Abstract
Many inherited diseases involve large genes with many different mutations. Identifying a wide spectrum of mutations requires an efficient gene-scanning method. By differentiating thermodynamic stability and mobility of heteroduplexes from heterozygous samples, temperature gradient capillary electrophoresis (TGCE) was used to scan the entire coding region of the cystic fibrosis transmembrane conductance regulator gene. An initial panel (29 different mutations) showed 100% agreement between TGCE scanning and previously genotyped results for heterozygous samples. Different peak patterns were observed for single base substitutions and base insertions/deletions. Subsequently, 12 deidentified clinical samples genotyped as wild type for 32 mutations were scanned for the entire 27 exons. Results were 100% concordance with the bidirectional sequence analysis. Ten samples had nucleotide variations including a reported base insertion in intron 14b (2789 + 2insA) resulting in a possible mRNA splicing defect, and an unreported missense mutation in exon 20 (3991 G/A) with unknown clinical significance. This methodology does not require labeled primers or probes for detection and separation through a temperature gradient eliminates laborious temperature optimization required for other technologies. TGCE automation and high-throughput capability can be implemented in a clinical environment for mutation scanning with high sensitivity, thus reducing sequencing cost and effort.
Detecting single nucleotide polymorphisms is increasingly important in molecular diagnostics to link DNA variation with complex inherited diseases. With the occurrence of single nucleotide changes/substitutions in the human population greater than 1%, technology detecting any sequence alteration is especially important for large genes containing many exons and multiple mutations.1 For example, the gene that encodes cystic fibrosis transmembrane conductance regulator (CFTR) consists of 27 exons and spans a region of 188,705 bp in chromosome 7. More than 1000 mutations have been reported that are associated with cystic fibrosis, a severe autosomal recessive disorder. Common symptoms include abnormal sweat electrolytes, pulmonary disease, exocrine pancreatic insufficiency, or male infertility (congenital bilateral absence of the vas deferens).2,3 A panel of 25 cystic fibrosis (CF) mutations is recommended by the American College of Medical Genetics for population carrier screening.4 However, this panel is designed only for the most common mutations found in the United States population. Other sequence alteration(s) unknown whether they are pathogenic or nonpathogenic require further characterization.5 Therefore, analyzing the entire mutation spectrum can improve correlation between genotypes and phenotypes, specifically in relation to atypical or mild forms of CF. Scanning the entire coding region of the target gene in a high-throughput format saves time and cost over full gene sequencing. Any technology used for scanning must have a high sensitivity for detecting any alteration.
Current technologies available for mutation detection have been reviewed.6,7 Basically, they can be categorized into two areas. The first category detects known mutations, and methods include real-time polymerase chain reaction (PCR) coupled with melting curve analysis (allele-specific hybridization probes),8,9 oligonucleotide arrays,10 minisequencing with primer extension, and enzymatic assays such as oligo ligation assay (Applied Biosystems, Foster City, CA) and restriction fragment length polymorphism. The second category detects unknown mutations, and technologies include direct sequencing and varied electrophoretic-based assays, such as single strand confirmation polymorphism,7 confirmation-sensitive gel electrophoresis,11 and constant denaturant capillary electrophoresis.12 Both categories have advantages and limitations. For example, an assay based on allele-specific hybridization probes is sensitive but is limited to detecting a single or few mutations. Methods for full gene analysis will detect a great number of mutations, but may be less sensitive overall. Direct sequencing is the current gold standard but is costly and labor intensive for analyzing a large multiexon gene.
A temperature-controlled ion-pair reverse-phase liquid chromatography has been used for unknown mutation discovery.13 This approach separates heteroduplex mutants from homoduplex wild types by specific melting temperatures (Tm) and different hydrophobicity in an alkylated C-18 column.13,14 Two research articles have been published for screening the CFTR gene using denaturing high-performance liquid chromatography and suggest this technology is suitable for large gene analysis with high accuracy.15,16 However, a limitation of denaturing high-performance liquid chromatography is that it requires intensive optimization to determine the best resolving temperature for each individual exon (amplicon) in a large gene. This limitation may decrease throughput for a high volume test. Recently, a technology using an automated temperature gradient capillary array electrophoresis (TGCE) provides another option for a rapid analysis of a large multiexon gene. This technology is similar to a method previously described17 but instead of using a denaturing agent, samples are resolved in a capillary array with a proprietary polymer matrix. This technology differentiates heteroduplex mutants from homoduplex wild types based on different mobility in a specific polymer matrix under a temperature gradient. Using a temperature gradient reduces Tm optimization for individual exons. Movement of different species of DNA is then captured by a charge-coupled device camera as image files for data analysis.18,19
The purpose of this study was to implement an automated analysis format using TGCE (model SCE 2410; SpectruMedix Inc., State College, PA) for mutation scanning of the CFTR gene. Exon-specific primers were designed so the entire coding regions as well as intron-exon boundaries of the 27 exons were optimally amplified with a standard PCR protocol. After amplification, PCR products were slowly cooled and subjected to the automated TGCE analysis. This technique showed 100% agreement with sequencing results for distinguishing between heterozygous mutants and wild types. We also detected one nucleotide alteration not previously reported [Cystic Fibrosis Genetic Analysis Consortium (CFGAC) database (http://www.genet.sickkids.on.ca/)]. It is desirable to detect single-base alteration in a wide range of DNA fragments, in terms of sizes and GC contents, using an automated high-throughput format. The method described in this study is capable of detecting single (or multiple) base alterations in varied length PCR products (175 bp to 834 bp). This technology has a great potential to be implemented in a high-throughput environment for mutation scanning of large multiexon genes.
Materials and Methods
Sample Collection for TGCE Scan
Forty-two previously characterized samples including 14 samples from Coriell Repository (Camden, NJ) and 28 deidentified clinical samples with known CF genotypes for 29 specific mutations were used to compare peak patterns of heterozygous samples to wild types using TGCE scanning (Table 1). Subsequently, 12 clinical specimens previously submitted to ARUP Laboratories (Salt Lake City, UT) were deidentified (according to an institutional review board-approved protocol) and used for full gene-scanning analysis (27 exons). These samples were previously genotyped as negative for a panel of 32 CF mutations by oligo ligation assay (Celera Diagnostics LLC., Alameda, CA).
Table 1.
Mutation Samples with Known Genotypes Scanned by TGCE*
| Exon | Mutation† | Amplicon size (bp) | Location of mutation from 5′ end (bp) | Base change | Detection‡ |
|---|---|---|---|---|---|
| 3 | G85E | 234 | 124 | G to A | 1/1 |
| 3 | 394delTT | 234 | 132 | del TT | 1/1 |
| 4 | R117H | 270 | 83 | G to T | 2/2 |
| 4 | I148T | 270 | 176 | T to C | 3/3 |
| Intron 4 | 621 + 1 G/T | 270 | 233 | G to T | 1/1 |
| 5 | 663delT/663delT | 186 | 75 | del T | 0/1 |
| Intron 5 | 711 + 1 G/T | 186 | 124 | G to T | 1/1 |
| 7 | R334W | 345 | 208 | C to T | 1/1 |
| 7 | R347P | 345 | 248 | G to C | 1/1 |
| 9 | A455E | 263 | 155 | C to A | 2/2 |
| 10 | I506V | 292 | 168 | A to G | 1/1 |
| 10 | ΔI507 | 292 | 171 | del ATC | 2/2 |
| 10 | ΔF508 | 292 | 174 | del TTT | 2/2 |
| 10 | ΔF508/ΔF508 | 292 | 174 | del TTT | 0/1 |
| 10 | F508C | 292 | 175 | T to G | 1/1 |
| 10 | V520F | 292 | 210 | G to T | 1/1 |
| Intron 10 | 1717–1 G/A | 175 | 50 | G to A | 1/1 |
| 11 | G542X | 175 | 90 | G to T | 2/2 |
| 11 | G542X/G542X | 175 | 90 | G to T | 0/1 |
| 11 | G551D | 175 | 118 | G to A | 3/3 |
| 11 | R553X | 175 | 123 | C to T | 3/3 |
| 11 | R560T | 175 | 145 | G to C | 2/2 |
| 13 | 2184delA | 834 | 356 | del A | 1/1 |
| Intron 14b | 2789 + 5G/A | 192 | 102 | G to A | 1/1 |
| Intron 16 | 3120 + 1G/A | 216 | 111 | G to A | 1/1 |
| 19 | R1162X | 322 | 68 | C to T | 1/1 |
| 19 | 3659delC | 322 | 111 | del C | 1/1 |
| 20 | W1282X | 206 | 154 | G to A | 1/1 |
| 21 | N1303K | 250 | 175 | C to G | 2/2 |
| Total exon/intron | Overall accuracy | ||||
| 17 | 93% |
Samples were compared with their respective wild-type control (confirmed by sequencing).
All genotypes were heterozygous except homozygous sample 663delT/663delT, ΔF508/ ΔF508, and G542X/G542X.
Number of samples scored (+) by TGCE/number of samples tested. Homozygous samples were detected only after mixing with a wild-type sample.
Primer Design, DNA Amplification, and TGCE Preconditioning
Genomic DNA of all specimen samples collected for this study was extracted by the MagNA Pure LC instrument (Roche Diagnostics, Indianapolis, IN) and 2 μl of each extracted DNA was used for PCR. Primers (except exon 9) specific to each exon were designed ∼20 to 100 bp (if applicable) upstream and downstream of each exon using Primer3 software20 so partial intron sequences and intron/exon boundaries will also be scanned. The primer pair specific to exon 9 was adapted from a published sequence15 to exclude a TG/T repeat polymorphic region in intron 8 that forms heteroduplexes in nearly all samples. The complete designed primers are listed in Table 2.
Table 2.
Primer Sets Used in This Study to Amplify Entire 27 Exons of the CFTR Gene Using the Same PCR Condition (Tm = 55°C, 30 Cycles)
| Exon | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| 1 | CAGCACTCGGCTTTTAAC | ATACACACGCCCTCCTCT |
| 2 | TCCAAATCTGTATGGAGACC | TGAATTTCTCTCTTCAACTAAACA |
| 3 | CAACTTATTGGTCCCACTTT | CACCTATTCACCAGATTTCG |
| 4 | TTGTAGGAAGTCACCAAAGC | TACGATACAGAATATATGTGCCA |
| 5 | TTGAAATTATCTAACTTTCCATTTT | CGCCTTTCCAGTTGTATAAT |
| 6a | GCTGTGCTTTTATTTTCCAG | ACTAAAGTGGGCTTTTTGAA |
| 6b | CTTAAAACCTTGAGCAGTTCT | CAATATTGAAATTATTGGAACAAC |
| 7 | AGATCTTCCATTCCAAGATC | TGCAGCATTATGGTACATTA |
| 8 | AAGATGTAGCACAATGAGAGTATAAA | GAAAACAGTTAGGTGTTTAGAGCAA |
| 9* | TGGGGAATTATTTGAGAAAG | CTTCCAGCACTACAAACTAGAAA |
| 10 | GCGTGATTTGATAATGACCT | TGGGTAGTGTGAAGGGTTC |
| 11 | AGATTGAGCATACTAAAAGTGAC | TGCTTGCTAGACCAATAATTAG |
| 12 | CCAGGAAATAGAGAGGAAATG | CATACCAACAATGGTGAACA |
| 13 | GCTAAAATACGAGACATATTGC | ATTCTGTGGGGTGAAATACC |
| 14a | GGTGGCATGAAACTGTACT | ATACATCCCCAAACTATCTTAAT |
| 14b | ATGGGAGGAATAGGTGAAGA | CAAAGTGGATTACAATACATACA |
| 15 | TGCCAAATAACGATTTCCTA | GTGGATCAGCAGTTTCATTT |
| 16 | TTGAGGAATTTGTCATCTTGT | GACTTCAACCCTCAATCAAA |
| 17a | CACTGACACACTTTGTCCAC | ATCGCACATTCACTGTCATA |
| 17b | ATTTGCAATGTTTTCTATGG | TGCTTAGCTAAAGTTAATGAGTTC |
| 18 | TTCATTTACGTCTTTTGTGC | GGTATATAGTTCTTCCTCATGC |
| 19 | GTGAAATTGTCTGCCATTCT | CAAGCAGTGTTCAAATCTCA |
| 20 | TGATCCCATCACTTTTACCT | TTTCTGGCTAAGTCCTTTTG |
| 21 | AGAACTTGATGGTAAGTACATG | CATTTCAGTTAGCAGCCTTA |
| 22 | TCTGAACTATCTTCTCTAACTGC | AATGATTCTGTTCCCACTGT |
| 23 | TTCTGTGATATTATGTGTGGTATT | AAGAATTACAAGGGCAATGA |
| 24 | CAGATCTCACTAACAGCCATT | TGTCAACATTTATGCTGCTC |
Primers adapted from Le Marechal et al.15
PCR was performed in a 50 μl reaction using the High Fidelity PCR master kit (Roche Diagnostics, Indianapolis, IN) following the manufacturer’s instructions. This enzyme mixture contains both TaqDNA polymerase and Tgo 3′ to 5′ exonuclease proofreading polymerase. The use of a high-fidelity PCR enzyme mixture was to minimize polymerization errors during PCR thus improving detection efficiency for genetic diversity.21 PCR was performed in a PTC-200 thermocycler (MJ Research, Waltham, MA). One standard PCR protocol was used to amplify all 27 exons of the CFTR gene simultaneously. PCR cycling conditions were 5 minutes at 95°C, 30 cycles of 94°C for 30 seconds, 55°C for 1 minute, 72°C for 1 minute, then 72°C for 5 minutes, and cooling to 4°C. After PCR, heteroduplexes were formed as recommended by the vender (SpectruMedix Inc.). PCR products were heated 5 minutes at 95°C, cooled slowly to 50°C in 1°C/minute intervals, held at 50°C for 20 minutes, and cooled to 25°C at the rate of 2.5°C/minute. To obtain the best peak resolution, treated PCR products were diluted with either 1× or 10× PCR buffer (Applied Biosystems, Foster City, CA) to ensure unsaturated fluorescent intensity and suitable salt contents, and injected into a TGCE equipped with 24 capillaries (model SLE 2410, SpectruMedix Inc.). Other parameters requiring optimization before scanning included injection time, range of temperature gradient, and ramping rate. Two injection conditions were tested, 3 kV for 20 seconds and 5 kV for 30 seconds. Five different temperature gradient ranges for capillary electrophoresis (CE) were also tested: 40 to 50°C, 50 to 55°C, 50 to 60°C, 55 to 60°C, and 60 to 65°C. The ramp period was always 21 minutes. The optimized dilution factor (1:4), injection time (3 kV for 20 seconds), and temperature gradient (50 to 55°C) were used for the rest of the study.
CFTR Full Gene-Scanning Setup
Based on the basic setup (24-capillary format) in our instrument, the full gene scanning of one patient sample requires one plate (24 exons), plus an additional three wells because of the 27 exons of the CFTR gene. Multiplexing several exons is feasible to fit a 24-well format, but requires further optimization to avoid interaction between multiplexed amplicons. In our design, each plate contained two exons for 12 patient samples. Thus the 27 exons of the 12 deidentified samples (324 PCR products) were scanned within three runs (14 plates; 6 plates per one complete run). These 12 samples constituted our wild-type controls, although 10 of 12 samples had variations in at least one exon. Ideally, peaks of additional samples will be compared to these original samples, thus reducing the need to run wild-type controls with every run.
Data Analysis
Data were analyzed using the Revelation 2.4 image analysis software (SpectruMedix Inc.). For graphical illustration, analyzed data (by Revelation 2.4) were exported to Microsoft Excel using time (seconds) as the x axis and fluorescent intensity as the y axis. After scanning and data analysis, for each individual exon, at least two samples possessing a single-sharp peak were assumed to be the wild types (negative control) and were sequenced for confirmation. The rest of the samples were compared to the confirmed wilt-type peaks for each exon. Samples possessing multiple peaks (2 to 4) or with any differences in peak shape when compared to the negative control were scored as positive. Positive samples were sequenced for the specific exon to identify the base alteration(s).
Sequencing
PCR samples showing an alteration by TGCE were sequenced using dideoxy terminator sequencing reactions (Applied Biosystems Inc.). In each reaction, 5 μl of Big Dye Terminator Ready Reaction mixture, 3 μl of undiluted and purified PCR product, and 4 μl of 0.8 pmol/μL primer (forward or reverse) were mixed and injected into the automated DNA sequencer (ABI Prism 3100 genetic analyzer, Applied Biosystems). Results were analyzed using both ABI Sequencing Analysis and Sequencher software (The BioCommons, Seattle, WA) to locate and identify alterations. The confirmed alteration(s) were compared to a current CF database (CFGAC) (http://www.genet.sickkids.on.ca/) for identification. The identified nucleotide changes were then classified into three categories: 1) known mutations with reported clinical significance, 2) known variants without clinical significance, and 3) sequence variants with unknown clinical significance (usually missense or intronic mutations). For alterations not previously reported, the laboratory must decide the classification, with deleterious (or suspected deleterious) mutations compromising insertions/deletions (frame shift), nonsense mutations, and predicted splice site mutations; or alterations with unknown significance include missense mutations and some intronic mutations.
Results
Peak Resolution
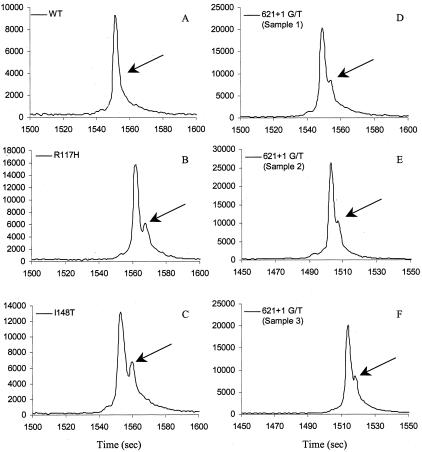
TGCE detects sequence alteration(s) based on the differentiation of different heteroduplex mobilities in a specifically designed polymer matrix. In theory, heteroduplexes move slower than homoduplexes because of the formation of bubble-like structures of strands with a single mismatch under a specific temperature (Tm). In addition to heteroduplex formation, the length of PCR amplicon, the GC content, the location and types of single nucleotide polymorphisms (A/T, A/C, A/G, C/T, C/G, or G/T) also affect the mobility of formed heteroduplexes under a partially denatured condition (temperature gradient). Thus, parameters that affect heteroduplex mobility in a capillary array will affect the performance of the TGCE. Parameters that need to be optimized for TGCE scanning include temperature gradient, sample injection time, and PCR buffer (salt concentration) used to dilute amplicons. Figure 1 shows the temperature gradient effect on peak resolution. The best temperature range for both heterozygous R117H (exon 4, 270 bp) (Figure 1; A to C) and the hard-to-discriminate mutation G551D heterozygous (exon 11, 175 bp) (Figure 1; D to F) is 50 to 55°C. This temperature range was used for all amplicons.
Figure 1.
Temperature gradient effect on peak resolution (in 100 seconds scale). A: Wild-type control (exon 4, 270 bp); B: R117H heterozygous (exon 4) with 50 to 55°C gradient; C: R117H heterozygous (exon 4) with 55 to 60°C gradient; D: wild-type control (exon 11, 175 bp); E: G551D heterozygous (exon 11) with 50 to 55°C gradient; F: G551D heterozygous (exon 11) with 55 to 60°C gradient.
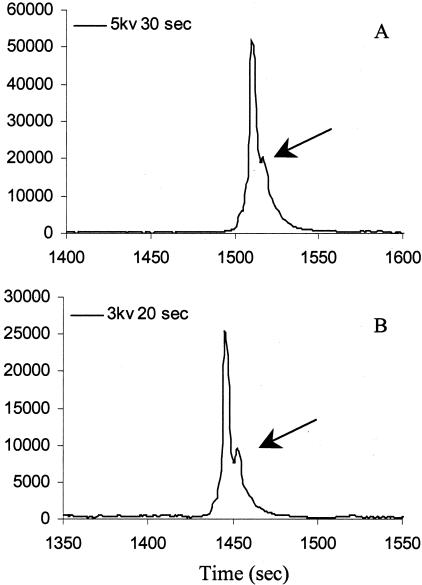
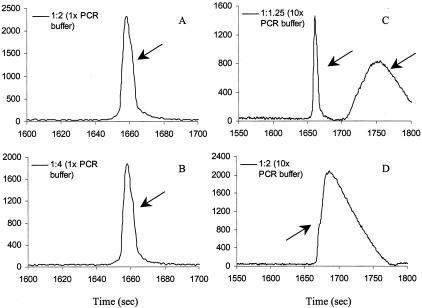
Another factor that can reduce peak resolution is saturated fluorescent signal (>60,000) (Figure 2, A and B). To avoid this, we decreased the sample injection time from the factory default (5 kV for 30 seconds) to 3 kV for 20 seconds for better resolution. Even with decreased injection time, some amplicons required dilution to avoid saturated signal. However, the salt and Mg+2 contents in the buffer may affect peak resolution. Figure 3 demonstrates the dilution effect using different buffers (1× and 10×) on peak resolution. In this figure, the hard-to-discriminate G551D heterozygous shows better resolution in both 1 to 2 and 1 to 4 dilutions using 1× PCR buffer (Figure 3, A and B). However, with higher salt and Mg+2 contents, peak resolution was disrupted and cannot be used for comparison (Figure 3, C and D). For consistency, all 27 amplicons of the CFTR gene were diluted 1 to 4 (using 1× PCR buffer) and injected at 3 kV for 20 seconds.
Figure 2.
Different sample injection times affect peak resolution. A: R117H heterozygous (exon 4, 270 bp) without dilution and injected at 5 kV for 30 seconds; B: R117H heterozygous (exon 4) without dilution and injected at 3 kV for 20 seconds.
Figure 3.
Effect of dilution with PCR buffers (1× buffer containing 1.5 mmol/L Mg+2; 10× buffer containing 15 mmol/L Mg+2) on peak resolution using amplicon G551D heterozygous (exon 11) as an example. A: Dilution (1 in 2) using 1× PCR buffer; B: 1 in 4 dilution using 1× PCR buffer; C: 1 in 1.25 dilution using 10× PCR buffer; D: 1 in 2 dilution using 10× PCR buffer.
Mutation Scanning of the CFTR Gene
In this study, no confirmed peak patterns for wild-type sequences of each exon were initially available, but samples possessing a single peak for each specific exon after TGCE scanning were considered as wild types and sequenced for confirmation. After sequencing confirmation, these samples were used as wild-type controls for peak comparison in the further studies.
Because of limited sample availability of rare CF mutations, detection of all 1291 reported mutations for accuracy study using TGCE is not feasible. Therefore, to test the accuracy of the TGCE protocol that we developed, we used 42 genotyped samples with 29 specific genotypes representing 27 mutations (Table 1). Each amplicon was injected in duplicate into the automated TGCE. Peak patterns were identical between duplicates (data not shown). Frame numbers of the same amplicon that appeared in the electropherogram were slightly different (±∼40 frames) between duplications. This is a normal phenomenon because each capillary acts independently.
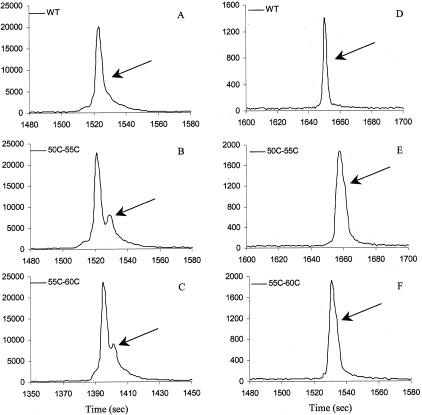
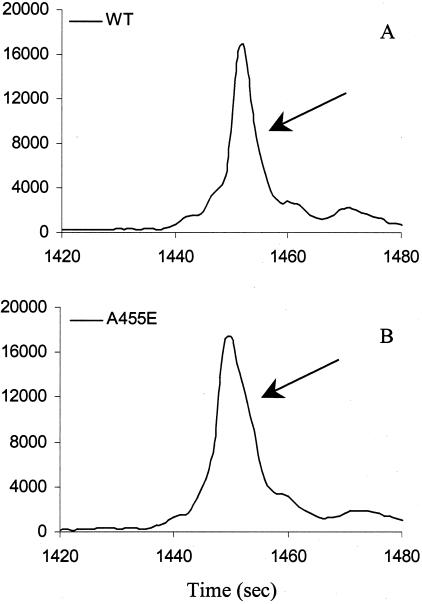
After comparison with each wild-type control, all heteroduplex mutations were identified correctly. Figure 4 demonstrates detection of different locations of a single base alteration in exon 4 fragment. In this 270-bp fragment, heterozygous R117H (G/T) is located 83 bp from the 5′ end, heterozygous I148T (T/C) is located in the middle of the fragment, and heterozygous 621 + 1 (G/T) is located at the end of the fragment (37 bp from the 3′ end) (Figure 4, A to D; Table 1). Heterozygous 621 + 1 (G/T) had the least distinct split-peak pattern when compared to heterozygous R117H and I148T (Figure 4; B to D). Different heterozygous 621 + 1 (G/T) samples showed similar patterns, with less peak resolution than the other exon 4 mutations (Figure 4; D to F). The reason for less peak resolution of heterozygous 621 + 1 (G/T) is not clear. A possible explanation is the nearest neighbor structure that helps to stabilize the mismatch, which results in similar mobility between the homoduplexes and heteroduplexes or the location close to the 3′ end. Another example of reduced resolution is heterozygous A455E in exon 9. A shoulder instead of a small peak was observed in heterozygous A455E when compared to the wild-type control (Figure 5, A and B). To clarify the detection limit of the TGCE, further studies should focus on the thermodynamic effect of amplicon length and type of single nucleotide polymorphisms on heteroduplex formation using either artificial templates or engineered plasmids as a study model.
Figure 4.
TGCE scan of samples containing single base changes in exon/intron 4 of the CFTR gene. A: Wild-type; B: R117H heterozygous (exon 4); C: I148T heterozygous (exon 4); D: 621 + 1 G/T heterozygous sample 1 (intron 4); E: 621 + 1 G/T heterozygous sample 2 (intron 4, from Coriell Repository); F: 621 + 1 G/T heterozygous sample 3 (intron 4, from Coriell Repository).
Figure 5.
TGCE scan of samples containing single base changes with reduced resolution. A: Wild-type (exon 9 of the CFTR gene); B: A455E heterozygous (exon 9 of the CFTR gene).
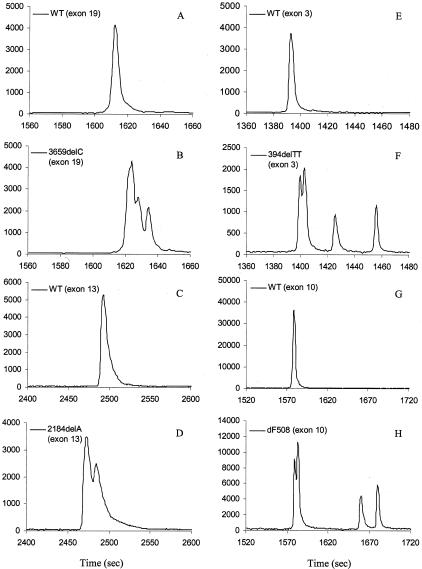
Figure 6 demonstrates examples of peak patterns for heterozygous base deletions. For a single base deletion, a pattern of two additional peaks associated with the main peak was observed in a shorter fragment (exon 19, 322 bp), and a pattern of one additional peak associated with the main peak was found in a longer fragment (exon 13, 834 bp) (Figure 6; A to D). This difference in peak resolution is possibly because of different deleted bases and different fragment lengths. Two or more deleted bases showed a distinct four-peak pattern in heterozygous 394delTT (exon 3, 234 bp) and heterozygous ΔF508 samples (exon 10, 292 bp) (Figure 6, E to H; Table 1).
Figure 6.
TGCE scan of samples containing base deletions in the CFTR gene (single base deletion, A to D; multiple base deletion, E to H). A: Wild-type control (exon 19); B: 3659delC heterozygous (exon 19); C: wild-type control (exon 13); D: 2184delA heterozygous (exon 13). E: wild-type control (exon 3); F: 394delTT heterozygous (exon 3); G: wild-type control (exon 10); H: ΔF508 heterozygous (exon 10).
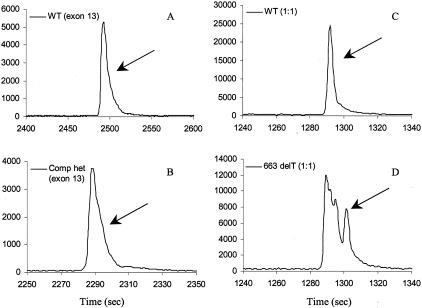
Compound heterozygotes and homozygous sequence alterations were also investigated by TGCE. Although compound heterozygotes in the same exon are rare, its mobility in TGCE is of interest. Figure 7, A and B, shows peak comparisons of a compound heterozygous mutation (2134 C/T plus 2151 A/G) with the wild-type control in exon 13 (834 bp). Homozygous mutations are difficult to discriminate in assays based on heteroduplex mobility separation because no heteroduplexes are formed. In our study, homozygous mutants 663 delT (exon 5), ΔF508 (exon 10), and G542X (exon 11) were not initially detected. However, on mixing with a wild-type sample in a 1:1 ratio, all three homozygotes were detected (Figure 7, C and D). As detailed in Table 1, the overall accuracy (sensitivity) of the TGCE scanning is 93% (without mixing with wild-type sample), 100% for heterozygotes detection, and 100% overall accuracy when mixing with wild-type samples (Table 1).
Figure 7.
Detection of compound heterozygous and homozygous mutation. A: Wild-type (exon 13); B: peak pattern of compound heterozygous 2134 C/T and 2151 A/G (exon 13); C: wild-type mixed with wild-type in 1:1 ratio (exon 5); D: homozygous 663 delT mixed with wild-type in 1:1 ratio (exon 5).
The image analysis software included with the TGCE system (Revelation 2.4) provides a convenient feature to generate a graphical report by computing differences of peak shape and peak area between an unknown sample and its wild-type control. Reports generated by Revelation were rapid and usually possessed >90% accuracy (confirmed by direct sequencing) depending on how stringent the parameters were set (data not shown). However, results generated were also reviewed manually for subtle changes not detected by analysis software.
After the initial study using genotyped samples, the entire 27 CF exons were amplified from each of 12 randomly selected genomic DNA samples (determined as wild types by the American College of Medical Genetics recommended panel) and scanned by TGCE. In the results, 17 exons showed a single peak pattern with no associated shoulder area, suggesting these exons did not have any sequence variation. Multiple peaks were found in various DNA samples in exons 1, 3, 6a, 6b, 14a, 14b, 15, 20, 21, and 24 (Table 3). These data suggest the presence of ∼5% sequence alterations in samples scored negative by our current CF 32-mutation panel. Samples with multiple peaks were sequenced for mutation identification and for final confirmation.
Table 3.
Entire 27 CF Exons of 12 Deidentified DNA Samples Scanned by TGCE Method
| Exon | Amplicon size (bp) | Alterations* | Confirmation by sequencing | Consequence† |
|---|---|---|---|---|
| 1 | 335 | 1 | 125 G/C | Sequence variation (5′ flanking) |
| 2 | 210 | 0 | ||
| 3 | 234 | 1 | 332 C/T | Change Pro to Leu at 67 |
| 4 | 270 | 0 | ||
| 5 | 186 | 0 | ||
| 6a | 248 | 1 | 875 + 40 A/G | Sequence variation in intron 6a |
| 6b | 239 | 4 | IVS6a (GATT)n‡ | Sequence variation in intron 6a |
| 7 | 345 | 0 | ||
| 8 | 233 | 0 | ||
| 9 | 263 | 0 | ||
| 10 | 292 | 0 | ||
| 11 | 175 | 0 | ||
| 12 | 250 | 0 | ||
| 13 | 834 | 0 | ||
| 14a | 248 | 5 | 2694 T/G | No change (Thr at 854) |
| 14b | 192 | 1 | 2789+ 2 ins A | Suspected deleterious |
| 15 | 322 | 1 | 3030 G/A | No change (Thr at 966) |
| 16 | 216 | 0 | ||
| 17a | 243 | 0 | ||
| 17b | 292 | 0 | ||
| 18 | 217 | 0 | ||
| 19 | 322 | 0 | ||
| 20 | 206 | 1 | 3991 G/A | Unknown mutation, change Gly to Arg at 1287 |
| 21 | 250 | 1 | 4029 A/G | No change (Thr at 1299) |
| 22 | 249 | 0 | ||
| 23 | 193 | 0 | ||
| 24 | 250 | 4 | 4521 G/A | No change (Gln at 1463) |
| Total | Amplicon analyzed | Potential SNP (%)§ | ||
| 27 | 324 | 5% |
Number of samples containing potential alterations (n = 12).
All mutations discovered by TGCE analysis were sequenced and compared to the CFTR database (http://www.genet.sickkids.on.ca/cftr/) for clinical significance.
IVS6a (GATT)n is a reported tetra nucleotide repeat, also known as short tandem repeat (STR).
STR is not included in the calculation.
Sequencing Confirmation
All multipeak samples (n = 20) and at least two single-peak samples (n = 54) per each exon were sequenced. In addition, we have sequenced exon 9 for all 12 samples because the mutation A455E is difficult to be detected in some systems. Results showed that TGCE had 100% concordance with direct sequencing: no sequence variation was found in samples with a single-peak pattern (true negatives) and one or more nucleotide changes were found in samples with multipeak pattern (true positives). No mutations were found in exon 9 amplicons and no homozygous mutations were found among all samples sequenced. So far, for all samples analyzed, we had 100% sensitivity and 100% specificity. However, a large study including sequence-confirmed positive and negative samples is needed to establish the true sensitivity and specificity of TGCE. In summary, sequence variations in the 12 deidentified DNA samples were characterized as following: 4 of 12 (33%) have STRs (short tandem repeats), 10 of 12 (83%) have single nucleotide changes, and 1 of 12 (8%) have either a nucleotide insertion or deletion. Only two DNA samples (17%) were identical to the published wild-type sequence for all 27 exons scanned. These data show the complex genetic diversity of the CFTR gene, which suggests the need of a reliable scanning method to analyze the full gene in a high-throughput format.
Nucleotide changes found in exon 14a, 15, 21, and 24 were reported sequence variations and did not change the encoding amino acid5 (Table 3). The single base changes found in exon 3 altered the coding amino acid, but no clinical significance has been reported to be associated with these changes (Table 3). Four of twelve samples contained a tetra nucleotide repeat IVS6a (GATT)n in intron 6a, which is a known genetic marker and has been used to trace the origin of different CF mutations.22,23 The primers for exon 6a were subsequently redesigned to exclude this polymorphism. An insertion of A 2 bases after exon 14b was found in one DNA sample. Previous literature reported this insertion might be a mild allele that contributes to the development of hypoplastic vas deferens phenotype in patients with ΔF508.24 In this study, we also found a single base alteration that has not been reported in the current CF database (CFGAC). This sequence variant is a G/A in position 3991 (exon 20) that changes the encoded amino acid (position 1287) from glycine to arginine. The significance of this alteration is unknown. To summarize, the additional 12 samples scanned added another nine different sequence alterations that were detected by our TGCE method (Tables 1and 3).
Discussion
More than 1000 mutations have been reported since the CFTR gene was cloned and characterized in 1989.23,25 Of these mutations, ΔF508 (a 3-base deletion) is the most frequent mutation and results in a defective cAMP-regulated chloride transport in epithelial cells.26 Other mutations in the CFTR gene such as G542X, G551D, and N1303K occur in greater than 1% in the CF population and are associated with severe pancreatic insufficiency.23,27 Recently, carriers of the I148T mutation have received more attention because I148T has been found in association with the 3199del6 mutation, which may be necessary for the classic CF phenotype.28 Because of the complexity of both the mutations and the phenotypes, a high-throughput mutation scanning method to screen the entire coding region of the CFTR gene may provide valuable clinical information regarding CF genotypes and respective phenotypes.
Electrophoresis-based methods such as denaturing gradient gel electrophoresis (DGGE) and confirmation sensitive gel electrophoresis have been widely used for mutation scanning with accuracy of up to 80%.6,19 However, for large gene analysis that is necessary for CFTR gene, these methods are labor-intensive and time consuming. A recent method using denaturing high-performance liquid chromatography to screen for sequence variations in the CFTR gene has been reported.15 This method is rapid (3 to 5 minutes per sample) but still requires intensive temperature (Tm) optimization for each exon to separate heteroduplexes from the wild types.
In comparison to these technologies, the approach we describe in this study possesses several advantages for scanning a large multiexon gene. This technology requires less DNA (50 to 500 pg/μl) and uses a temperature gradient rather than a fixed temperature.19 Another advantage is that this method does not require labeled primers, probes, or specific dyes. The experimental protocol for using TGCE is simple, and by replacing the polymer matrix after each use, cross contamination is minimized, thus eliminating the residual interference left from a previous experiment. Finally, instruments with 192 capillaries are available for higher throughput applications. Using the CFTR gene (27 exons) as an example, a total of 30 patient samples (one wild-type plus five patient samples per plate) can be scanned in one complete run (6- of 192-well plate per one complete run) without operator intervention. The disadvantages with this method are inherent with all scanning techniques. First, sample mixing is required to distinguish homoduplex mutants from wild-type, and every TGCE run currently requires a wild-type control as a reference for automatic heteroduplex calling, resulting in reduced throughput. Second, PCR primer placement is a concern for all PCR-based assays, especially for mutations located deep inside the introns. The mutation 3849 + 10 kb C/T would not be detected in this study because its location was out of the primer scanning range. Therefore, primers must be designed to cover the specific regions of interest. However, one testing scenario would test for common mutations first and reflex to full gene analysis only in cases of clinically diagnosed CF patients with zero or one identified mutation.
TGCE peak patterns could indicate the type of mutation present. In general, a shoulder or an additional peak was usually associated with a single base substitution. Samples with more than two peaks were found to be a sequence insertion or deletion. Because we did not have samples with mutations in every exon, the sensitivity has not been determined for these particular exons. Additional samples with mutations in these exons are needed to complete and validate this work. Because sensitivity and specificity is exon-dependent and not completely known at this time, full gene sequencing is required for CF patients if scanning results are negative.
Two factors that influence mutation detection efficiency in both TGCE and other scanning methods are heteroduplex formation and GC content of a target amplicon. In addition, according to the literature, a minimum amount of 10 to 20% of minority DNA species (sequence varied from wild-type) is required to form enough heteroduplexes in PCR products.13 The nearest neighbor (stacking structure) also affects the heteroduplex formation. Because of the limited data available for longer DNA fragments, we are continuing to investigate how the locations of mutation site, mismatch stability, as well as fragment length, influence the detection range of our TGCE method. These factors may have combined effects and potentially decrease the sensitivity. Amplicons with extreme GC contents (>64% or <27%) sometimes escape detection if not using an optimized melting temperature (Tm).13 In our approach, even with a temperature gradient, we designed each amplicon to have a GC content ∼40 ± 10% except for exon 1 (57.3%) and exon 21 (27.6%). Thus all 27 amplicons theoretically are resolved within our 50 to 55°C gradient.
A main challenge for CFTR mutation scanning is test result interpretations. Determining whether an alteration is a sequence variation or a disease-causing mutation is often challenging without clear clinical data. Interpretation is even more difficult when a disease-causing mutation is associated with mutations of other sites that may be thousands of bases away. This level of complexity is particularly true for cystic fibrosis with severe and mild, atypical forms. For this reason, scanning the CFTR gene should be used cautiously in a clinical setting and only for disease confirmation or highly suspected cases of CF. Its use for general population screening is limited because of the cost and the uncertainty in interpreting alterations in carriers at this time. However, high-throughput full gene analysis can help link different mutations, thus providing valuable information for clinical significance. The approach demonstrated here can be applied not only to CFTR gene but also to other large multiexon genes.
Acknowledgments
We thank SpectruMedix (State College, PA) for initial instrument placement, reagent supply, and technical assistance.
Footnotes
Supported by the Institute for Clinical and Experimental Pathology, ARUP Laboratories.
References
- Wang DG, Fan J-B, Siao C-J, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, Kruglyak L, Stein L, Hsie L, Topaloglou T, Hubbell E, Robinson E, Mittmann M, Morris MS, Shen N, Kilburn D, Rioux J, Nusbaum C, Rozen S, Hudson TJ, Lipshutz R, Chee M, Lander ES. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- Noone PG, Knowles MR. Review: “CFTR-opathies”: disease phenotypes associated with cystic fibrosis transmembrane regular gene mutations. Respir Res. 2001;2:328–332. doi: 10.1186/rr82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratjen F, Döring G. Cystic fibrosis. Lancet. 2003;361:681–689. doi: 10.1016/S0140-6736(03)12567-6. [DOI] [PubMed] [Google Scholar]
- Grody WW, Cutting GR, Klinger KW, Richards CS, Watson MS, Desnick RJ. Laboratory standards and guidelines for population-based cystic fibrosis carrier screening. Genet Med. 2001;3:149–154. doi: 10.1097/00125817-200103000-00010. [DOI] [PubMed] [Google Scholar]
- Bombieri C, Giorgi S, Carles S, de Cid R, Belpinati F, Tandoi C, Pallares-Ruiz N, Lazaro C, Ciminelli BM, Romey MC, Casals T, Pomper F, Gandini G, Claustres M, Estivill X, Pignatti PF, Modiano G. A new approach for identifying non-pathogenic mutations. An analysis of the cystic fibrosis transmembrane regulator gene in normal individuals. Hum Genet. 2000;106:172–178. doi: 10.1007/s004390051025. [DOI] [PubMed] [Google Scholar]
- Kirk BW, Feinsod M, Favis R, Kliman RM, Barany F. Survey and summary: single nucleotide polymorphism seeking long term association with complex disease. Nucleic Acids Res. 2002;30:3295–3311. doi: 10.1093/nar/gkf466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen VN, Kelefiotis D, Kristensen T, Borresen-Dale AL. High throughput methods for detection of genetic variation. BioTechniques. 2001;30:318–332. doi: 10.2144/01302tt01. [DOI] [PubMed] [Google Scholar]
- Witter CT, Herrmann MG, Moss AA, Rasmussen RP. Continuous fluorescent monitoring of rapid cycle DNA amplification. BioTechniques. 1997;22:130–138. doi: 10.2144/97221bi01. [DOI] [PubMed] [Google Scholar]
- Lyon E. Mutation detection using fluorescent hybridization probes and melting curve analysis. Exp Rev Mol Diagn. 2001;1:17–26. doi: 10.1586/14737159.1.1.92. [DOI] [PubMed] [Google Scholar]
- Cronin MT, Fucini RV, Kim SM, Masino RS, Wespi RM, Miyada CG. Cystic fibrosis mutation detection by hybridization to light-generated DNA probe arrays. Hum Mutat. 1996;7:244–255. doi: 10.1002/(SICI)1098-1004(1996)7:3<244::AID-HUMU9>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Ganguly A. An update on conformation sensitive gel electrophoresis. Hum Mutat. 2002;19:334–342. doi: 10.1002/humu.10059. [DOI] [PubMed] [Google Scholar]
- Li-Sucholeiki XC, Thilly WG. A sensitive scanning technology for low frequency nuclear point mutations in human genomic DNA. Nucleic Acids Res. 2000;28(9):E44. doi: 10.1093/nar/28.9.e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Smith DI, Rechtzigel KJ, Thibodeau SN, James CD. Denaturing high performance liquid chromatography (DHPLC) used in the detection of germline and somatic mutations. Nucleic Acids Res. 1998;26:1396–1400. doi: 10.1093/nar/26.6.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oefner PJ, Underhill PA. DNA mutation detection using denaturing high-performance liquid chromatography (DHPLC). Curr Prot Hum Genet. 1998;7:10.1–10.12. doi: 10.1002/0471142905.hg0710s48. [DOI] [PubMed] [Google Scholar]
- Le Marechal C, Audrezet MP, Quere I, Raquenes O, Langonne S, Ferec C. Complete and rapid scanning of the cystic fibrosis transmembrane conductance regulator (CFTR) gene by denaturing high-performance liquid chromatography (D-HPLC): major implications for genetic counselling. Hum Genet. 2001;108:290–298. doi: 10.1007/s004390100490. [DOI] [PubMed] [Google Scholar]
- Ravnik-Glavac M, Atkinson A, Glavac D, Dean M. DHPLC screening of cystic fibrosis gene mutations. Hum Mutat. 2002;19:374–383. doi: 10.1002/humu.10065. [DOI] [PubMed] [Google Scholar]
- Gelfi C, Righetti PG, Cremonesi L, Ferrari M. Detection of point mutations by capillary electrophoresis in liquid polymers in temporal thermal gradients. Electrophoresis. 1994;15:1506–1511. doi: 10.1002/elps.11501501215. [DOI] [PubMed] [Google Scholar]
- Gao Q, Yeung ES. High-throughput detection of unknown mutations by using multiplexed capillary electrophoresis with poly(vinylpyrrolidone) solution. Anal Chem. 2000;72:2499–2506. doi: 10.1021/ac991362w. [DOI] [PubMed] [Google Scholar]
- Li Q, Liu Z, Monore H, Culiat CT. Integrated platform for detection of DNA sequence variants using capillary array electrophoresis. Electrophoresis. 2002;23:1499–1511. doi: 10.1002/1522-2683(200205)23:10<1499::AID-ELPS1499>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. Krawetz S, Misener S, editors. Totowa: Humana Press,; Bioinformatics Methods and ProtocolsMethods in Molecular Biology. 2000:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Malet I, Belnard M, Agut H, Cahour A. From RNA to quasispecies: a DNA polymerase with proofreading activity is highly recommended for accurate assessment of viral diversity. J Virol Methods. 2003;109:161–170. doi: 10.1016/s0166-0934(03)00067-3. [DOI] [PubMed] [Google Scholar]
- Chehab EF, Johnson J, Louie E, Goossens M, Kawasaki E, Erlich H. A dimorphic 4-bp repeat in the cystic fibrosis gene is in absolute linkage disequilibrium with the ΔF508 mutation: implications for prenatal diagnosis and mutation origin. Am J Hum Genet. 1991;48:223–226. [PMC free article] [PubMed] [Google Scholar]
- Mateu E, Calafell F, Lao O, Bonne-Tamir B, Kidd JR, Pakstis A, Kidd KK, Bertranpetit J. Worldwide genetic analysis of the CFTR region. Am J Hum Genet. 2001;68:103–117. doi: 10.1086/316940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezequel P, Dubourg C, Le Lannou D, Odent S, Le Gall JY, Blayau M, Le Treut A, David V. Molecular screen of the CFTR gene in men with anomalies of the vas deferens: identification of three novel mutations. Mol Hum Reprod. 2000;6:1063–1067. doi: 10.1093/molehr/6.12.1063. [DOI] [PubMed] [Google Scholar]
- Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, Drumm ML, Iannuzzi MC, Collins FS, Tsui LC. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Sermet-Gaudelus I, Vallee B, Urbin I, Torossi T, Marianovski R, Fajac A, Feuillet MN, Bresson JL, Lenoir G, Bernaudin JF, Edelman A. Normal function of the cystic fibrosis conductance regulator protein can be associated with homozygous ΔF508 mutation. Pediatr Res. 2002;52:628–635. doi: 10.1203/00006450-200211000-00005. [DOI] [PubMed] [Google Scholar]
- Walkowiak J, Herzig KH, Witt M, Pogorzelski A, Piotrowski R, Barra E, Sobczynska-Tomaszewska A, Trawinska-Bartnicka M, Strzykala K, Cichy W, Sands D, Rutkiewicz E, Krawczynski M. Analysis of exocrine pancreatic function in cystic fibrosis: one mild CFTR mutation does not exclude pancreatic insufficiency. Eur J Clin Invest. 2001;31:796–801. doi: 10.1046/j.1365-2362.2001.00876.x. [DOI] [PubMed] [Google Scholar]
- Rohlfs EM, Zhou Z, Sugarman EA, Heim RA, Pace RG, Knowles MR, Silverman LM, Allitto BA. The I148T CFTR allele occurs on multiple haplotypes: a complex allele is associated with cystic fibrosis. Genet Med. 2002;4:319–323. doi: 10.1097/00125817-200209000-00001. [DOI] [PubMed] [Google Scholar]