Abstract
The identification of intragenic rearrangements is important for a comprehensive understanding of mutations that occur in some clinically important genes. Single nucleotide polymorphism haplotypes obtained from clinical sequence data have been used to identify patients at high risk for rearrangement mutations. Application of this method identified a novel 26-kb deletion of BRCA1 exons 14 through 20 in patients from multiple families with hereditary breast and ovarian cancer. Clinical sequence data from 5911 anonymous patients were screened for genotypes that were inconsistent with known pairs of canonical haplotypes in BRCA1 that could be explained by hemizygous deletions involving exon 16. Long-range polymerase chain reaction demonstrated that two of six samples identified by this search contained a deletion in the expected region encompassing exons 14 through 20. The breakpoint was fully characterized by DNA sequencing and demonstrated that the deletion resulted from Alu-mediated recombination. This mutation was also identified twice in a set of 982 anonymous specimens that had negative clinical test results, but uninformative haplotypes. Three additional occurrences of this mutation were found by testing 10 other patients with the indicative genotype. An assay for this mutation was added to a comprehensive clinical breast/ovarian cancer test and eight more instances were found in 20,649 probands. This multiexon deletion has therefore been detected in 15 different North American families with hereditary breast/ovarian cancer. In conclusion, this primarily computational approach is highly effective and identifies specimens using existing data that are enriched for deletion mutations.
Deleterious mutations in two tumor suppressor genes, BRCA1 and BRCA2, confer high risk for breast and/or ovarian cancer and are responsible for the majority of hereditary families. Recently, comprehensive clinical DNA sequencing revealed deleterious mutations in BRCA1 or BRCA2 in 17.2% of 10,000 patients. Women with a history of breast cancer or ovarian cancer had deleterious mutation rates of 20% and 34%, respectively.1 The discovery of large genomic rearrangements within BRCA1 may partially explain the apparent discrepancy between expected mutation frequency and practical clinical sensitivity because existing clinical protocols using polymerase chain reaction (PCR)-based techniques designed to scan coding regions generally cannot detect these mutations.2,3 The population distributions of rearrangement mutations mirror those of point mutations where some, such as the duplication of exon 13, are highly prevalent, whereas others appear restricted to certain ethnic groups or limited to single families.3,4,5
Because both genes exhibit mutations distributed throughout their coding regions, clinical genetic tests have focused on mutation scanning encompassing their entire open reading frames. This approach has generated substantial data on the genetic variation within the coding regions of these genes (Breast Cancer Information Core, http://research.nhgri.nih.gov/bic/). Analyses of the common polymorphisms in BRCA1 have shown that several prevalent haplotypes exist.6 Some canonical haplotypes harbor increased incidence of mutations.7 Recently, haplotype pair analysis applied to clinical polymorphism data for BRCA1 identified patients at 50% risk for carrying novel large deletion mutations.8 A subsequent study used this method to show a high percentage of rearrangement mutations in northern Italian breast/ovarian cancer families.9 An important advantage of this primarily computational approach includes the potential to identify undetected mutations using existing data. This study confirms the utility of haplotype analysis for mutation detection by finding a recurrent novel deletion in BRCA1.
Materials and Methods
Haplotype Analysis
A group of 5911 anonymous specimens from patients with family histories of breast/ovarian cancer that had previously tested negative by clinical full sequence testing for BRCA1/BRCA2 were identified. Haplotypes were assigned to this sample set using an automated program using an expectation-maximization algorithm.10,11 The common polymorphisms used to designate haplotypes included: 18,202C->T; 31,385delT; 34,268A->G (Q356R); 35,278G->A (D693N); 35,283C->T (S694S); 35,512T->C (L771L); 35,813C->T (P871L); 36,314A->G (E1038G); 36,320G->A (S1040N); 36,749A->G (K1183R); 37,240A->G (R1347G); 46,278T->C (S1436S); 57,655A->G (S1613G); 57,774G->A (M1652I). Unless otherwise indicated all numeric base designations throughout this report conform to GenBank accession L78833. The polymorphisms above that reside in coding regions are also listed as more commonly referenced, by codon-position and amino acid change, in parentheses.
Nucleic Acid Biochemistry
Genomic DNA was isolated from peripheral blood mononuclear cells using commercial kits (Qiagen, Valencia, CA). PCR used a commercially available kit and followed the manufacturer’s suggested protocols (LA Taq; Takara Mirus Bio, Madison, WI). Specifically, cycling profiles involved annealing temperatures of 62°C for 30 seconds with 6-minute extensions (1 minute per kb) at 68°C. PCR primers for these reactions annealed upstream from exon 13 (forward, 5′-GCAACCATTGCTGTTCCTTCTAAA) and downstream from exon 21 (reverse, 5′-CGTGGGATCTTGCTTATAATACTCCA). DNA sequencing used commercial reagents detected on automated instruments (Big Dye primer and model 377; Applied Biosystems, Inc., Foster City, CA). For clinical testing, mutation-specific PCR was developed using a technique previously reported.4 Primers are designed around the recombination region such that a specific product is generated only in samples with the mutation. To facilitate high-throughput screening, fluorescently labeled primers were used and results were visualized on automated instruments (model 377; Applied Biosystems, Inc.).
Specimens
Several specimen sets were used for these experiments. Haplotype analysis was performed on the clinical genotype data for common polymorphisms found in 5911 anonymous specimens from breast/ovarian cancer families that were previously found to be negative for mutations by clinical DNA sequencing. Another set consisting of 982 anonymous genomic DNA samples were derived from hereditary cancer patients with negative clinical test results, but with single nucleotide polymorphism (SNP) genotypes that suggest canonical haplotype pairs. From a database of ∼20,000 breast cancer patients that had previously received negative clinical tests results, 14 patients were identified with the genotypes that could indicate the deletion mutation described here. Ten of these patients submitted specimens for testing after they were contacted through their physicians and offered the opportunity to participate in clinical research testing for the deletion. After the initial characterization of this deletion, an assay for this specific mutation was added to an existing panel of recurrent rearrangement mutations as part of a comprehensive clinical test for breast/ovarian cancer risk (BRACAnalysis; Myriad Genetic Laboratories, Inc., Salt Lake City, UT). A group of 20,649 patients that were tested for this deletion of exons 14 to 20 in this way are reported.
Results
The first objective of this project involved the identification of patient specimens possessing genotypes that could not be explained as a pair of known canonical haplotypes for BRCA1 and might therefore represent deletion mutations. A group of 14 polymorphisms located in or around BRCA1 exons 4 through 16, and that are routinely observed during clinical testing by DNA sequencing were used to assign haplotypes.6 By definition, none of these polymorphisms are present on the consensus allele (allele 1) and eight of these polymorphisms (31,385delT; 35,283C->T; 35,512T->C; 35,813C->T; 36,314A->G; 36,749A->G; 46,278T->C; 57,655A->G) occur together on the most prevalent nonconsensus allele (allele 2). A computer program assigned haplotypes using SNP genotype data from 5911 clinical specimens. The haplogroup containing allele 2 in non-African samples was detected with a frequency of 34% in agreement with a previous report.7 An unusual genotype was identified in six of these specimens and selected for additional analysis. This genotype is best explained by a haplotype pair consisting of allele 1 and allele 2 with the exception that only G was detected at position 57,655, the base contributed by allele 2, instead of the expected A/G heterozygous polymorphism. If this genotype is explained by a genomic deletion, it must reside downstream from the SNP at position 46,278 in exon 13, because this SNP was heterozygous, and would involve exon 16 because that SNP was missing. The consensus allele would be affected by the deletion because its contribution to the SNP in exon 16 was absent. Because there was no information for SNPs downstream from exon 16 in this data set, it was impossible to predict a 3′ limit to a putative deletion.
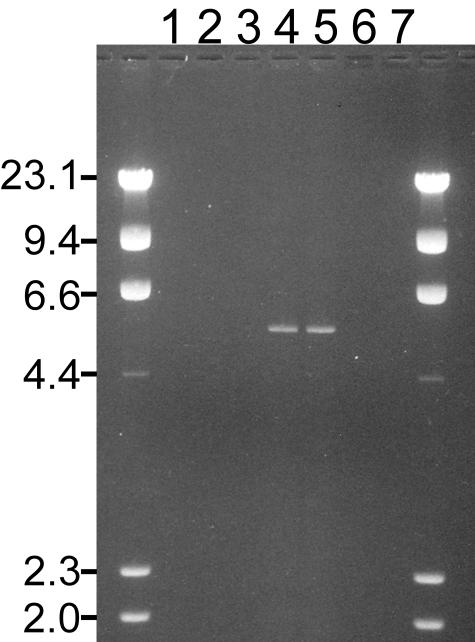
The hypothesis that this genotype resulted from a deletion was confirmed by long-range PCR experiments. A single forward primer was selected upstream from exon 13 where the heterozygous SNP of the suspect genotype placed a 5′ limit on any potential deletion. Because no data were available regarding the 3′ limit of potential deletions, a series of primers were designed that walked downstream from exons 16 through 24. On controls, none of these primer pairs amplified fragments because the annealing sites are too distant for amplification under the thermal cycling conditions used. However, a fragment was generated in two of the six patient specimens with the suspect genotype using primers that annealed at exons 13 and 21 (Figure 1). Because a PCR fragment ∼5 kb long was generated by primers that should have been spaced 31 kb apart, a deletion of ∼26 kb was suspected. Restriction mapping of this PCR fragment confirmed a 26-kb deletion with breakpoints close to the 5′ ends of exons 14 and 21 (Figure 2). PCR-based DNA sequencing provided for the complete characterization of the breakpoint (Figure 3). The deletion involves 26,454 bp and removes exons 14 through 20. The breakpoint region contains 54 bp of identity between the two Alu sequences that may have mediated a translocation. If transcripts from this allele were stable and spliced according to the remaining exons, the normal amino acid sequence would stop at residue 1452. An additional 69 abnormal residues from frame three of exons 20 to 24 would precede a new stop codon. This deletion certainly represents a clinically significant mutation because deleterious protein truncating mutations are known to occur 3′ of codon 1452 and the deleted region contains known deleterious missense mutations.12,13 Also, both BRCT domains are removed by the deletion.14
Figure 1.
Detection of a large deletion mutation in patients by PCR. A fragment of ∼5 kb in length was produced in two (lanes 4 and 5) of six patients (lanes 1 through 6) with an unusual BRCA1 haplotype. PCR primers for these reactions annealed upstream from exon 13 (forward) and downstream from exon 21 (reverse). Lane 7 contains the PCR reaction from a control specimen with no family history of cancer. The lanes containing PCR product (lanes 1 to 7) are flanked on both sides by a HindIII digest of λ DNA as a size marker (left-hand column indicates marker fragment lengths in kb).
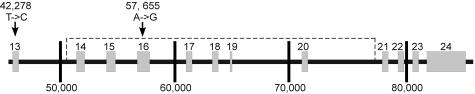
Figure 2.
Schematic diagram of relevant regions of the BRCA1 locus. BRCA1 structure is depicted to scale as a horizontal black line (introns) with numbered gray boxes (exons 13 through 24). Distance is marked every 10 kb by vertical lines and the numbering conforms to GenBank accession no. L78833. Vertical arrows indicate the locations for 2 of the 14 polymorphisms used to define haplotypes that are assayed during clinical testing in exons 13 and 16 along with their base positions and the base change (consensus -> polymorphism). The dashed line depicts the region of BRCA1 encompassed by this deletion.
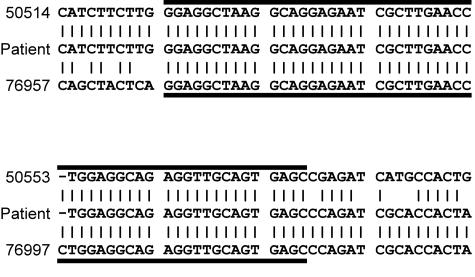
Figure 3.
The sequence of the deletion breakpoint. Eighty bases centered about the breakpoint in the patient are flanked vertically by the wild-type sequences (numeric designations and sequence from GenBank accession no. L78833). Horizontal lines denote the region of identity (54 bases) between the sequences that have recombined.
It is reasonable to conclude that these specimens came from unrelated individuals because both patients received comprehensive clinical tests (whole gene BRCA1 and BRCA2 testing) that are normally administered to the single most suitable patient in a hereditary family. Although this conclusion cannot be tested formally because the specimens are anonymous, this conclusion reflects the ascribed use of this clinical test.
More specimens were tested for this mutation to provide further confirmation that it occurs in unrelated patients. First, two additional instances of the mutation were detected in 982 anonymous specimens with familial cancer, negative comprehensive clinical tests, and genotype data that conforms to canonical haplotype pairs (homozygous for allele 1 or heterozygous for the mutant allele and allele 1). These results further support the conclusion that this mutation occurs in unrelated patients. Next, a large clinical data set of ∼20,000 specimens was screened to identify patients with the suspicious genotype, allele 1 and allele 2 with the exception of only G at position 57,655. Five deletion mutations were found in the 10 patients that elected to submit a specimen to be assayed for the 14 to 20 deletion from a total of 14 patients identified with the appropriate genotype. The result represents at least three additional families with the 14 to 20 deletion because this set of patients has some overlap with the anonymous specimens used to identify the mutation. Because this mutation was recurrent in this laboratory’s patients, an assay for this mutation was added to an existing test panel for prevalent rearrangement mutations in BRCA1 as a routine component of a comprehensive clinical test for breast/ovarian cancer risk. Eight more instances of the 14 to 20 deletion mutation were detected in probands and another eight instances in family members from the first 20,649 patients after its introduction. Thus, the 14 to 20 deletion occurs in 0.039% of these patients and represents 0.31% of all deleterious mutations reported in probands.
For the 21 patients that have been identified with this mutation, the reported ancestries included 18 Western European, 1 Irish, and 2 that were not specified. The self-reported cancer incidence in these patients included nine with breast cancers, one with bilateral breast cancer, one with ductal carcinoma in situ, one with breast and ovarian cancer, two with ovarian cancers, and seven unaffected.
Discussion
Herein, a novel recurrent mutation where BRCA1 exons 14 to 20 are deleted through Alu-mediated recombination is described. This mutation is certainly deleterious because it truncates the protein and removes regions of the gene known to encode important functional domains. Also, 14 of 21 patients with the mutation are affected with cancer. So far, its incidence has been restricted to patients that claim Western European ancestry, but the patient populations examined here are biased toward this group and additional evaluations of other patient populations could be useful in this respect. Because the breakpoint for the mutation is solved, researchers with access to additional samples can easily address this question.
Only 7 of 16 specimens with the suspect haplotype from the groups described here were shown to carry this deletion mutation. It is possible that the genotypes for the other nine specimens result from point mutations or recombination. However, it is also possible that these specimens could harbor mutations that were not detected because they extend beyond the regions examined here by PCR. This question is currently being addressed in these specimens with quantitative PCR approaches that determine gene dosage in these specimens downstream from exon 13.
This project corroborates previous conclusions that haplotype analysis of clinical genotypes is useful to identify patients at increased risk for deletion mutations.8,11 BRCA1 is an especially suitable target for this approach because it is represented by a small group of 10 prevalent haplotypes in which highly informative SNPs are detected routinely in clinical tests. Of course, this approach may have application to other genes. This technique is attractive because it is primarily computational and uses existing data to greatly refine specimen sets to only those samples highly predisposed to contain deletion mutations. Information is also provided regarding the location of the deletion and equally important, regions that are indeed diploid.
References
- Frank TS, Deffenbaugh AM, Reid JE, Hulick M, Ward BE, Lingenfelter B, Gumpper KL, Scholl T, Tavtigian SV, Pruss DR, Critchfield GC. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20:1480–1490. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, Weber B, Lenoir G, Chang-Claude J, Sobol H, Teare MD, Struewing J, Arason A, Scherneck S, Peto J, Rebbeck TR, Tonin P, Neuhausen S, Barkardottir R, Eyfjord J, Lynch H, Ponder BAJ, Gayther SA, Birch JM, Lindblom A, Stoppa-Lyonnet D, Bignon Y, Borg A, Hamann U, Haites N, Scott RJ, Maugard CM, Vasen H, Seitz S, Cannon-Albright LA, Schofield A, Zelada-Hedman M, Breast Cancer Linkage Consortium Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson BC, Deffenbaugh AM, Gaglio CA, Judkins T, Chen S, Ward BE, Scholl T. Prevalence results for five recurrent BRCA1 rearrangement mutations in 7570 analyses. Am J Hum Genet. 2003;73:253. [Google Scholar]
- The BRCA1 Exon 13 Duplication Screening Group The exon 13 duplication in the BRCA1 gene is a founder mutation present in geographically diverse populations. Am J Hum Genet. 2000;67:207–212. [PMC free article] [PubMed] [Google Scholar]
- Petri-Bosch A, Peelen T, van Vliet M, van Eijk R, Olmer R, Drusedau M, Hogervorst FBL, Hageman S, Arts PJW, Ligtenberg MJL, Meijers-Heijboer H, Klijn JGM, Vasen HFA, Cornelisse CJ, van’t Veer LJ, Bakker E, van Ommen G-JB, Devilee P. BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients. Nat Genet. 1997;17:341–345. doi: 10.1038/ng1197-341. [DOI] [PubMed] [Google Scholar]
- Shattuck-Eidens D, Oliphant A, McClure M, McBride C, Gupte J, Rubano T, Pruss D, Tavtigian SV, Teng DH-F, Adey N, Staebell M, Gumpper K, Lundstrom R, Hulick M, Kelly M, Holmen J, Lingenfelter B, Manley S, Fujimara F, Luce M, Ward B, Cannon-Albright L, Steele L, Offit K, Gilewski T, Norton L, Brown K, Schulz C, Hampel H, Schluger A, Giulotto E, Zoli W, Ravaioli A, Nevanlinna H, Pyhonen S, Rowley P, Loader S, Osborne MP, Daly M, Tepler I, Weinstein PL, Scalia JL, Michaelson R, Scott RJ, Radice P, Pierotti MA, Garber JE, Isaacs C, Peshkin B, Lippman ME, Dosik MH, Caligo MA, Greenstein RM, Pilarski R, Weber B, Burgermeister R, Frank TS, Skolnick MH, Thomas A. BRCA1 sequence analysis in women at high risk for susceptibility mutations, risk factor analysis and implications for genetic testing. JAMA. 1997;278:1242–1250. [PubMed] [Google Scholar]
- Orsoria A, de la Hoya M, Rodriguez-Lopez R, Granizo JJ, Diez O, Vega A, Duran M, Carracedo A, Baiget M, Caldes T, Benitez J. Over-representation of two specific haplotypes among chromosomes harbouring BRCA1 mutations. Eur J Hum Genet. 2003;11:489–492. doi: 10.1038/sj.ejhg.5200969. [DOI] [PubMed] [Google Scholar]
- Hendrickson BC, Pruss D, Lyon E, Scholl T. Application of haplotype pair analysis for the identification of hemizygous loci. J Med Genet. 2003;40:346–347. doi: 10.1136/jmg.40.5.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagna M, Dalla Palma M, Menin C, Agata S, De Nicolo A, Chieco-Bianchi L, D’Andrea E. Genomic rearrangements account for more than one-third of the BRCA1 mutations in northern Italian breast/ovarian cancer families. Hum Mol Genet. 2003;12:1055–1061. doi: 10.1093/hmg/ddg120. [DOI] [PubMed] [Google Scholar]
- Ceppellini R, Siniscalco M, Smith CAB. The estimation of gene frequencies in a random mating population. Ann Hum Genet. 1955;20:97–115. doi: 10.1111/j.1469-1809.1955.tb01360.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Altschul SF, Bork P. BRCA1 protein products: functional motifs. Nat Genet. 1996;13:266–268. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- Vallon-Christersson J, Cayanan C, Haraldsson K, Loman N, Bergthorsson JT, Brondom-Nielsen K, Gerdes A-M, Moller P, Kristoffersson U, Olsson H, Borg A, Monteiro ANA. Functional analysis of BRCA1 C-terminal missense mutations identified in breast and ovarian cancer families. Hum Mol Genet. 2001;10:353–360. doi: 10.1093/hmg/10.4.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyton T, Bates PA, Zhang X, Sternberg MJW, Freemont PS. The BRCA1 C-terminal domain: structure and function. Mutat Res. 2000;460:319–332. doi: 10.1016/s0921-8777(00)00034-3. [DOI] [PubMed] [Google Scholar]