biotin-labeled aRNA was generated with a Bioarray High Yield RNA transcription kit (Enzo Diagnostics, Farmingdale, NY), purified on RNeasy affinity columns (Qiagen, Valencia, CA), and quantified on a Nanodrop ND-1000 spectrophotometer. To assess aRNA quality, 200 to 500 ng was electrophoresed on a 1% agarose gel stained with SYBR Gold.
Small Sample RNA
Breast 1:
The first round amplification was performed on 100 ng of total RNA using the MessageAmp aRNA kit (Ambion Inc., Austin, TX) following the manufacturer’s protocol. The in vitro transcription was performed at 37°C for 10 hours and 30 minutes. Between 300 ng to 2 μg aRNA was used in the second round amplification, whose first step uses random-priming rather than oligo-dT. After second round double-stranded cDNA synthesis and purification (MessageAmp aRNA kit, Ambion), in vitro transcription for small C and E was performed using the same reagents, supplemented with biotin-11-CTP and biotin-16-UTP (Enzo Diagnostics) according to the manufacturer’s protocol. For the other four small samples (small A, B, D, F), in vitro transcription was performed using the Bioarray High Yield RNA transcription kit (Enzo Diagnostics).
Breasts 2, 3, 4:
For first round amplification, the MessageAmp aRNA kit was used in breast 2 and the RiboAmp OA RNA amplification kit (Arcturus) was used in breast 3 and 4. All second round amplifications were performed using the MessageAmp aRNA kit (double-stranded cDNA) and Enzo Diagnostics reagents (in vitro transcription), see above section for details.
Hybridization
For each hybridization, 20 μg of biotin-labeled aRNA was fragmented to an average size of 35 to 200 bases by incubating in 40 mmol/L Tris-acetate, pH 8.1, 100 mmol/L KOAc, 30 mmol/L MgOAc, for 35 minutes at 94°C. Ten μg of fragmented RNA, along with hybridization controls supplied by Affymetrix (Santa Clara, CA), was hybridized to each U133A GeneChip array containing probes for 22,283 human genes (Affymetrix). The arrays were hybridized for 16 hours at 45°C and 60 rpm, and then washed and stained according to the standard antibody amplification for eukaryotic targets protocol (Affymetrix). The stained arrays were scanned at 488 nm using a G2500 Scanner (Agilent, Palo Alto, CA).
Data Analysis
The images from the scanned chips were quantified and scaled by using Affymetrix Microarray Suite 5.0 (for detailed information see the statistical algorithms description document at http://www.affymetrix.com/support/technical/whitepapers/sadd_whitepaper.pdf; Affymetrix). Signal intensities from the 22 probes for each gene were used to determine an overall expression level for that gene as well as a detection confidence score. The gene expression levels were linearly scaled to an average of 500 units on each chip. We used the detection confidence score, which is a measure of the sequence specificity of the hybridization intensities, to eliminate genes that are not expressed or not detected in any of the samples and this reduced our dataset from 22,283 genes to 14,377. Studentized extreme-value test statistics, correlation coefficients (r) and χ2-test statistics (X2) were calculated with Excel (Microsoft Corp., Redmond, WA). Principal components analysis of gene z-scores was performed with DecisionSite (Spotfire, Inc., Somerville, MA).
Results and Discussion
RNA, aRNA, and Array Quality
SYBR Gold-stained denaturing agarose gels were run to examine the quality of input RNA and aRNA. Anti-sense RNA generated from the standard samples had an average size of 1000 nucleotides, whereas aRNA from all small samples had an average size of 500 nucleotides (data not shown), indicating that good quality RNA can be obtained reliably from LCM-captured cells.
We compared several parameters of RNA and hybridization quality among our 14 samples, as shown in Table 1. The yield of biotinylated aRNA was, in all cases, sufficient to hybridize to the oligonucleotide array. The ratios of the hybridization intensity from the 3′ and 5′ ends of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript were generally low, although higher in small samples compared with standard. This is consistent with the shorter biotinylated aRNAs from small samples seen on the agarose gels, and is caused by the amplification creating targets that are skewed to the 3′ end of the transcript. We examined raw Q, scaling factor, and percent present call (% present) to evaluate whether the small sample procedure produces high-quality aRNA.14 As seen in Table 1, these measures were similar among standard and small samples, and all indicated good quality targets and hybridizations. None of the yields or scaling factor values were significant outliers (P > 0.05, Studentized extreme-value test), although the 3′/5′ ratio from breast 2N were significantly higher than the other samples (P < 0.05, Studentized extreme-value test). As expected, ∼50% of probesets were called present in most samples. Although the percentage of genes reliably detected in the samples obtained via LCM without a cap (small C and D) are somewhat lower than the other samples, neither of these values are significant outliers relative to the other 12 samples (P > 0.05, Studentized extreme-value test). The percent present probesets appears to vary slightly between specimens and between tissue types. This may reflect biological difference between samples or variable quality of input RNA. In sum, these results indicate that multiple, independently obtained, laser-captured, small sample RNAs can reliably produce sufficient satisfactory aRNA and generate good quality oligonucleotide hybridizations.
Table 1.
Comparison of Samples
| IVT yield (μg)* | 3′/5′ ratio (GAPDH)† | Noise (raw Q)‡ | SF§ | % Probesets present¶ | |
|---|---|---|---|---|---|
| Breast 1 | |||||
| Standard A | 67.8 | 2.68 | 2.74 | 3.7 | 54.9 |
| Standard B | 62.4 | 1.48 | 2.54 | 5.1 | 53.1 |
| Small A | 38.2 | 4.25 | 2.98 | 3.2 | 50.8 |
| Small B | 32.6 | 3.12 | 2.80 | 2.6 | 50.5 |
| Small C | 35.8 | 4.10 | 2.85 | 3.9 | 46.8 |
| Small D | 31.2 | 3.37 | 3.32 | 6.2 | 40.2 |
| Small E | 24.0 | 5.97 | 3.17 | 2.3 | 51.3 |
| Small F | 35.6 | 5.82 | 2.46 | 3.0 | 51.3 |
| Breast 2 | |||||
| Breast 2N | 54.2 | 17.12 | 2.19 | 4.8 | 44.3 |
| Breast 2C | 38.2 | 7.33 | 2.10 | 3.5 | 49.2 |
| Breast 3 | |||||
| Breast 3N | 45.5 | 3.37 | 2.15 | 4.4 | 46.5 |
| Breast 3C | 50.7 | 3.60 | 2.06 | 4.8 | 44.7 |
| Breast 4 | |||||
| Breast 4N | 24.2 | 5.64 | 2.06 | 4.0 | 50.6 |
| Breast 4C | 25.4 | 8.26 | 1.99 | 3.7 | 51.2 |
IVT yield is the amount of purified biotin-labeled aRNA generated.
3′/5′ ratio is the ratio of 3′ probeset signal intensity and 5′ probeset signal intensity of GAPDH transcript.
Noise (raw Q) is the degree of pixel-to-pixel variation among the probe cells used to calculate the background.
Scaling factor (SF) is the multiplier used to adjust the trimmed mean signal of a probe array to a selected target signal value (all arrays were scaled to 500 relative signal intensity units).
% Probesets present is the percentage of probe sets scored detected by Affymetrix Microarray suite 5.0 (MAS.5.0).
N, normal epithelium; C, carcinoma in situ.
Comparison of Replicate Samples (Breast 1)
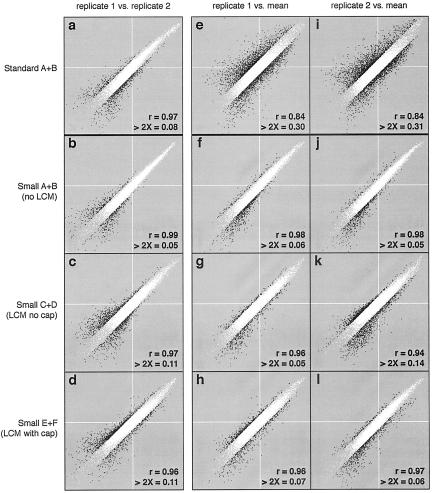
We performed replicates of each standard and small sample to test reproducibility of gene expression measurements in each of the four sample isolation and processing protocols. Independent RNA isolations followed by independent aRNA syntheses and hybridizations were performed (see Materials and Methods: Study Design). Scatterplots show the probeset hybridization intensities for each pair of replicate samples (Figure 2; a to d). The correlation between the replicates of the standard sample (r = 0.97) is similar to the correlation between replicates of any small samples (r = 0.96 to 0.99) (despite standard B’s high raw Q metric). We also determined the fraction of probesets that differ between replicates by more than twofold. This fraction was similar in standard sample replicates (8%) and in small sample replicates (5 to 11%). The majority of probesets with a more than twofold difference between replicates have lower than average hybridization intensity; these differences could primarily be because of the poor signal-to-noise characteristics of weak hybridization intensity measurements. These data demonstrate that pairs of replicates are alike despite differences in sample isolation and processing.
Figure 2.
Comparison of hybridization signal intensities. White lines indicate the mean scaled hybridization intensity. Black dots are probesets that change more than twofold between the samples being compared. The fraction of probesets that change more than twofold, and the correlation coefficient (r), are listed at the bottom right of each graph. The left column of scatterplots (a–d) compares hybridization intensity between replicates within each condition. The middle and right columns compare hybridization intensity of individual samples versus the mean intensity of small samples. Each y axis is the log intensity of the single sample indicated. Each x axis is the log mean intensity of all small samples (except for the small sample on the y axis).
Comparison between Small Samples (Breast 1)
Reproducibility in the expression measurements obtained from the six small samples was evaluated by plotting the signal intensities of probesets from each small sample as a function of the mean signal intensity of all remaining small samples. These results are shown in Figure 2; g to l. The correlation coefficients are high (r = 0.94 to 0.98) and the fraction of probesets showing a more than twofold difference is low (5 to 14%). Of the probesets exhibiting a more than twofold difference in any individual small sample relative to the average of the other small samples 68.4% have a more than twofold lower intensity. The percentage (68.4%) of probesets in any individual sample with a more than twofold lower-than-average intensity is significantly higher than 50% (P ≪ 10−5, X2). This finding suggests that technical variability is not random, and that many of the more than twofold changes within the small-sample group might be because of decreased amplification among low abundance transcripts.
Overall, the variability between all six small samples of breast 1 is similar to the variability between replicates, suggesting that any small sample is as similar to another as each small sample is to its replicate. Thus, the process of LCM, use of independent RNA isolations, and of different kits to amplify and label the RNA do not appear to alter the quality of the isolated RNA in a way that affects the gene expression measurements.
Comparison of Standard versus Small Samples (Breast 1)
To determine the fidelity of hybridization intensity measurements between small sample and standard sample RNA, we compared the hybridization intensity for each probeset from the standard samples with the mean hybridization intensity of the same probeset from all small samples. The results are shown in Figure 2, e and i. The standard and small samples are less correlated (r ∼ 0.84) and ∼31% of genes show more than twofold difference in hybridization intensity. The differences between the standard samples and the small samples appear fairly evenly spread out across the intensity spectrum. These data are consistent with that of others4,8,15,16 and suggest that gene expression measurements obtained from standard and small-sample material cannot be directly compared.
The results from breast 1 lead us to propose that much of the sample-to-sample variation among the small samples is the result of amplification failure among low-abundance transcripts. From our data we cannot determine whether the variability between standard sample replicates also results from amplification failure of low-abundance transcripts. However, because replicate-to-replicate variability is similar between standard and small samples, it is reasonable to suggest that both arise via the same mechanism. The compound effect of amplification failures during two rounds of amplification is likely responsible for a fraction of the probesets that have more than twofold lower hybridization intensity in small compared to standard samples. Additional differences in hybridization intensity between the small and standard samples (Figure 2, e and i), may be because of sequence-specific differences in amplification efficiency resulting from the small sample protocol’s incorporation of a random-primed reverse transcription step (see Materials and Methods).
Comparison of Technical and Biological Variability
Encouraged by the high degree of reproducibility from microdissected and amplified small samples, we wished to determine whether this level of reproducibility is sufficient to permit detection of variation in gene expression because of biological differences between samples. We performed six hybridizations using pairs of normal epithelium and DCIS from three independent specimens (breasts 2, 3, 4). These six samples were obtained using the same methods as small E and F. Typical of in vivo tissues, lesion size and total RNA quantity were limited, so replicates could not be performed.
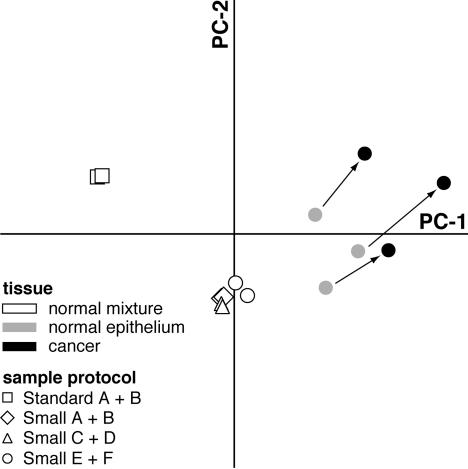
We used principal components analysis to identify the major axes of variability among all 14 arrays. Of the total variation observed among the samples 50.2% is accounted for in the first two principal components of variation. As shown in Figure 3, all six small samples from breast 1 are similar to each other and dissimilar from the two standard samples (which are also similar to each other). The tight clustering of the six small samples from breast 1 indicates that the different sample preparation protocols result in similar measurements of gene expression. The tight clustering is consistent with the low number of differences and high degree of correlation seen between all six small samples (Figure 2; f to h and j to l). The separation between the six small samples and the two standard samples is consistent with the large number of hybridization intensity differences and low degree of correlation seen between the standard and small samples (Figure 2, e and i).
Figure 3.
Principal components analysis. The two major axes along which the 14 samples vary in gene expression hybridization intensity were determined by principal components analysis. Shown here are the distribution of samples along these two axes, which account for 50.2% of the total observed variation. The scale of each axis is equivalent with respect to the variability represented. Shading reflects tissue type and shape reflects sample protocol. Breast 1 samples are represented in white with black borders; breasts 2, 3, and 4 normal epithelial samples are represented in gray, and cancers are in black. This graph shows that measurements of gene expression from breast 1 small samples, that differ only in the technical details of their processing, are much more similar to each other than are any small samples from breasts 2, 3, or 4, which differ from each other biologically. Each specimen’s normal/cancer pair is joined by an arrow.
Importantly, the six small samples from breast 1 are also easily distinguished from each of the small samples of normal tissue from breasts 2, 3, 4. Some of these differences are likely because of the distinct composition of breast 1 (a mixture of cell types) compared to breasts 2, 3, 4 (epithelium). However, breasts’ 2, 3, and 4 normal epithelia are also distinguishable from each other. We posit that much of the difference between breasts’ 2, 3, and 4 normal epithelia results from biological differences between the three specimens, based on the low degree of technical variability seen in independent preparations of a single tissue (ie, breast 1’s small samples). The potential source(s) of this specimen effect on gene expression are numerous (eg, clinical, environmental, genetic).
Consistent with these results, the variation in gene expression within the three DCIS samples also appears larger than what would be expected because of technical variability. The distribution of the DCIS samples relative to each other along the two major axes of variation is similar to the distribution of matched normal epithelial samples from these specimens. This suggests that the specimen effect is consistent between normal epithelium and DCIS. In addition to this specimen effect, a cancer effect distinguishes the normal epithelial samples from the DCIS samples. With enough cases, cancer-specific differences in gene expression could be detected with a paired-sample t-test. These findings are consistent with those of McClintick and colleagues,17 who found biological variability to be greater than technical variability when using the standard protocol in rat liver.
In sum, these findings suggest that technical variability associated with independent LCM preparations, sample processing, and array hybridizations is small relative to differences because of biological variability between specimens and disease states. Thus, oligonucleotide array experiments comparing small sample aRNA between different samples within a specimen, or similar samples between specimens, will likely generate useful, functionally important data that reflect the biological differences between samples.
Summary
The purpose of this study is to test the reliability, reproducibility, and potential utility of gene expression measurements made with oligonucleotide microarrays using small sample amplified RNA obtained from microdissected clinical specimens. In a series of eight oligonucleotide arrays hybridized with RNA isolated from a single human breast specimen, we found that independent microdissections, RNA isolations, and amplifications reliably produced reproducible gene expression measurements. The results obtained with these highly processed independent samples correlated with each other as well as results obtained using the standard protocol that requires 50 times more RNA as starting material. As expected, gene expression measurements from small sample RNA do not mirror measurements from standard sample RNA. Both amplification failure among low-abundance transcripts and sequence-specific differences in amplification efficiency may account for the differences. In a second series of six hybridizations using three normal/cancer pairs, we found specimen-specific and disease state-specific differences in gene expression that appear larger than differences because of technical variability. These findings suggest that the technical variability introduced by LCM, small sample amplification, and array hybridization is smaller than the biological variability between human breast samples. These results indicate that it will be possible to use LCM-isolated primary tissues with oligonucleotide microarrays to uncover gene expression differences associated with homogeneous cell populations in the breast in vivo.
Footnotes
Supported by NIH PHS CA081078, and the Department of Defense Breast Cancer Research Program, DAMD17-01-1- 0159.
C.K. and N.G. contributed equally to this work.
References
- Van Gelder RN, Von Zastrow ME, Yoll A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci USA. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leethanakul C, Patel V, Gillespie J, Pallente M, Ensley JF, Koontongkaew S, Liotta LA, Emmert-Buck M, Gutkind JS. Distinct pattern of expression of differentiation and growth-related genes in squamous cell carcinomas of the head and neck revealed by the use of laser capture microdissection and cDNA arrays. Oncogene. 2000;19:3220–3224. doi: 10.1038/sj.onc.1203703. [DOI] [PubMed] [Google Scholar]
- Luo L, Salunga RC, Guo H, Bittner A, Joy KC, Galindo JE, Xiao H, Rogers KE, Wan JS, Jackson MR, Erlander MG. Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat Med. 1999;5:117–122. doi: 10.1038/4806. [DOI] [PubMed] [Google Scholar]
- Zhao H, Hastie T, Whitfield ML, Borresen-Dale A-L, Jeffrey SS. Optimization and evaluation of T7 based RNA linear amplification protocols for cDNA microarray analysis. BMC Genomics. 2002;3:31. doi: 10.1186/1471-2164-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X-J, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang BM, Zhou Y-X, Varnholt H, Smith B, Gadd M, Chatfield E, Kessler J, Baer TM, Erlander MG, Sgroi DC. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzzi V, Mahadevappa M, Raja R, Warrinto JA, Watson MA. Accurate and reproducible gene expression profiles from laser capture microdissection, transcript amplification, and high density oligonucleotide microarray analysis. J Mol Diagn. 2003;5:9–14. doi: 10.1016/S1525-1578(10)60445-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama H, Zhang X, Kohno Y, Alevizos I, Posner M, Wong DT. Laser capture microdissection-generated target sample from high-density oligonucleotide array hybridization. Biotechniques. 2000;29:530–536. doi: 10.2144/00293st05. [DOI] [PubMed] [Google Scholar]
- Scherer A, Krause A, Walker JR, Sutton SE, Seron D, Raulf F, Cooke MP. Optimized protocol for linear RNA amplification and application to gene expression profiling of human renal biopsies. Biotechniques. 2003;34:546–556. doi: 10.2144/03343rr01. [DOI] [PubMed] [Google Scholar]
- Aoyagi K, Tatsuta T, Nishigaki M, Akimoto S, Tanabe C, Omoto Y, Hayashi S-I, Sakamoto H, Sakamoto M, Yoshida T, Terada M, Sasaki H. A faithful method for PCR-mediated global mRNA amplification and its integration into microarray analysis on laser-captured cells. Biochem Biophys Res Commun. 2003;300:915–920. doi: 10.1016/s0006-291x(02)02967-4. [DOI] [PubMed] [Google Scholar]
- Alevizos I, Mahadevappa M, Zhang X, Ohyama H, Kohno Y, Posner M, Gallagher GT, Varvares M, Cohen D, Kim D, Kent R, Donoff RB, Todd R, Yung CM, Warrington JA, Wong DT. Oral cancer in vivo gene expression profiling assisted by laser capture microdissection and microarray analysis. Oncogene. 2001;20:6196–6204. doi: 10.1038/sj.onc.1204685. [DOI] [PubMed] [Google Scholar]
- Luzzi V, Holtschlag V, Watson MA. Expression profiling of ductal carcinoma in situ by laser capture microdissection and high-density oligonucleotide arrays. Am J Pathol. 2001;158:2005–2010. doi: 10.1016/S0002-9440(10)64672-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidl SJ, Nilsson S, Kalen M, Hellstrom M, Takemoto M, Hakansson J, Lindahl P. mRNA expression profiling of laser microbeam microdissected cells from slender embryonic structures. Am J Pathol. 2002;160:801–813. doi: 10.1016/S0002-9440(10)64903-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- Affymetrix Palo Alto: Affymetrix; Affymetrix Technical NoteGeneChip eukaryotic small sample target labeling assay version II. 2002 [Google Scholar]
- Baugh LR, Hill AA, Brown EL, Hunter CP. Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res. 2001;29:e29. doi: 10.1093/nar/29.5.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affymetrix Palo Alto: Affymetrix; Affymetrix Technical NoteGeneChip eukaryotic small sample target labeling. 2001 [Google Scholar]
- McClintick JN, Jerome RE, Nicholson CR, Crabb DW, Edenberg HJ. Reproducibility of oligonucleotide arrays using small samples. BMC Genomics. 2003;4:4. doi: 10.1186/1471-2164-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]