Abstract
Sufficient quantity of genomic DNA can be a bottleneck in genome-wide analysis of clinical tissue samples. DNA polymerase Phi29 can be used for the random-primed amplification of whole genomes, although the amplification may introduce bias in gene dosage. We have performed a detailed investigation of this technique in archival fresh-frozen and formalin-fixed/paraffin-embedded tumor DNA by using cDNA microarray-based comparative genomic hybridization. Phi29 amplified DNA from matched pairs of fresh-frozen and formalin-fixed/paraffin-embedded tumor samples with similar efficiency. The distortion in gene dosage representation in the amplified DNA was nonrandom and reproducibly involved distinct genomic loci. Regional amplification efficiency was significantly linked to regional GC content of the template genome. The biased gene representation in amplified tumor DNA could be effectively normalized by using amplified reference DNA. Our data suggest that genome-wide gene dosage alterations in clinical tumor samples can be reliably assessed from a few hundred tumor cells. Therefore, this amplification method should lend itself to high-throughput genetic analyses of limited sources of tumor, such as fine-needle biopsies, laser-microdissected tissue, and small paraffin-embedded specimens.
The availability of a simple and reliable method for the amplification of entire genomes from limited sources of DNA would lend itself to high-throughput genetic analyses in virtually all areas of translational research. High-throughput genomic profiling, for example cDNA microarray-based comparative genomic hybridization (array-CGH),1 requires μg quantities of genomic DNA. A particular challenge for translation of array-CGH methodology to clinical application is to link it to a robust up-front technology that allows reliable and unbiased amplification of limited sources of DNA, for example from fine-needle biopsies. Much effort has been invested in developing methods for whole genome amplification, for example polymerase chain reaction-based methods.2,3,4 In particular, the pitfalls of substantial variation in the extent of amplification occurring between different markers, incomplete representation, and inadequate average DNA size have limited the use of most existing amplification methods, making them particularly unsuitable for genomic applications.2,3,4
Bacteriophage Phi29 DNA polymerase random-primed DNA amplification5,6,7 is based on an isothermal strand displacement amplification reaction in which random hexamer primers anneal to the genomic template at multiple sites and Phi29 initiates replication at these sites on the denatured linear DNA. As synthesis proceeds, strand displacement of complementary DNA generates new single-stranded DNA available to be primed by additional primers. The subsequent strand displacement replication of this DNA leads to the formation of double-stranded DNA. The presence of an associated proofreading activity of Phi29 ensures a high sequence accuracy of the amplified DNA, indicating its suitability for genotyping.8,9,10 The utility of Phi29-based whole genome amplification has been shown for plasmid and bacteriophage DNA11 as well as human DNA from whole blood, buccal swabs, buffy coats, and cultured cells.6,12,13,14 However, this technique results in biased gene representation.6,12 The nature and mechanism for this misrepresentation in amplified human and yeast genomes has remained uncertain.14 Compared to genotyping, array-CGH analysis is more demanding because it requires not only high sequence accuracy of the amplification product but also representative gene dosage coverage across the entire genome.
Here we have examined in detail the suitability of Phi29 for the whole genome amplification of archival fresh-frozen and formalin-fixed/paraffin-embedded (FFPE) clinical solid tumor samples, and its utility for subsequent global gene dosage assessments. The utilization of 43,000-element cDNA microarray-based array-CGH technology provided a high-resolution means to measure genome-wide representational bias and to map gene dosage alterations in the amplified DNA on a gene-by-gene basis. We provide conclusive mechanistic explanation for the varying gene dosage representation in the amplified DNA. We describe a reliable way how to normalize biased gene representations to generate highly precise and comprehensive genomic profiles in both archival fresh-frozen and FFPE tumor tissue from few hundred cells.
Materials and Methods
Tumors, Cell Lines, and Reference DNA
Matched pairs of archival fresh-frozen and FFPE samples from several glioblastomas were obtained from the Stanford Brain Tumor Tissue Bank with institutional review board approval. Fresh-frozen specimens had been stored at −80°C and FFPE had been fixed in 10% buffered formalin, embedded in paraffin, and archived for 2 to 3 years. Breast cancer cell lines BT474 and MCF-7 were obtained from the American Type Culture Collection, Rockville, MD, and grown in RPMI 1640 media (Invitrogen, Carlsbad, CA), supplemented with 10% fetal calf serum (Hyclone, Logan, UT) and 1% penicillin-streptomycin (Invitrogen). Reference genomic DNA was prepared from male and female whole blood donors or was purchased from Promega (Madison, WI).
Isolation of DNA
Ten 4-μm FFPE tissue sections were deparaffinized in 3 × 1 ml xylene and 2 × 1 ml 100% ethanol for 10 minutes each. After air-drying, samples were suspended in 1 ml of DNA extraction buffer, composed of 900 μl of ATL buffer and 100 μl of proteinase K (both DNeasy tissue kit by Qiagen, Valencia, CA), and were incubated at 55°C overnight. Additional proteinase K (50 μl) was added 24 hours and 48 hours later for a total incubation time of 72 hours. Twenty-five mg of fresh-frozen glioblastoma tissue were digested in DNA extraction buffer at 55°C overnight. Silica gel-membrane extraction of DNA was performed in both fresh-frozen and FFPE samples using the DNeasy tissue kit following the manufacturer’s protocol. DNA concentration was determined by spectrophotometric absorption at 260 and 280 λ and the purity was calculated by the A260/A280 ratio. DNA size was estimated by agarose gel electrophoresis with ethidium bromide staining (Invitrogen). DNA from BT474 and MCF-7 cell lines and from whole blood from male and female donors was extracted using the blood and cell culture DNA maxi kit (Qiagen) according to the manufacturer.
Phi29-Based Amplification of Genomic DNA
Components of the GenomiPhi DNA amplification kit (Amersham Biosciences, Piscataway, NJ) were used for the amplification reactions. Various amounts of purified genomic DNA (0.25 to 30 ng) in a total volume of 1 μl were added to 9 μl of sample buffer and heated to 95°C for 3 minutes to denature the template DNA. After cooling on ice for 5 minutes, samples were mixed with 10 μl of a preprepared reaction mix (9 μl of reaction buffer and 1 μl of Phi29 per reaction), and were incubated at 30°C for several hours (2 to 18 hours). After amplification, Phi29 was heat-inactivated at 65°C for 10 minutes and samples were cooled to room temperature. Amplification products were purified by ethanol precipitation using 1.5 mol/L sodium acetate/250 mmol/L ethylenediamine tetraacetic acid buffer (pH > 8.0). Twenty μl of nuclease-free water, 4 μl of sodium acetate/ethylenediamine tetraacetic acid buffer, and 100 μl of 100% ethanol were added sequentially to 20 μl of amplification product and the mixture was centrifuged for 15 minutes at 12,000 rpm. The supernatant was removed and DNA pellets were washed in 400 μl of 70% ethanol for 2 minutes. After centrifugation at 3200 × g for 5 minutes, the supernatant was removed and DNA pellets were dried and resuspended in 10 μl of 1× Tris-ethylenediamine tetraacetic acid (TE) buffer (Sigma, St. Louis, MO). In each experiment, 10 ng of control λ DNA were co-amplified to assess the efficiency of each amplification reaction. At least three independent experiments were concurrently performed per template amplification. Reaction products were quantified spectrophotometrically and analyzed by agarose gel electrophoresis. BT474 and MCF-7 DNAs (along with corresponding normal male/female reference DNAs) were amplified essentially as above by Molecular Staging Inc.
Digestion and Purification of DNA
For labeling reactions, 6 μg each of nonamplified genomic DNA and amplification product were digested separately with DpnII restriction enzyme (New England Biolabs, Beverly, MA) at 37°C for 1.5 hours (total volume of 40 μl, 1.5 μl DpnII, and 6 μl DpnII buffer). After DpnII inactivation by heating at 65°C for 20 minutes, samples were snap-cooled on ice for 2 minutes. Digests were purified using the QIAquick polymerase chain reaction purification kit (Qiagen), following the manufacturer’s instructions. Samples were resuspended in 50 μl of EB buffer and digestion products were quantified by spectrophotometric absorption at 260 and 280 λ and stored at −80°C.
Labeling of DNA
For microarray hybridization, 2 μg each of nonamplified and amplified digested DNA in a volume of 22.5 μl were separately labeled using random primers (Bioprime labeling kit, Invitrogen), modified to include a 10× dNTP mix composed of 1.2 mmol/L each of dATP, dGTP, and dTTP and 0.6 mmol/L dCTP. To each sample, 20 μl of 2.5× random primers were added, the mixture was boiled for 5 minutes at 100°C, and snap-cooled on ice for 5 minutes. After adding 5 μl of 10× dNTP mix, 3 μl of Cy3-dCTP and Cy5-dCTP fluorescent dye to paired hybridization samples (Amersham Biosciences), and 1 μl of concentrated Klenow enzyme, samples were incubated for 2 hours at 37°C. Reactions were stopped by adding 5 μl of stop buffer, placed on ice for 5 minutes, and centrifuged at 18,000 × g for 2 minutes. Dye switch between amplified and nonamplified DNA was performed to rule out a dye bias effect.
CGH of Microarrays
Labeled products were purified using YM-30 microcon filters (Millipore Corporation, Bedford, MA). Corresponding Cy3- and Cy5-labeled probes were combined to the centrifugal filter unit, 400 μl of 1× TE buffer (pH = 7.4) were added, and the mixture was inverted several times and centrifuged at 13,800 × g for 7 minutes. After two additional washes with 450 μl of 1× TE, 380 μl of 1× TE, 20 μl of 5 μg/μl yeast tRNA (Invitrogen), 50 μl of 1 μg/μl human Cot-1 DNA (Invitrogen), and 2 μl of 10 μg/μl poly(dA-dT) (Sigma) were added to block nonspecific binding, hybridization to repetitive elements, and undesired hybridization to extended poly(A) tails, respectively. The mixture was concentrated to <32 μl by centrifugation at 12,000 × g for 12 to 14 minutes. Probes were recovered by inverting filters into a new microcon tube and centrifugation at 14,000 × g for 2 minutes. After adjusting the volume to 32 μl with 1× TE, 6.8 μl of 20× standard saline citrate (SSC) (Invitrogen) and 1.2 μl of 10% sodium dodecyl sulfate (Sigma) were added and the mixture was denatured at 100°C for 2 minutes. After a 30-minute Cot-1 preannealing step at 37°C, probes were hybridized to cDNA microarrays containing more than 43,000 cDNA sequences (manufactured by the Stanford Functional Genomics Facility) under a 22 × 60-mm glass coverslip and incubated in a hybridization chamber at 65°C for 15 to 18 hours.
Washing of Microarrays
After overnight hybridization, coverslips were removed by briefly dipping microarrays into a 65°C 2× SSC, 0.03% sodium dodecyl sulfate washing solution. To remove unbound labeled DNA, microarrays were sequentially washed in 2× SSC, 0.03% sodium dodecyl sulfate at 65°C for 5 minutes, rinsed in 2× SSC at 65°C, followed by shaking washes of 5 minutes each at room temperature in 1× SSC (one wash) and 0.2× SSC (two washes). Microarrays were centrifuged dry at 500 rpm for 5 minutes and placed into a light-protected box.
Imaging of Microarrays and Data Reduction
Microarrays were immediately scanned in dual wavelengths on a GenePix 4000B scanner (Axon Instruments, Union City, CA). Fluorescence intensity ratios for each cDNA element on the microarray were calculated after background subtraction using GenePix Pro 5.1 software. Array spots with overlying fluorescent debris were excluded, as were spots either <25% or >400% in average spot size. To correct for differences in DNA labeling efficiency between samples, fluorescence ratios were normalized to achieve an average log ratio of 0 using the Stanford Microarray Database. Measurements with consistent (regression correlation, >0.6) and sufficient fluorescent intensities in either wavelength channel (signal, >2.0 above background) were considered reliable. When indicated, gene dosage ratios were reported as symmetric five-nearest neighbors moving average.1
Data Analysis and Map Positions
The GoldenPath Human Genome Assembly (http://genome.ucsc.edu, National Center for Biotechnology Information build 34) was used to map fluorescent ratios of the arrayed human cDNAs to chromosomal positions. The CaryoScope software (http://genome-www5.stanford.edu/cgi-bin/caryoscope/nph-aCGH-dev_update.pl) was used to display moving average gene dosage ratios on a gene-by-gene basis along the human chromosomes.
Analysis of GC Content and GC Heterogeneity
The fractional GC content of 2.5 Mb-terminal chromosomal regions was analyzed using the geecee function in EMBOSS suite.15 The freely distributed program draw_chromosome_gc.pl (http://genomat.img.cas.cz) was used to analyze and plot the genome-wide GC content in windows of 100-kb size along the chromosomes as described by Paces and colleagues.16 Sequence data were obtained from the Human Genome Assembly, National Center for Biotechnology Information build 34.
Statistics
If not otherwise indicated, statistical analyses were performed in R.17
Results
Phi29-Based Amplification of Fresh-Frozen and Paraffin-Embedded Tumor DNA
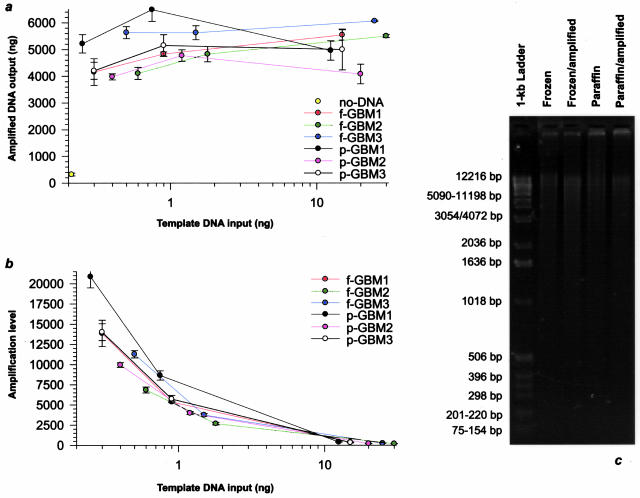
We examined in detail the efficiency of Phi29 in amplifying genomic DNA from clinical solid tumor tissues using matched pairs of archival fresh-frozen and FFPE glioblastoma samples. The amplification of less than a ng of genomic template both from fresh-frozen as well as FFPE tissue sources generated μg amounts of amplification product (Figure 1a). For the same template, there was high consistency with regard to the yield of amplified DNA between independent experiments. Increasing amounts of template DNA only slightly increased the yield of amplification product (Figure 1a), which, in turn, was paralleled by a related decrease in fold-amplification (Figure 1b). Gel electrophoresis disclosed a comparable average molecular weight (>12 kb) for amplified and nonamplified DNA from matched pairs of fresh-frozen and FFPE tumor (Figure 1c).
Figure 1.
Phi29-based amplification of fresh-frozen and FFPE tumor DNA. a: Graphical depiction of the relationship between genomic template and amplification product. The amplification of as low as ∼250 pg of template DNA both from matching fresh-frozen (f) and FFPE (p) glioblastoma (GBM) generated μg quantities of amplification product, whereas negligible amounts (<5%) of product were generated by Phi29 without added DNA template. Sets of three independent amplifications were performed for each sample with overnight 16-hour incubation at 30°C to evaluate the consistency in the amount of generated DNA. In each of the tumor samples there was high concordance with regard to the yield of amplified DNA between independent experiments. Comparable yields of amplification product were obtained with corresponding amounts of starting genomic template from fresh-frozen and FFPE tumor. Higher amounts of template DNA resulted in negligible increases in amplified DNA output, suggesting either limited primer availability or decreasing polymerase activity during the course of the amplification reaction. b: Graph displaying the amplification level corresponding to the data of a. A steady decrease in fold-amplification, as measured by the fold-change of amplified to template DNA, was noted as the amount of starting genomic template was increased both in the fresh-frozen and FFPE tumor. c: Analysis by gel electrophoresis of DNA from a matched pair of fresh-frozen and FFPE tumors with and without amplification. Fresh-frozen and FFPE DNA demonstrated a comparable average molecular weight as did nonamplified tumor DNA and the amplification product (>12 kb).
Genome-Wide Gene Dosage Representation in Phi29-Amplified DNA
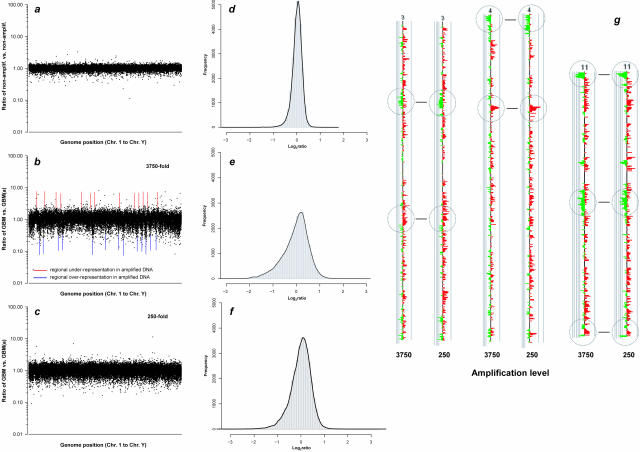
We then assessed on a gene-by-gene basis the gene dosage representation across the human genome in tumor DNA amplified at various amplification fold-levels versus nonamplified template tumor DNA by CGH on 43,000-element cDNA microarrays (Figure 2). This analysis showed considerable, amplification level-dependent scatter of the raw red/green (R/G) fluorescent ratios, signifying substantial clonal misrepresentation (Figure 2, b and c). When the R/G ratios for all analyzed clones were plotted along the genome, most of the overrepresented as well as most of the underrepresented clones clustered together in sizable subgroups at various chromosomal map coordinates, as indicated by vertical upward and downward peaks in Figure 2b. In FFPE tissue, a greater scatter of these ratios was noted as compared to fresh-frozen tissue.
Figure 2.
Array-CGH-based genome-wide assessment of gene dosage representation in Phi29-amplified DNA. a: Graph depicting the signal intensity ratios of a nonamplified male genomic versus nonamplified female genomic DNA array-CGH experiment. Signal intensity ratios were generated by hybridizing equal amounts of DpnII-digested, purified, and fluorescent dye-labeled nonamplified template and amplification product to a 43,000-element microarray. Each dot signified the raw intensity ratio for a single clone on the microarray, which in turn indicated how this clone was represented in the amplified DNA relative to the nonamplified DNA. Ratios were plotted against the order of the genes in the human genome, starting from chromosome 1 to chromosome Y. b: Array-CGH result of a corresponding nonamplified versus amplified (a) fresh-frozen glioblastoma (GBM) DNA, the latter having been amplified 3750-fold. The considerably more profound scatter of the ratios from the ideal 1.0 value, as compared to a, indicated significant misrepresentations of many clones in the amplified DNA. Red and blue lines indicate genomic regions of higher-than-average underrepresentation and overrepresentation, respectively, as evidenced by clusters of clones aligned as vertical upward and downward peaks at the same chromosome map coordinates. c: Signal intensity ratios in a nonamplified versus amplified DNA hybridization experiments of the same GBM as in b, in which the amplification product had been amplified 250-fold. Although the scatter of the ratios was less than in the experiment plotted in b, compared to a, considerable clonal misrepresentation was apparent with distinct genomic regions demonstrating more distortion in representation than others. d to f: Histogram plots of log2 ratio distributions of data points corresponding to the experiments shown in a to c. g: CaryoScope plots comparing clonal representations for chromosomes 3, 4, and 11 of the experiments shown in b and c. Red and green bars indicate that a clone is overrepresented and underrepresented in the amplified DNA, respectively. Despite a 15-fold difference in the amplification level between the two experiments, the regional pattern of misrepresentation was highly consistent.
Pattern and Reproducibility of Clonal Misrepresentations in Phi29-Amplified DNA
We then performed a detailed analysis of the genomic pattern of sequence variation using the CaryoScope software. Although some of the underrepresented loci were scattered across various intrachromosomal regions, the most significant underrepresentations mapped to terminal chromosomal regions (Figure 3a). In repeat experiments, there was high consistency among loci underrepresented in amplified tumor DNA even when a varying amplification level was chosen (Figure 2g). The consistent distortion in sequence representation in amplified tumor DNA highly matched patterns of regional misrepresentation in amplified normal genomic DNA (Figure 3d).
Figure 3.
Pattern of regional misrepresentation and GC content in amplified DNA. a to c: Graph interrelating the genome-wide pattern of misrepresentation along the chromosomes in an amplified (a) glioblastoma (GBM) DNA versus nonamplified GBM DNA array-CGH experiment and as displayed as a moving average by CaryoScope analysis (symmetric five-nearest neighbors) (a); corresponding raw intensity ratios for underrepresented clones plotted against the genomic order of genes (b); and average GC content measurements in selected terminal chromosomal regions (c). Clusters of highly underrepresented clones in the raw-intensity-ratio diagram were linkable to ∼50% of terminal chromosomal regions. Six chromosomal termini demonstrating maximal underrepresentation in the amplified DNA are circled in orange (5pter, 7pter, 9qter, 16pter, 17qter, 20qter). Six chromosomal termini with normo-representation are circled in blue (1qter, 3qter, 4qter, 12pter, 18pter, 20pter). Comparative assessment of the fractional GC content of these 12 chromosomal ends within a terminal length of 2.5 Mb revealed a significant difference in average GC content between the normo-represented (42.3%; range, 39 to 45%) and underrepresented (54.7%; range, 51 to 57%) termini (P = 0.002, two-sample Wilcoxon test), with the average genomic GC content of 41% indicated by the brown line. d: Comparison of terminal chromosomal and intrachromosomal clonal representations of selected regions in an amplified normal male DNA versus nonamplified female DNA and an amplified tumor DNA versus nonamplified tumor DNA array-CGH experiment, exemplifying the high concordance in the representational patterns of amplified normal and amplified tumor DNA.
Deterministic Mechanism for Biased Gene Representation in Phi29-Amplified DNA
Notably, only a subset (∼50%) of the terminal chromosomal regions was underrepresented in the amplified tumor and amplified normal DNA (100% concordance). Because there is evidence that the GC content of DNA can influence polymerase processivity as well as DNA priming, we examined the relationship between regional GC content and regional amplification efficiency. Comparison of the GC content of the six most underrepresented terminal loci versus six loci with average gene representation in amplified genomic DNA disclosed a significant difference in average GC content within a 2.5-Mb terminal region (54.7% versus 42.3%, respectively; P = 0.002, two-sample Wilcoxon test) (Figure 3c).
We then extended this comparative analysis to the whole genome and obtained chromosome-wide GC content profiles by averaging the GC content in a 100-kb window moved across the chromosomes in 10-kb steps. Figure 4 depicts the fixed length, moving-window plot of compositional GC heterogeneity mapped along with regional gene dosage representation in amplified versus nonamplified normal human DNA for chromosomes 3, 16, and 20. This graphical portrayal displays the profound variation of average GC content along the chromosomes and discloses a significant relationship between regional amplification efficiency and GC content across the human genome both in terminal chromosomal and intrachromosomal regions.
Figure 4.
Graphical portrayal of regional GC content heterogeneity versus regional amplification efficiency on chromosomes 3, 16, and 20, which were exemplarily selected because of their varying clonal representation pattern toward the chromosomal ends. a: Gene-by-gene display of regional amplification efficiency as depicted by the moving average ratios of amplified versus nonamplified normal human genomic DNA. b: Color-coded, fixed-length, moving-window plot depicting the variation in GC content across 100-kb windows. Substantial variation in GC levels between these windows was apparent, with particularly high GC content toward the end of chromosomal arms 16p, 16q, and 20q. Notably, regional underrepresentation along the chromosomes—and thus regional amplification efficiency—closely followed regional GC levels. Toward the end of chromosomal arms 16p, 16q, and 20q, where gene density is greatest,18 both clonal underrepresentation in the amplified DNA as well as GC content reached a maximum.
Genome-Wide Compensation for Distortion in Gene Representation in Phi29-Amplified DNA
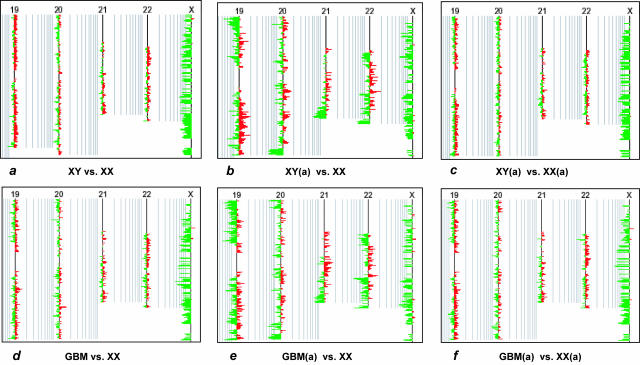
Because of the link between regional GC levels and amplification efficiency, we tested whether overall clonal distortions in the amplified tumor DNA could be balanced out by using human reference DNA amplified under exactly the same conditions. We therefore performed sets of array-CGH hybridizations including 1) nonamplified male DNA versus nonamplified female DNA, 2) amplified male DNA versus nonamplified female DNA, 3) amplified male DNA versus amplified female DNA, 4) nonamplified tumor DNA versus nonamplified normal human DNA, 5) amplified tumor DNA versus nonamplified human DNA, and 6) amplified tumor DNA versus amplified human DNA. Significant, albeit reproducible regional heterogeneity in gene representation was observed in the amplified sample versus nonamplified sample experiments (Figure 5). By contrast, the representation profiles obtained in hybridization experiments in which both test DNA and reference DNA had been amplified were remarkably similar to those obtained when hybridizing both nonamplified test and reference DNA (Figure 5). This compensation for misrepresentation was uniformly observed in amplified normal human DNA and amplified DNA from both fresh-frozen and FFPE tumor (Figures 5and 6, b and c).
Figure 5.
Compensation for representational distortion in amplified DNA. a to c: CaryoScope plots (moving window size, five clones) showing three independent array-CGH experiments using normal genomic DNA, including nonamplified male DNA versus nonamplified female DNA, amplified (a) male DNA versus nonamplified female DNA, and amplified male DNA versus amplified female DNA hybridizations, respectively. d to f: CaryoScope plots showing corresponding hybridization experiments with male glioblastoma (GBM) DNA, specifically nonamplified tumor DNA versus nonamplified normal female DNA, amplified tumor DNA versus nonamplified female DNA, and amplified tumor DNA versus amplified female DNA, respectively. As internal control, the ratio values for X-linked genes indicated the expected 0.5 dosage of these genes in the male test DNA versus the female reference DNA. The hybridization of either amplified normal DNA or amplified tumor DNA against nonamplified reference DNA revealed a reproducible pattern of misrepresentation, as indicated by a considerable difference in the clonal representation profiles between a and b and between d and e, respectively. As evidenced by almost similar representation profiles between a and c and between d and f, the use of reference DNA, amplified under exactly identical experimental conditions, remarkably compensated for clonal misrepresentations in the amplified study DNA by balancing out regional differences in amplification efficiency.
Figure 6.
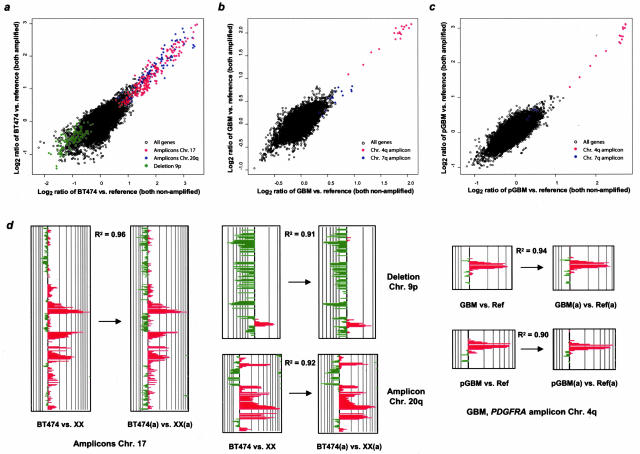
Representation of genetic alterations in amplified tumor cell line, fresh-frozen, and FFPE (p) tumor DNA. a to c: Scatter plots interrelating the moving average log2 hybridization ratios of independent experiments in which tumor and reference DNA were either both amplified (y axis) or both nonamplified (a) (x axis). In addition to showing the correlation between all genes on the microarray, signature genetic alterations—including the ERBB2/TOP2A amplicon on chromosome 17q11-22 in BT474 cells and the Platelet-derived growth factor receptor A (PDGFRA) amplicon on chromosome 4q12 in the glioblastoma (GBM)—are indicated separately. A strong concordance for all genes between the nonamplified and amplified experiments was apparent. Clones that belonged to one amplicon closely clustered together. d: CaryoScope plot depiction of the high degree of preservation of major genetic alterations—both gene amplifications and deletions—in the amplified DNA in tumor cell lines and fresh-frozen and FFPE tumor. Concordances between the two data sets for signature changes on chromosomes 17, 9p, and 20q in BT474 and on chromosome 4 in fresh-frozen and FFPE tumor were R2 = 0.96, R2 = 0.91, R2 = 0.92, R2 = 0.94, and R2 = 0.90, respectively.
Compensation for Distortion in Gene Representation at Tumor Loci with Genomic Alterations
We then evaluated the efficiency of compensating for misrepresentations in genomic tumor regions with genetic aberrations. In addition to analyzing fresh-frozen and FFPE glioblastoma samples, well-characterized BT474 and MCF-7 breast cancer cell lines demonstrating numerous gene copy number alterations were studied. Hybridization ratios of experiments in which study and reference DNA had been either both amplified or both nonamplified were plotted against each other. Figure 6 shows the representation of all genes and of signature genetic alterations in the BT474 cell line (including the ERBB2/TOP2A amplicon on 17q11-q22) and in a matched pair of fresh-frozen and FFPE glioblastoma (including the PDGFRA amplicon on 4q12). A high degree of concordance was seen between the whole nonamplified and amplified data sets. Clones belonging to one amplicon closely clustered together in the scatter plots (Figure 6; a to c). Corresponding CaryoScope plots depict the reliable preservation of genetic signatures in the amplified tumor DNA-to-amplified normal DNA hybridization experiments both in tumor cell lines and fresh-frozen and FFPE tumor. Concordances for representative amplifications and deletions on chromosomes 17, 9p, and 20q in BT474 were R2 = 0.96, R2 = 0.91, and R2 = 0.92, respectively (Figure 6d). A similar degree of concordance was revealed for the characteristic PDGFRA amplicon on chromosome 4 in fresh-frozen and FFPE glioblastoma (R2 = 0.94 and R2 = 0.90, respectively).
Calculation of Confidence Limits after Compensation for Representational Distortion
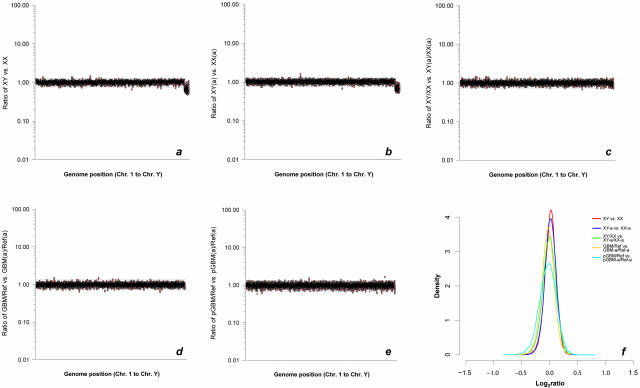
To estimate the amount of remaining bias that could not be compensated by using an amplified reference, we separately plotted the signal intensity ratios of the corresponding nonamplified male DNA versus nonamplified female DNA and amplified male DNA versus amplified female DNA hybridizations against the order of the genes in the human genome (Figure 7, a and b). The confidence limits for 99.9% of the data for autosomal genes calculated for the amplified DNA experiment were remarkably similar to the nonamplified DNA experiment (0.782 and 1.271 versus 0.763 and 1.273, respectively). Because such plotting of intensity ratios generated by two independent experiments only considered the overall variance of the intensity ratios without recognizing differences in ratios between the experiments on a clone-by-clone basis, data of both experiments were plotted as combined ratio of nonamplified male DNA/nonamplified female DNA versus amplified male DNA/amplified female DNA along the human genome (Figure 7c). In this model, the extent of deviation from the ideal ratio of 1 indicated for each single clone the representational distortion at the corresponding genomic map position in the amplified experiment relative to the nonamplified experiment. Again, the 99.9% confidence limits (0.741 and 1.329) were comparable to the single nonamplified and amplified experiments. Similar plots were then generated for the corresponding nonamplified tumor DNA versus nonamplified reference DNA and amplified tumor DNA versus amplified reference DNA experiments both in fresh-frozen and FFPE tumor (Figure 7, d and e). Despite the increased variation of the intensity ratios in each of the experiments because of genetic differences between tumor and control DNA, the confidence bounds for 99.9% of data calculated for the fresh-frozen (0.719 and 1.325) as well as paraffin-embedded tumor experiments (0.663 and 1.349) were almost comparable to those of the corresponding plot of ratios in the normal human DNA. Accordingly, probability density estimate plots demonstrated similar distribution spreads of data points in these experiments (Figure 7f).
Figure 7.
Confidence limits after compensation for representational distortion. a and b: Comparative signal intensity ratio-to-genome position plots of two array-CGH experiments in which either both a nonamplified male and female DNA (a) or both amplified (a) male and female DNA (b) were hybridized against each other. Moving average signal intensity ratios were ordered according to genome position. Ratio values for X-linked genes signified the expected 0.5 dosage of these genes in the male DNA. Confidence limits for 99.9% of data for autosomal genes expressed as linear ratios are indicated by horizontal lines. The confidence bounds calculated for both experiments were almost identical (0.763 and 1.273 and 0.782 and 1.271, respectively). c: Graphical display of clone-by-clone comparison of intensity ratios in experiments shown in a and b, expressed as a ratio of five-nearest neighbor averaged intensity ratios of the nonamplified experiment versus intensity ratios of the amplified experiment. The resultant ratio for each clone therefore indicated the representation of that clone in the amplified experiment relative to the nonamplified experiment. The calculated 99.9% confidence limits (0.741 and 1.329) were similar to those of the separate nonamplified and amplified experiment plots. d and e: Same graphical model as c, for corresponding nonamplified glioblastoma (GBM) DNA versus nonamplified reference DNA and amplified GBM DNA versus amplified reference DNA array-CGH experiments in fresh-frozen (d) and FFPE (p) tumor (e). The confidence bounds for 99.9% of data for both the fresh-frozen tumor (0.719 and 1.325) and FFPE tumor (0.663 and 1.349) experiments were comparable to those of the corresponding normal DNA experiments. f: Probability density estimate plots showing similar distribution spreads of data points corresponding to graphs a to e.
Discussion
We have successfully linked Phi29 to accurate and comprehensive high-resolution analysis of gene copy number alterations in fresh-frozen and FFPE tumor samples. Our initial studies have shown that Phi29-amplified human tumor genomes demonstrate a reproducible gene representation bias. Genomic loci of significant clonal underrepresentation mapped primarily toward the ends of chromosomal arms, but were also present as clusters in various intrachromosomal regions. Terminal underrepresentations were 1000-fold larger than those reported for the yeast genome.14
We found a striking relationship between clonal representation and local GC content across the human genome. Previous analyses have revealed substantial variation in average GC content among chromosomal fragments and the existence of GC-rich and GC-poor regions in the human genome.18,19,20 It has been suggested that the human genome can be partitioned into mosaics of fairly homogeneous GC content, which have been referred to as isochores.20 Such a mosaic organization is at variance with an alternative hypothesis that that GC levels drift more or less continuously throughout the human genome.19 Analysis of the draft genome sequence has revealed regions of varying size with GC levels far beyond the average of 41%.19,20 This high GC content is partly attributable to transposable element insertions and regional gene density.19,21,22
It has been noted that the GC content of DNA can significantly affect polymerase chain reaction amplification efficiency, sometimes causing premature chain termination at the beginning of G(C)-rich regions.23,24,25,26 Although the exact mechanism of how GC content may affect DNA polymerase function is not well understood, it has been hypothesized that amplification of GC-rich templates can be hampered because of the formation of secondary structures such as hairpins and by higher melting temperatures, which can inhibit primer extension by DNA polymerases as well as enzyme processivity.24,27,28,29 These variables may also significantly affect the displacement of the complementary DNA strand in the branching reaction. If one considers that only a small fraction (∼3 to 4%) of human DNA is highly GC-rich, but that more than a quarter of the genes are located in these regions,18 considerable amounts of data would be lost in gene dosage investigations of Phi29-amplified DNA, if such regions are not properly represented. The highest gene density in the human genome has been reported to be in the telomeric bands of metaphase chromosomes, those regions that were partly highly misrepresented in our amplified DNA.18
We have shown that the reproducible variation in regional amplification efficiency could be effectively normalized when test and reference genomes were amplified under identical conditions and compared by array-CGH. Analogous observations have been made in classical CGH experiments30,31 and have been successfully extrapolated to the random-primed amplification of yeast and human genomic DNA using the strand-displacing Bacillus stearothermophilus polymerase.14 It has been hypothesized that genome-wide adjustments in regional priming frequency between the study and reference DNA may have compensated for regional misrepresentations.14 While possible, our observations rather suggest that the use of amplified reference DNA might effectively balance out distortions in regional gene representation by adjusting not only for regional variation in DNA priming but also for varying polymerase efficiency because of substantial GC heterogeneity across the human genome.
The proper representation of gene dosages in genetically altered chromosomal regions is of crucial importance in functional genomics studies. We could show that even small gene dosage alterations, such as amplifications and deletions that are limited to just a few clones, were readily detectable in the bias-adjusted amplified tumor genomes. Genome-wide gene dosage profiles could also be generated by array-CGH from Phi29-amplified DNA originating from FFPE tumor, archived for several years. It has been previously suggested that, owing to priming frequency, the yield of Phi29-amplified DNA is a direct function of the molecular weight of the starting genomic template.14 It was therefore concluded that the random-primed amplification of DNA by strand-displacing polymerases, such as Phi29, may not be ideal for analysis of FFPE tissue sources.14 In contrast to a previous observation13 of almost complete failure of Phi29 to amplify DNA from FFPE sample, we have demonstrated similar amplification efficiencies in DNA from pairs of archival fresh-frozen and FFPE tumor and a similar average product length of DNA amplified from corresponding tissue specimens.
An important factor that may contribute to failure of proper amplification of FFPE DNA is the formation of secondary structures between DNA and proteins in paraffin-embedded DNA because of the formalin fixation.32 Formylation of DNA has been known to produce Schiff bases on free amino groups of nucleotides. Exposure of nucleo-proteins to formalin causes cross-linking between DNA and proteins. However, both processes are reversible.32 To adjust for these potential confounding variables, we have used a modified version of a previously described DNA extraction protocol,33 which includes an extended 3-day digestion with proteinase K. This extended digestion period has been shown to produce high-molecular weight DNA33 and may have successfully reduced the known pitfalls of FFPE DNA in our studies, which can be detrimental to reliable downstream molecular analyses.
In summary, our studies have demonstrated the suitability of Phi29 for the amplification of whole tumor genomes from fresh-frozen and FFPE clinical specimens. We have shown that the distortion in gene representation in Phi29-amplified DNA is nonrandom and reproducible across the human genome, indicating a mechanism for amplification bias that leads to regional but reproducible sequence distortions. Varying amplification efficiency is significantly linked to regional GC content of the genomic template and can be effectively normalized by using amplified reference DNA. Our data also suggest that gene-dosage alterations in both fresh-frozen and paraffin-embedded clinical tumor DNA can be reliably assessed by array-CGH from only few hundred tumor cells. Therefore, this amplification method should be highly valuable for the preparation of study DNA for genome-wide gene dosage assessments from very small amounts of archival tissue.
Acknowledgments
We thank Tina Hernandez-Boussard (Stanford Microarray Database) for assistance with sequence alignments and Jan Pa[caron]ces (Institute of Molecular Genetics, Academy of Science of the Czech Republic) for help with GC content analysis.
Footnotes
Supported by the National Institutes of Health (grant CA 92474 to B.I.S. and grant CA 97139 to J.R.P.) and by the Emmy-Noether-Programme of the German Research Society (to M.B.).
References
- Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BA, Tunnacliffe A. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc Natl Acad Sci USA. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, Nelson SF. Whole genome amplification using a degenerate oligonucleotide primer allows hundreds of genotypes to be performed on less than one nanogram of genomic DNA. Proc Natl Acad Sci USA. 1996;93:14676–14679. doi: 10.1073/pnas.93.25.14676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco L, Bernad A, Lazaro JM, Martin G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, Driscoll M, Song W, Kingsmore SF, Egholm M, Lasken RS. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- Barker DL, Hansen MS, Faruqi AF, Giannola D, Irsula OR, Lasken RS, Latterich M, Makarov V, Oliphant A, Pinter JH, Shen R, Sleptsova I, Ziehler W, Lai E. Two methods of whole-genome amplification enable accurate genotyping across a 2320-SNP linkage panel. Genome Res. 2004;14:901–907. doi: 10.1101/gr.1949704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez JG, Lin M, Beroukhim R, Lee JC, Zhao X, Richter DJ, Gabriel S, Herman P, Sasaki H, Altshuler D, Li C, Meyerson M, Sellers WR. Genome coverage and sequence fidelity of phi29 polymerase-based multiple strand displacement whole genome amplification. Nucleic Acids Res. 2004;32:e71. doi: 10.1093/nar/gnh069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook MS, Delach SM, Deyneko G, Worlock A, Wolfe JL. Whole genome amplification of DNA from laser capture-microdissected tissue for high-throughput single nucleotide polymorphism and short tandem repeat genotyping. Am J Pathol. 2004;164:23–33. doi: 10.1016/S0002-9440(10)63092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono S, Faruqi AF, Dean FB, Du Y, Sun Z, Wu X, Du J, Kingsmore SF, Egholm M, Lasken RS. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 2003;13:954–964. doi: 10.1101/gr.816903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Brennan C, Rook M, Wolfe JL, Leo C, Chin L, Pan H, Liu WH, Price B, Makrigiorgos GM. Balanced-PCR amplification allows unbiased identification of genomic copy changes in minute cell and tissue samples. Nucleic Acids Res. 2004;32:e76. doi: 10.1093/nar/gnh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage JM, Leamon JH, Pejovic T, Hamann S, Lacey M, Dillon D, Segraves R, Vossbrinck B, Gonzalez A, Pinkel D, Albertson DG, Costa J, Lizardi PM. Whole genome analysis of genetic alterations in small DNA samples using hyperbranched strand displacement amplification and array-CGH. Genome Res. 2003;13:294–307. doi: 10.1101/gr.377203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Paces J, Zika R, Paces V, Pavlicek A, Clay O, Bernardi G. Representing GC variation along eukaryotic chromosomes. Gene. 2004;333:135–141. doi: 10.1016/j.gene.2004.02.041. [DOI] [PubMed] [Google Scholar]
- Ikaha R, Gentleman RR. A language for data analysis and graphics. J Comp Graph Stat. 1996;5:299–314. [Google Scholar]
- Saccone S, De Sario A, Della Valle G, Bernardi G. The highest gene concentrations in the human genome are in telomeric bands of metaphase chromosomes. Proc Natl Acad Sci USA. 1992;89:4913–4917. doi: 10.1073/pnas.89.11.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Pavlicek A, Paces J, Clay O, Bernardi G. A compact view of isochores in the draft human genome sequence. FEBS Lett. 2002;511:165–169. doi: 10.1016/s0014-5793(01)03283-5. [DOI] [PubMed] [Google Scholar]
- Zoubak S, Clay O, Bernardi G. The gene distribution of the human genome. Gene. 1996;174:95–102. doi: 10.1016/0378-1119(96)00393-9. [DOI] [PubMed] [Google Scholar]
- Saccone S, Caccio S, Kusuda J, Andreozzi L, Bernardi G. Identification of the gene-richest bands in human chromosomes. Gene. 1996;174:85–94. doi: 10.1016/0378-1119(96)00392-7. [DOI] [PubMed] [Google Scholar]
- Bachmann HS, Siffert W, Frey UH. Successful amplification of extremely GC-rich promoter regions using a novel ‘slowdown PCR’ technique. Pharmacogenetics. 2003;13:759–766. doi: 10.1097/00008571-200312000-00006. [DOI] [PubMed] [Google Scholar]
- Woodford K, Weitzmann MN, Usdin K. The use of K(+)-free buffers eliminates a common cause of premature chain termination in PCR and PCR sequencing. Nucleic Acids Res. 1995;23:539. doi: 10.1093/nar/23.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadaraj K, Skinner DM. Denaturants or cosolvents improve the specificity of PCR amplification of a G + C-rich DNA using genetically engineered DNA polymerases. Gene. 1994;140:1–5. doi: 10.1016/0378-1119(94)90723-4. [DOI] [PubMed] [Google Scholar]
- Arezi B, Xing W, Sorge JA, Hogrefe HH. Amplification efficiency of thermostable DNA polymerases. Anal Biochem. 2003;321:226–235. doi: 10.1016/s0003-2697(03)00465-2. [DOI] [PubMed] [Google Scholar]
- McDowell DG, Burns NA, Parkes HC. Localised sequence regions possessing high melting temperatures prevent the amplification of a DNA mimic in competitive PCR. Nucleic Acids Res. 1998;26:3340–3347. doi: 10.1093/nar/26.14.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usdin K, Woodford KJ. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 1995;23:4202–4209. doi: 10.1093/nar/23.20.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Q. Minimizing deletion mutagenesis artifact during Taq DNA polymerase PCR by E. coli SSB. Nucleic Acids Res. 1992;20:4371. doi: 10.1093/nar/20.16.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Schantz SP, Rao PH, Mo J, McCormick SA, Chaganti RS. Improving degenerate oligonucleotide primed PCR-comparative genomic hybridization for analysis of DNA copy number changes in tumors. Genes Chromosom Cancer. 2000;28:395–403. [PubMed] [Google Scholar]
- Voullaire L, Wilton L, Slater H, Williamson R. Detection of aneuploidy in single cells using comparative genomic hybridization. Prenat Diagn. 1999;19:846–851. [PubMed] [Google Scholar]
- Dubeau L, Chandler LA, Gralow JR, Nichols PW, Jones PA. Southern blot analysis of DNA extracted from formalin-fixed pathology specimens. Cancer Res. 1986;46:2964–2969. [PubMed] [Google Scholar]
- Isola J, DeVries S, Chu L, Ghazvini S, Waldman F. Analysis of changes in DNA sequence copy number by comparative genomic hybridization in archival paraffin-embedded tumor samples. Am J Pathol. 1994;145:1301–1308. [PMC free article] [PubMed] [Google Scholar]