Abstract
Epidermolysis bullosa (EB) is an inherited mechano-bullous disorder of the skin, and is divided into three major categories: EB simplex (EBS), dystrophic EB, and junctional EB (JEB). Mutations in the plectin gene (PLEC1) cause EBS associated with muscular dystrophy, whereas JEB associated with pyloric atresia (PA) results from mutations in the α6 and β4 integrin genes. In this study, we examined three EB patients associated with PA from two distinct families. Electron microscopy detected blister formation within the basal keratinocytes leading to the diagnosis of EBS. Surprisingly, immunohistochemical studies using monoclonal antibodies to a range of basement membrane proteins showed that the expression of plectin was absent or markedly attenuated. Sequence analysis demonstrated four novel PLEC1 mutations. One proband was a compound heterozygote for a nonsense mutation of Q305X and a splice-site mutation of 1344G→A. An exon-trapping experiment suggested that the splice-site mutation induced aberrant splicing of the gene. The second proband harbored a heterozygous maternal nonsense mutation, Q2538X and homozygous nonsense mutations R1189X. Analysis of the intragenic polymorphisms of PLEC1 suggested that R1189X mutations were due to paternal segmental uniparental isodisomy. These results indicate that PLEC1 is a possible causative gene in this clinical subtype, EBS associated with PA. Furthermore, two patients out of our three cases died in infancy. In terms of clinical prognosis, this novel subtype is the lethal variant in the EBS category.
Epidermolysis bullosa (EB) comprises a group of genetically determined skin fragility disorders characterized by blistering of the skin and mucous membrane. EB has traditionally been divided into three main categories on the basis of the level of tissue separation within the cutaneous basement membrane zone (BMZ); tissue separations in EB simplex (EBS), dystrophic EB and junctional EB (JEB) occur in the basal keratinocytes, the dermis, and the lamina lucida of basement membrane, respectively (Table 1).1 Recent advances in EB research, have allowed the identification of mutations in 10 different genes, which account for the clinical heterogeneity in EB (Table 1).2 Dominantly inherited EBS results from mutations in the basal keratinocyte-specific keratin 5 and 14 genes, whereas mutations in the plectin gene (PLEC1) can cause recessive EBS complicated by muscular dystrophy (EBS-MD). Dystrophic EB is characterized by severe blistering and scarring, and is due to mutations in the type VII collagen gene. The defective genes in JEB include the three genes encoding for the laminin 5 chains, the type XVII collagen gene, or the genes encoding the hemidesmosome-associated integrin α6 and β4 subunits (ITGA6 and ITGB4).
Table 1.
Epidermolysis Bullosa (EB) Classification and the Causative Genes1
| Major EB type | Major EB subtypes | Involved genes/protein |
|---|---|---|
| EB simplex (EBS) | Dowling-Meara EBS | K5, K14 |
| Koebner EBS | K5, K14 | |
| Weber-Cockayne EBS | K5, K14 | |
| EBS with muscular dystrophy | Plectin | |
| Junctional EB (JEB) | Herlitz JEB | Laminin 5 |
| Non-Herlitz JEB | Laminin 5, BPAG2 | |
| JEB with pyloric atresia | α6β4 integrin | |
| Dystrophic EB (DEB) | Dominant DEB | Type VII collagen |
| Hallopeau-Siemens recessive DEB | Type VII collagen | |
| Non-Hallopeau-Siemens recessive DEB | Type VII collagen |
The subtype of JEB involving α6 and β4 integrins is associated with congenital pyloric atresia (PA).3,4,5 Staining with specific antibodies to the α6 or β4 integrin subunits in the JEB-PA skin reveals reduced or absent staining.6,7 JEB-PA is usually lethal, but non-lethal variants have also been reported. The affected individuals with the lethal forms usually die within the first weeks or months after birth, whereas in the non-lethal variants, the clinical severity tends to improve with age.3,5,8,9,10
However, there have been several recent reports demonstrating EB cases associated with PA showing an intraepidermal level of cleavage consistent with the diagnosis of EBS.11,12,13,14 In this study, we have encountered three similar cases of EBS associated with PA that demonstrated an abnormal epidermal expression of plectin. Furthermore, we have identified novel four mutations, Q305X, 1344G→A, R1189X, Q2538X in the PLEC1 gene of those cases. Thus, this study furthers our understanding of the possible range of pathophysiology in EB and of the biology of the cutaneous basement membrane.
Materials and Methods
Electron Microscopy
For electron microscopic examination, skin specimens were fixed in 5% glutaraldehyde and post-fixed in 1% osmium tetroxide, stained en-block in uranyl acetate. They were dehydrated in a graded series of ethanol solutions, then embedded in Araldite 6005 (NEM, Tokyo, Japan). Ultra-thin sections were cut, stained with uranyl acetate and lead citrate. The sections were examined with a transmission electron microscope (H-7100, Hitachi; Tokyo, Japan) at 75kv.
Immunofluorescence Studies
Direct immunofluorescence analysis using a series of antibodies against BMZ antigens and cryostat skin sections was performed as described previously.6,15,16 The following monoclonal antibodies (mAbs) against BMZ components were used: mAbs HD1–121, K15, 10F6 and 5B3 against the rod domain of plectin; mAbs GoH3 and 3E1 (Chemicon International, CA) against the α6 and β4 integrins, respectively; mAb GB3 (Sera-lab, Cambridge, UK) against laminin 5 antibody; mAb LH7.2 (Sigma, St. Louis, MO) against type VII collagen; mAb S1193 and HDD20 against BPAG1 and BPA2, respectively. The antibodies GoH3, S1193, and HDD 20 were kind gifts from Dr. A. Sonnenberg, the Netherlands Cancer Institute. The antibodies HD1–121 and K15 were kind gifts from Dr. K. Owaribe, Nogoya University.
Mutation Detection
Genomic DNA was obtained from both patients and the parents. The mutation detection strategy was performed after polymerase chain reaction (PCR) amplification of all exons and intron-exon borders, followed by direct automated nucleotide sequencing (Applied Biosystems, Foster City, CA). The genomic DNA nucleotides, the cDNA nucleotides and the amino acids of the protein were numbered based on the previous sequence information (GenBank Accession No. AH003623).17 In particular, PCR amplification of exon 9, 12, 27, and one part of exon 32 was performed using following primers. Primers 5′-GTCGCTGTATGACGCCATGC-3′and 5′-TGGCTGGTAGCTCCATCTCC-3′ for exon 9 produced a 387-bp fragment of the genomic DNA extending from g.2717 to g.3103, primers 5′-CCCACTCGCCTTAGGACAGT-3′ and 5′-AAACCAACTCTGCCCAAAGC-3′ for exon 12 synthesized a 428-bp fragment from g.3571 to g.3998, primers 5′-TTTCGAGGCTGGGGCTTCAT-3′ and 5′-GCCTGGGTGATGGTGTGGTC-3′ for exon 27 synthesized a 771-pb fragment from g.9681 to g.10451, and primers 5′-TCTGCTTTGGTGGGTGATGG-3′and 5′-AGCCTCTGGTTCTCCTCAGC-3′ for a single part of exon 32 synthesized a 422-bp fragment from g.15326-g.15747. The PCR conditions of the amplification were 5 minutes at 94°C for one cycle, followed by 38 cycles of 45 seconds at 94°C, 30 seconds at 57°C or 60°C, and 1 minute at 72°C. The informed consents both for studies and publication of the photographs were obtained from both families in this study.
Verification of Mutations
Each mutation was confirmed by restriction enzyme digestion of PCR products. The Q305X and 1395G->A mutations resulted in the loss of a restriction site for the PstI and HphI, respectively. The R1189X mutation caused the generation of a new restriction enzyme site for Tsp45I.
There was no proper restriction enzyme to verify the Q2538X mutation. PCR amplification was carried out using the following PCR primers, 5-TCTGCTTTGGTGGGTGATGG-3 and 5-CTCCAGCTTGGCCTTCTCCA-3 for generation of a 225-bp product. We changed the last base of the latter primer (underlined) from the original sequence, so that the combination of this change and the upstream sequence created a new AluI site in the PCR product. Since the Q2538X mutation was also located just one base upstream from that primer, Q2538X abolished this AluI site.
Exon-Trapping Experiments
Exon trapping system (Invitrogen, Carlsbad, CA) is an approach used for the direct isolation of transcribed sequences from genomic DNA.18 To generate a PLEC1 genomic fragment extending from intron 10 to intron 13, we synthesized two primers, 5′-AAACTCGAGGGCTGTCCCAGGTCTGGT-3′ and 5′-ATTGGATCCTGGGGCCGTGTGTACCTG-3′ which contained the following restriction enzyme sites, XhoI and BamHI, respectively. PCR was performed using genomic DNA from the proband 1 as a template. The DNA fragment was digested with XhoI and BamHI and subcloned into the multi-cloning site of a pSPL3 expression vector, which contained a portion of the HIV-1 tat gene, an intron, splice donor and acceptor sites, and some flanking exon sequences. Sequence analysis selected constructs with or without the splice site mutation 1344G→A. The constructs were transfected into normal human epidermal keratinocytes using N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate (Roche Molecular Biochemicals). Total RNA was extracted from the culture cells and RT-PCR was performed using the trapping vector-specific oligonucleotide primers. The samples without transfection of the pSPL3 were used as controls. The PCR products were subcloned into a TA cloning vector pCR II (Invitrogen) and sequenced.
Results
Clinical Features of EB Associated with PA
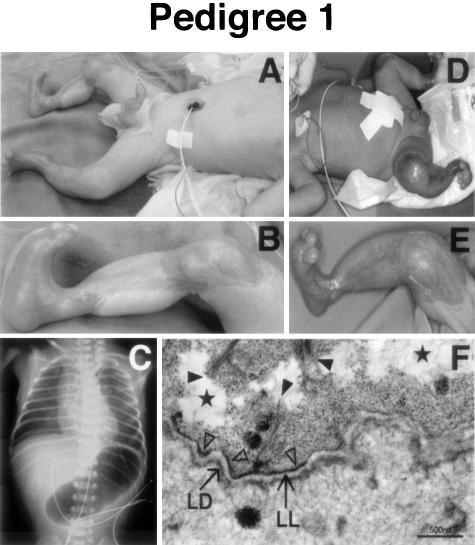
The proband 1 was a male, born by spontaneous vaginal delivery after a 39-week gestation. His parents were unrelated and clinically normal. Pyloric atresia (PA) and polyhydramnios were suspected antenatally after ultrasound examination. Immediately after delivery, he presented with blisters and widespread ulcers on his trunk, genitalia, and legs. Findings from a routine abdominal radiograph led to a clinical diagnosis of EB associated with PA (Figure 1, A to C). Laparotomy revealed a severely distended stomach with a membrane at the pylorus. The membrane was excised and a pyloroplasty was performed. The histopathology of the septum showed re-epithelization of the epithelium, an atrophic lamina muscularis mucosae, and edematous submucosal tissue. He survived the operation, but still required intensive care at the age of 16 months. His elder brother, born 2 years before the proband 1, was also diagnosed as suffering from PA by routine abdominal X-ray. He manifested with multiple blisters and ulcers on his scalp, genitalia, and extremities, identical to those seen in the proband 1 (Figure 1, D to E). On the second day after birth, pyloroplasty and skin grafting were performed, but he later died of sepsis 4 months after birth.
Figure 1.
Clinical, X-ray, and ultrastructural findings of proband 1 in pedigree 1. A: Sharply demarcated erosions and ulcers on the trunk, genitalia, and lower extremity. B: Marked ulcers on the lower extremities. C: A single abdominal bubble of gas in an abdominal X-ray of proband 1. D and E: The same clinical manifestation of his elder brother. F: Electron microscopy of the skin from proband 1 shows tissue separation occurring at the base of the basal keratinocytes (stars). Keratin filaments are sparse and thin and not well associated with the hemidesmosome (HD) inner plaque (full arrowheads). Reduced numbers of hypoplastic HDs are recognized (triangles) and occasional HDs can be observed associated with thin bundles of keratin filaments within the basal keratinocyte (open arrow). The lamina densa (LD) and lamina lucida (LL) are present in the papillary dermis. Bar, 500 nm.
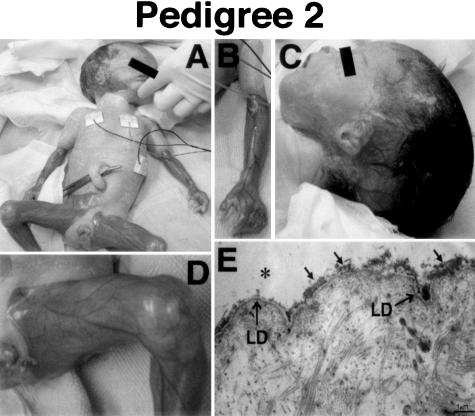
The proband 2 was a female, born by induced vaginal delivery after a 36-week gestation, as the second child of non-consanguineous, healthy parents. She had a 1-year-old brother who was clinically unaffected. Antenatal ultrasonography had suggested a diagnosis of PA and polyhydramnios. At birth, she presented with blisters and ulcers on the scalp, trunk, and extremities (Figure 2, A to D). Routine abdominal X-rays demonstrated a single abdominal bubble of gas, resulting in a clinical diagnosis of EB associated with PA. A laparotomy on the second day after birth showed a hugely distended stomach with a membrane at the pylorus. A gastroduodenostomy was subsequently performed. Histopathological analysis of the septum showed atrophic changes in general, normal appearance of the desquamative epithelium, and no increase in collagen fibers numbers. Due to her poor respiratory condition, she died of dyspnea at 31 days of age.
Figure 2.
Clinical and ultrastructural findings of proband 2 in pedigree 2. A: Clearly demarcated blisters and ulcers on the scalp, abdomen, and extremities. B: Ulcers on the left arm. C: Localized erosions on the scalp. D: Ulcers on the left leg. E: Electron microscopy of the skin shows basal cell debris including flat electron densities (representing remnant hemidesmosome outer plaques (arrows) can be seen at the base of the intraepidermal split (asterisk). Bar, 1 μm.
Skin Separation in Basal Keratinocytes
First, we examined the skin separation level within the epidermal BMZ. Electron microscopy of the skin samples from proband 1 revealed that the tissue separation was localized at the base of the basal keratinocytes and that hemidesmosomes (HDs) were reduced in frequency and hypoplastic (Figure 1F). In the skin sample from the proband 2 there was a reduction in HD numbers and many of the HDs appeared to be small or hypoplastic. The plane of separation was consistently within the basal keratinocyte, just above the HD inner plaque.19 The majority of HD outer plaques remained attached to the underlying plasma membrane and extracellular electron-dense lamina densa. Keratin filaments within the basal cells appeared to form poor attachment to these rudimentary HDs, which remained firmly attached to the base of the separation (Figure 2E). The ultrastructural findings in both cases were consistent with these patients being classified as suffering from EBS.
Abnormal Expression of Plectin
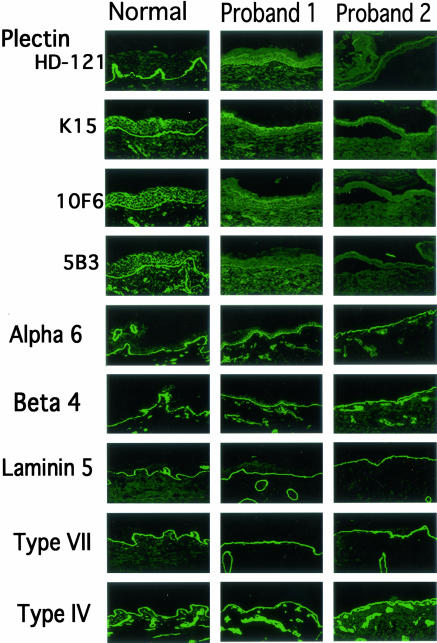
An immunohistochemical study using monoclonal antibodies (mAbs) to a range of BMZ component proteins was performed (Figure 3). Immunoreactivity using the mAb HD1–121 against plectin was markedly attenuated in the proband 1 and completely lost in the proband 2. Analysis using other plectin mAbs K15, 10F6, and 5B5 revealed similar results, in which proband 1 showed markedly reduced expression while no immunoreactivity was detected in proband 2. Immunostaining for other BMZ proteins including the α6 and β4 integrins, laminin 5, type VII and IV collagens were normal (Figure 3). BPAG1 and BPAG2 also showed normal, bright, linear labeling of the BMZ in both probands (data not shown).
Figure 3.
Immunofluorescence using antibodies against basement membrane zone components. Staining using HD1–121, K15, 10F6, and 5B3 monoclonal antibodies (mAbs) for plectin were markedly attenuated in the proband 1 and completely loss in the proband 2. Immunostaining for other proteins including α6 and β4 integrins, laminin 5, type VII and IV collagens were normal in both probands and controls.
PLEC1 Mutations in Proband 1
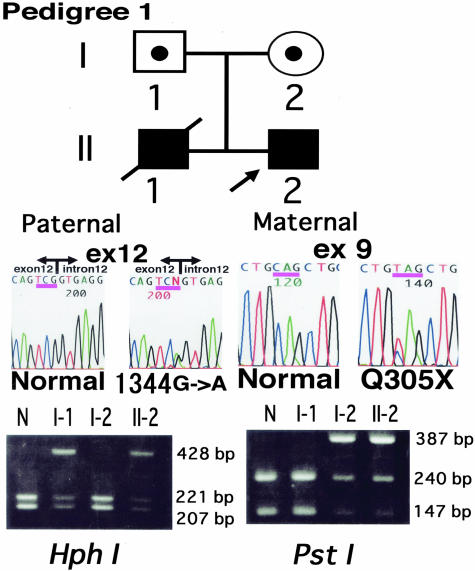
Since abnormal expression of plectin was observed in both probands, we performed direct mutational analysis of the entire plectin gene. In this study, the cDNA nucleotides and the amino acids of the protein were numbered based on the previous sequence information (GenBank Accession No. AH003623).17 Direct nucleotide sequencing of PLEC1 from proband 1 demonstrated nonsense and splice-site mutations. The nonsense mutation was a C→T transition at nucleotide c.913 of cDNA in exon 9, resulting in the substitution of a glutamine (CAG) at position 305 with a stop codon (TAG) (Q305X). The splice-site mutation was a G→A substitution at nucleotide c.1344 in the cDNA that was located at the 3′ end of exon 12 (1344G→A) (Figure 4). The Q305X mutation was maternal and the 1344G→A mutation was paternal. These mutations were confirmed by restriction endonuclease analysis (Figure 4).
Figure 4.
Mutation detection in pedigree 1. The proband harbored a G→A transition at position c.1344 in exon 12 within an intron-exon border (middle-right panel). He also possessed a heterozygous transition 913C→T (exon 9), leading to the substitution of glutamine 305 with a nonsense codon (Q305X). HphI digestion of the 428-bp fragment with and without the 1344G->A mutation product resulted in single band of 428 bp and double bands of 221 and 207 bp, respectively (left panel). The 387-bp PCR fragment containing the Q305X mutation was not digested by PstI whereas the digestion of the fragment without the mutation showed two bands of 240 and 147 bp (right panel). The 1344G→A mutation was paternal and the Q305X mutation was maternal.
Since the splice site mutation of 1344G→A might just be a polymorphism, we examined 120 unrelated alleles as control and were unable to detected the same nucleotide change. Furthermore, we performed an exon-trapping experiment, as keratinocytes from the proband 1 or the parents were unavaliable. We inserted the genomic fragments with or without 1344G→A mutation into this vector, transfected these constructs into normal cultured human epidermal keratinocytes and prepared total RNA from the keratinocytes. We then synthesized cDNA, and amplified the extracted exons by PCR. Agarose gel electrophoresis showed strong 460-bp upper and weak lower 305-bp bands from the samples of the constructs containing the mutation, although a single 554-bp band was amplified from that construct without the mutation (data not shown).
Sequence analysis revealed that the upper 460-bp band contained exons 11 and 13 while the lower 305-bp band contained exon 11 alone. Conversely, exons 11, 12, and 13 were normally extracted from the DNA fragment without the mutation, resulting in a 554-pb band. Deletion of exon 12, which was 94 bp in size, caused a change in the mRNA coding frame and generation of a premature downstream stop codon. However, the combined size of exons 12 and 13 was 249 bp, so the deletion of these exons did not alter the coding frame and restored the translation of a polypeptide that was encoded by the downstream exons.
PLEC1 Mutations in Proband 2
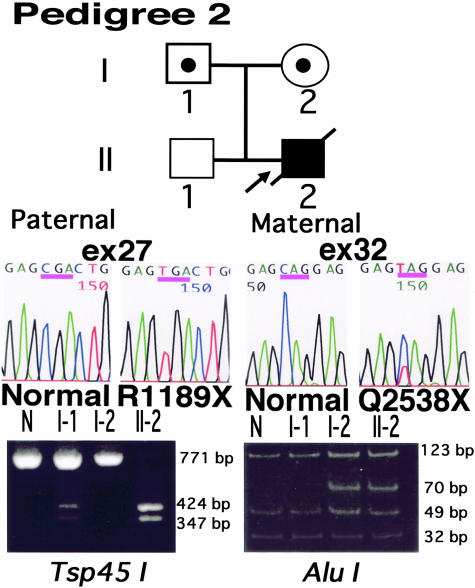
Sequence analysis of the proband 2 revealed an unusual finding of both homozygous and heterozygous mutations. The homozygous mutation was a C→T transition at cDNA position c.3565 in exon 27, resulting in a substitution of an arginine (CGA) at residue 1189 with a stop codon (TGA) (R1189X). Conversely, the heterozygous mutation was a C→T transition at position c.7612 in exon 32, leading to an alteration of a glutamine (CAG) at 2538 to a stop codon (TAG) (Q2538X) (Figure 5). Nucleotide sequences of her parents disclosed that the father and mother were heterozygous for R1189X and Q2538X, respectively. These mutations were confirmed by restriction endonuclease analysis (Figure 5).
Figure 5.
Mutation detection in pedigree 2. The proband 2 harbored a homozygous transition 3565C→T (exon 27) at codon 1189 producing a nonsense codon instead of arginine (R1189X) (middle panel). She also harbored a heterozygous C→T (c.7612) substitution (exon 32) in codon 2538, replacing glutamine with a nonsense codon (Q2538X). The R1189X mutation caused the generation of site for the Tsp45I restriction enzyme. The 771-bp PCR product with the mutation was digested by Tsp45I resulting in 424-bp and 347-bp bands (left panel). Since no proper restriction enzyme site was found around the Q2538X mutation, we changed one base of the PCR primer from the original sequence to create a site for AluI (see Materials and Methods). The digestion of 225-bp PCR product with Q2538X produced 70-bp band (right panel). The father and the mother are heterozygous for R1189X and Q2538X mutations, respectively.
To eliminate a possibility that a sequence variation under one of the primers caused failure to amplify a maternal allele, we investigated by using a second set of primers for exon 27/intron 28. This also confirmed that the R1189X mutation was homozygous (data not shown).
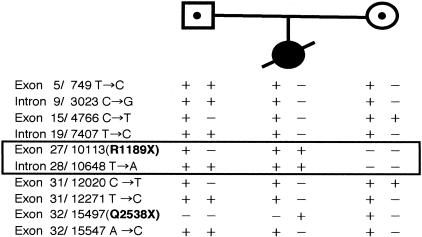
Furthermore, comparison of 8 intragenic polymorphisms showed that the father and mother were heterozygous for 2 of 8 and 6 of 8 markers, respectively (Figure 6). The mutation R1189X and intron 28/10648 T→A were informative for the absence of a maternal allele in the proband 2. These results indicated that the mutations R1189X and their adjacent sequences were of paternal origin, suggesting possibility of segmental paternal uniparental isodisomy over this short area.
Figure 6.
Detection of uniparental isodisomy. Comparison of 8 intragenic polymorphisms around the homozygous R1189 mutation showed that the father and the mother were heterozygous for 2 of 8 and 6 of 8 markers. The mutation R1189X and intron 28/10648 T→A were informative for the absence of a maternal allele in the proband.
Discussion
Plectin, a component of the hemidesmosome (HD) inner plaque, is involved in the attachment and cross-linking of the cytoskeleton and intermediate filaments to specific membrane complexes.20,21 Each monomer comprises a central rod domain of an α-helical coiled-coil structure and flanking amino-and carboxyl-terminal large globular domains.22,23 It is widely distributed in almost all tissues and the exact function in each tissue is still unclear.24,25 Although a family with EBS Ogna was reported to result from plectin mutation,26 mutations in the plectin gene (PLEC1) generally cause EBS associated with muscular dystrophy (EBS-MD), which is characterized by generalized blistering, and a late-onset muscular dystrophy.15,16,17,27,28,29
The two probands in this study were both microscopically classified and clinically diagnosed as suffering from EBS associated with PA. We speculated at first, whether the causative mutations might lie in either ITGA6 or ITGB4 genes, known to cause PA-JEB. Surprisingly, however, the tissue separation was not within the lamina lucida as might be expected for JEB-PA, but was higher within basal keratinocyte, consistent with EBS. Immunohistochemical examination subsequently revealed abnormal plectin expression, leading us to speculate that mutations in PLEC1 might be the underlying cause of this condition.
Indeed, mutational analysis detected novel nonsense and splice-site mutations on each PLEC1 allele of the first proband (Figure 4). The splice-site mutation is thought to affect the splicing of the PLEC1 gene, since the G nucleotide in the GT splice donor site in many human genes is relatively conserved within 78% of all splice junctions.30 We first examined 120 unrelated alleles and could not detect the same nucleotide change. Furthermore, the exon-trapping experiments revealed that this mutation caused aberrant splicing of this region and suggested that this splice site mutation was likely to be pathogenic.
Immunofluorescence staining using the plectin antibodies, HD1–121, K15, 10F6, and 5B3, detected some plectin expression in proband 1 (Figure 3). HD1–121 recognizes multiple epitopes on the plectin rod domain.31 In addition, the 10F6 and 5B3 epitopes were located in the central and carboxyl-terminal parts of the rod, respectively.32 The 1344G→A mutation lies upstream of the region encoding the rod domain. As shown in the exon-trapping experiment, the presence of the low weaker band suggested some expression of the plectin rod domain, which is expected to be insufficient for the normal function of plectin.
Although we found nonsense mutations of both paternal and maternal origins in proband 2, it was unusual that R1189X mutation was homozygous. To further understand the mechanism causing this defect, we searched for several intragenic polymorphisms within the PLEC1 gene (Figure 6). The results suggested that the R1189X mutations and their adjacent sequences had originated from one paternal allele. Uniparental disomy refers to the situation in which two copies of a chromosome come from the same parent.33 The possibility of isodisomy can be hypothesized if two identical segments from one parental homologue are present in the offspring, whereas sequences of both homologues from the transmitting parent are present in the normal cause, heterodisomy. Angelman syndrome and Prader-Willi syndrome are examples of disorders caused by uniparental disomy.34 This study indicates that the R1189X mutations and its adjacent sequences are likely to be derived from one paternal homologue and suggested possibility of segmental uniparental paternal isodisomy.
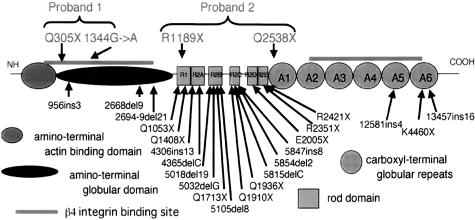
The next point we should discuss was how PLEC1 mutation led to PA. We have reviewed the previously reported cases of plectin mutations in EBS-MD. In total, there were 22 mutations, of those, 16 mutations resided within the region encoding the rod domain (Figure 7). Of the 4 novel mutations from our cases, roughly, Q305X, 1344G->A, and Q2538X were not within the rod domain region and R1189X was in the very end of this domain. Plectin interacts with the cytoplasmic tail of β4 integrin via its carboxyl as well as amino-terminal domains (Figure 7),23,35 and mutations in the β4 integrin gene result in PA. Therefore, we postulated that the development of PA in our cases is closely linked with the functions of plectin and the β4 integrin subunit. Interestingly, the mutations Q305X and 1344G→A in the first proband resided in the amino-terminal binding site for β4 integrin (Figure 7). However, the mutations of the second proband were not in the β4 integrin-binding site.
Figure 7.
Database showing the position of mutations in PLEC1. Each functional domain is shown in the schematic model of plectin structure. The cDNA and the amino acids of the protein are numbered based on the previous sequence information (GenBank Accession No. AH003623).17 Amino-terminal actin binding domain, amino-terminal globular domain, β4 integrin binding sites, rod domain, and carboxyl-terminal globular domain are shown.
The level of tissue separation in EBS due to plectin gene mutations is different from that due to K5 or K14 gene mutations. In the latter cases there is a global cytolysis of the basal keratinocytes, while in case of the patients with PLEC1 mutations the tissue separation is very low at the level of hemidesmosomes. Also, molecular consequences of deletion of the cytoplasmic domain of β4 integrin subunit or the type XVII collagen caused predominant features of EBS with intraepidermal separation.36,37 Therefore, some investigators have proposed hemidesmosomal variants that include EBS with muscular dystrophy, JEB with pyloric atresia and generalized atrophic benign EB2. In this aspect, EBS with pyloric atresia in this study would belong to the variants.
Molecular mechanism of PA development even in JEB-PA has not yet been fully elucidated although phenotype and genotype information of this disease has been accumulated. Targeted ablation of both the ITGA4 and ITGB6 genes in mice clearly induced separation of the epidermis from the dermis as seen in the skin of JEB-PA.38,39,40 In contrast, those model mice could not show straightforward evidence concerning mechanism of PA. Further studies are needed to clarify the pathogenesis of PA in all patients with EBS-PA as well as those with JEB-PA.
In this study, we have demonstrated for the first time that PLEC1 mutations induce novel subtype of EB, EBS-PA. No definite conclusion has yet been reached regarding the occurrence of late onset muscular dystrophy phenotype of our three cases, since two patients have already died and one patient is still a neonate. However, this study furthers our understanding of the possible range of pathophysiology in EB and of the biology of the cutaneous basement membrane. In the previously reported four cases of EBS-PA,11,12,13,14 two cases11,12 also died shortly after birth although a search for plectin mutations were not carried out. In terms of clinical prognosis, this novel subtype EBS-PA is the lethal variant in the EBS category and will become a real target for gene therapy in the near future.
Acknowledgments
We thank the patients and their family for their interest in our study and the referring physicians for providing clinical information.
Footnotes
Supported by a Grant-In-Aid for Scientific Research (A) from the Japanese Society for the Promotion of Science (project no. 13357008 to H.S.), by a Health and Labor Sciences Research Grant (Research on Specific Diseases to H.S.) from the Ministry of Health Labor and Welfare of Japan, by a grant from the Japanese Society for the Promotion of Science (JSPS grant no. 00345 to J.R.M.), and by a grant-in-aid for JSPS fellows’ research expenses (no. 00345).
References
- Fine JD, Eady RA, Bauer EA, Briggaman RA, Bruckner-Tuderman L, Christiano A, Heagerty A, Hintner H, Jonkman MF, McGrath J, McGuire J, Moshell A, Shimizu H, Tadini G, Uitto J. Revised classification system for inherited epidermolysis bullosa: report of the Second International Consensus Meeting on diagnosis and classification of epidermolysis bullosa. J Am Acad Dermatol. 2000;42:1051–66. [PubMed] [Google Scholar]
- Pulkkinen L, Uitto J. Mutations analysis and molecular genetics of epidermolysis bullosa. Matrix Biol. 1999;18:29–42. doi: 10.1016/s0945-053x(98)00005-5. [DOI] [PubMed] [Google Scholar]
- Vidal F, Aberdam D, Miquel C, Christiano AM, Pulkkinen L, Uitto J, Ortonne JP, Meneguzzi G. Integrin beta 4 mutations associated with junctional epidermolysis bullosa with pyloric atresia. Nat Genet. 1995;10:229–234. doi: 10.1038/ng0695-229. [DOI] [PubMed] [Google Scholar]
- Ruzzi L, Gagnoux-Palacios L, Pinola M, Belli S, Meneguzzi G, D’Alessio M, Zambruno G. A homozygous mutation in the integrin alpha6 gene in junctional epidermolysis bullosa with pyloric atresia. J Clin Invest. 1997;99:2826–2831. doi: 10.1172/JCI119474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulkkinen L, Kimonis VE, Xu Y, Spanou EN, McLean WH, Uitto J. Homozygous alpha6 integrin mutation in junctional epidermolysis bullosa with congenital duodenal atresia. Hum Mol Genet. 1997;6:669–674. doi: 10.1093/hmg/6.5.669. [DOI] [PubMed] [Google Scholar]
- Shimizu H, Suzumori K, Hatta N, Nishikawa T. Absence of detectable alpha 6 integrin in pyloric atresia-junctional epidermolysis bullosa syndrome: application for prenatal diagnosis in a family at risk for recurrence. Arch Dermatol. 1996;132:919–925. [PubMed] [Google Scholar]
- Brown TA, Gil SG, Sybert VP, Lestringant GG, Tadini G, Caputo R, Carter WG. Defective integrin alpha 6 beta 4 expression in the skin of patients with junctional epidermolysis bullosa and pyloric atresia. J Invest Dermaol. 1996;132:919–925. doi: 10.1111/1523-1747.ep12363370. [DOI] [PubMed] [Google Scholar]
- Nissen CM, van der Raaij-Helmer MH, Hulsman EH, van der Neut R, Jonkman MF, Sonnenberg A. Deficiency of the integrin beta 4 subunit in junctional epidermolysis bullosa with pyloric atresia: consequences for hemidesmosome formation and adhesion properties. J Cell Sci. 1996;109:1695–1706. doi: 10.1242/jcs.109.7.1695. [DOI] [PubMed] [Google Scholar]
- Mellerio JE, Pulkkinen L, McMillan JR, Lake BD, Horn HM, Tidman MJ, Harper JI, McGrath JA, Uitto J, Eady RA. Pyloric atresia-junctional epidermolysis bullosa syndrome: mutations in the integrin β4 gene (ITGB4) in two unrelated patients with mild disease. Br J Dermatol. 1998;139:862–871. doi: 10.1046/j.1365-2133.1998.02515.x. [DOI] [PubMed] [Google Scholar]
- Ashton GH, Sorelli P, Mellerio JE, Keane FM, Eady RA, McGrath JA. Alpha6 beta4 integrin abnormalities in junctional epidermolysis bullosa with pyloric atresia. Br J Dermatol. 2001;144:408–414. doi: 10.1046/j.1365-2133.2001.04038.x. [DOI] [PubMed] [Google Scholar]
- Cowton JA, Beattie TJ, Gibson AA, Mackie R, Skerrow CJ, Cockburn F. Epidermolysis bullosa in association with aplasia cutis congenital and pyloric atresia. Acta Paediatr Scand. 1982;71:155–160. doi: 10.1111/j.1651-2227.1982.tb09391.x. [DOI] [PubMed] [Google Scholar]
- Gire C, Nicaise C, Minodier P, Meneguzzi G, Prost C, Souia F. Epidermolysis bullosa and pyloric atresia. Acta Paediatr. 1999;88:105–106. doi: 10.1111/j.1651-2227.1999.tb01281.x. [DOI] [PubMed] [Google Scholar]
- Kim DK, Kim SC, Chang SN, Kim SY. Epidermolysis bullosa simplex (Dowling-Meara type) associated with pyloric atresia and congenital urologic abnormalities. Yonsei Med J. 2000;41:411–415. doi: 10.3349/ymj.2000.41.3.411. [DOI] [PubMed] [Google Scholar]
- Morrell DS, Rubenstein DS, Briggaman RA, Fine JD, Pulkkinen L, Uitto J. Congenital pyloric atresia in a newborn with extensive aplasia cutis congenital and epidermolysis bullosa simplex. Br J Dermatol. 2000;143:1342–1343. doi: 10.1046/j.1365-2133.2000.03929.x. [DOI] [PubMed] [Google Scholar]
- Smith FJ, Eady RA, Leigh IM, McMillan JR, Rugg EL, Kelsell DP, Bryant SP, Spurr NK, Geddes JF, Kirtschig G, Milana G, de Bono AG, Owaribe K, Wiche G, Pulkkinen L, Uitto J, Mclean WH, Lane EB. Plectin deficiency results in muscular dystrophy with epidermolysis bullosa. Nat Genet. 1996;13:450–457. doi: 10.1038/ng0896-450. [DOI] [PubMed] [Google Scholar]
- Shimizu H, Takizawa Y, Pulkkinen L, Murata S, Kawai M, Hachisuka H, Udono M, Uitto J, Nishikawa T. Epidermolysis bullosa simplex associated with muscular dystrophy: phenotype-genotype correlation and review of the literature. J Am Acad Deramtol. 1999;41:950–956. doi: 10.1016/s0190-9622(99)70252-5. [DOI] [PubMed] [Google Scholar]
- McLean WH, Pulkkinen L, Smith FJ, Rugg EL, Lane EB, Bullric F, Burgeson RE, Amano S, Hudson DL, Owaribe K, McGrath JA, McMillan JR, Eady RA, Leigh IM, Christiano AM, Uitto J. Loss of plectin causes epidermolysis bullosa with muscular dystrophy: cDNA cloning and genomic organization. Genes Dev. 1996;10:1724–1735. doi: 10.1101/gad.10.14.1724. [DOI] [PubMed] [Google Scholar]
- Buckler AJ, Chang DD, Graw SL, Brook JD, Haber DA, Sharp PA, Housman DE. Exon amplification: a strategy to isolate mammalian genes based on RNA splicing. Proc Natl Acad Sci USA. 1991;88:4005–4009. doi: 10.1073/pnas.88.9.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan JR, McGrath JA, Tidman MJ, Eady RA. Hemidesmosomes show abnormal association with the keratin filament network in junctional forms of epidermolysis bullosa. J Invest Dermatol. 1998;110:132–137. doi: 10.1046/j.1523-1747.1998.00102.x. [DOI] [PubMed] [Google Scholar]
- Klatte DH, Kurpakus MA, Grelling KA, Jones JCR. Immunochemical characterization of three components of the hemidesmosome and their expression in cultured epithelial cells. J Cell Biol. 1989;109:3377–3390. doi: 10.1083/jcb.109.6.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borradori L, Sonnenberg A. Structure and function of hemidesmosome: more than simple adhesion complexes. J Invest Dermatol. 1999;112:411–41. doi: 10.1046/j.1523-1747.1999.00546.x. [DOI] [PubMed] [Google Scholar]
- Liu CG, Maercker C, Castanon MJ, Hauptmann R, Wiche G. Human plectin: organization of the gene, sequence analysis, and chromosome localization (8q24). Proc Natl Acad Sci USA. 1996;93:4278–4283. doi: 10.1073/pnas.93.9.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiche G. Role of plectin in cytoskeleton organization and dynamics. J Cell Sci. 1998;111:2477–2486. doi: 10.1242/jcs.111.17.2477. [DOI] [PubMed] [Google Scholar]
- Wiche G, Krepler R, Artlieb U, Pytela R, Denk H. Occurrence and immunolocalization of plectin in tissues. J Cell Biol. 1983;97:887–901. doi: 10.1083/jcb.97.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitto J, Pulkkinen L, Smith FJ, McLean WHI. Plectin and human genetic disorders of the skin and muscle: the paradigm of epidermolysis bullosa with muscular dystrophy. Exp Dermatol. 1996;5:237–246. doi: 10.1111/j.1600-0625.1996.tb00124.x. [DOI] [PubMed] [Google Scholar]
- Koss-Harnes D, Hoyheim B, Anton-Lamprecht I, Gjesti A, Jorgensen RS, Jahnsen FL, Olaisen B, Wiche G, Gedde-Dahl T., Jr A site-specific plectin mutation causes dominant epidermolysis bullosa simplex Ogna: two identical de novo mutations. J Invest Dermatol. 2002;118:87–93. doi: 10.1046/j.0022-202x.2001.01591.x. [DOI] [PubMed] [Google Scholar]
- Chavanas S, Pulkkinen L, Gache Y, Smith FJ, Mclean WH, Uitto J, Ortonne JP, Meneguzzi G. A homozygous nonsense mutation in the PLEC1 gene in patients with epidermolysis bullosa simplex with muscular dystrophy. J Clin Invest. 1996;98:2196–2200. doi: 10.1172/JCI119028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gache Y, Chavanas S, Lacour JP, Wiche G, Owaribe K, Meneguzzi G, Ortonne JP. Defective expression of plectin/HD1 in epidermolysis bullosa simplex with muscular dystrophy. J Clin Invest. 1996;97:2289–2298. doi: 10.1172/JCI118671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulkkinen L, Smith FJD, Shimizu H, Murata S, Yaoita H, Hachisuga H, Nishikawa T, McLean WHI, Uitto J. Homozygous deletion mutations in the plectin gene (PLEC1) in patients with epidermolysis bullosa simplex associated with late-onset muscular dystrophy. Hum Mol Genet. 1996;5:1539–1546. doi: 10.1093/hmg/5.10.1539. [DOI] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura M, Uematsu J, Hirako Y, Nishizawa Y, Shimizu H, Kido N, Owaribe K. Identification of the hemidesmosomal 500 kDa protein (HD1) as plectin. J Biochem. 1999;126:1144–1150. doi: 10.1093/oxfordjournals.jbchem.a022560. [DOI] [PubMed] [Google Scholar]
- Foisner R, Feldman B, Sander L, Seifert G, Artlieb U, Wiche G. A panel of monoclonal antibodies to rat plectin: distinction by epitope mapping and immunoreactivity with different tissues and cell lines. Acta Histochem. 1994;96:421–438. doi: 10.1016/S0065-1281(11)80029-2. [DOI] [PubMed] [Google Scholar]
- Kotzot D. Complex and segmental uniparental disomy (UPD): review and lessons from rare chromosomal complements. J Med Genet. 2001;38:497–507. doi: 10.1136/jmg.38.8.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy SB, Schwartz S. Prader-Willi and Angelman syndromes: disorders of genomic imprinting. Medicine. 1998;77:140–151. doi: 10.1097/00005792-199803000-00005. [DOI] [PubMed] [Google Scholar]
- Schaapveld RQ, Borradori L, Geerts D, van Leusden MR, Kuikman I, Nievers MG, Niessen CM, Steenbergen RD, Snijders PJ, Sonnenberg A. Hemidesmosome formation is initiated by the beta4 integrin subunit, requires complex formation of beta4 and HD1/plectin, and involves a direct interaction between beta4 and the bullous pemphigoid antigen 180. J Cell Biol. 1998;142:271–284. doi: 10.1083/jcb.142.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontao L, Tasanen K, Huber M, Hohl D, Koster J, Bruckner-Tuderman L, Sonnenberg A, Borradori L. Molecular consequences of deletion of the cytoplasmic domain of bullous pemphigoid 180 in a patient with predominant features of epidermolysis bullosa simplex. J Invest Dermatol. 2004;122:65–72. doi: 10.1046/j.0022-202X.2003.22125.x. [DOI] [PubMed] [Google Scholar]
- Jonkman MF, Pas HH, Nijenhuis M, Kloosterhuis G, Steege G. Deletion of a cytoplasmic domain of integrin beta4 causes epidermolysis bullosa simplex. J Invest Dermatol. 2002;119:1275–1281. doi: 10.1046/j.1523-1747.2002.19609.x. [DOI] [PubMed] [Google Scholar]
- van der Neut R, Krimpenfort P, Calafat J, Niessen CM, Sonnenberg A. Epithelial detachment due to absence of hemidesmosomes in integrin beta 4 null mice. Nat Genet. 1996;13:366–369. doi: 10.1038/ng0796-366. [DOI] [PubMed] [Google Scholar]
- Georges-Labouesse E, Messaddeq N, Yehia G, Cadalbert L, Dierich A, Le Meur M. Absence of integrin alpha 6 leads to epidermolysis bullosa and neonatal death in mice. Nat Genet. 1996;13:370–373. doi: 10.1038/ng0796-370. [DOI] [PubMed] [Google Scholar]
- Murgia C, Blaikie P, Kim N, Dans M, Petrie HT, Giancotti FG. Cell cycle and adhesion defects in mice carrying a targeted deletion of the integrin 4 cytoplasmic domain. EMBO J. 1998;17:3940–3951. doi: 10.1093/emboj/17.14.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]