Abstract
A significant fraction of hereditary nonpolyposis colorectal cancer cases with defective mismatch repair (ie, Lynch syndrome) have large genomic deletions or duplications in the mismatch repair genes, hMLH1 and hMSH2, which can be challenging to detect by traditional methods. For this study, we developed and validated a novel Southern blot analysis method that allows for ascertainment of the extent of the dosage alterations on an exon-by-exon basis and compared this method to a second novel technique, multiplex ligation-dependent probe amplification (MLPA). From a total of 254 patients referred for Lynch syndrome testing, 20 of the 118 MLH1 cases and 42 of the 136 MSH2 cases had large genomic alterations, as detected by Southern blot. MLPA and Southern blot results were concordant with the exception of three major discrepancies: one because of a lack of MLPA probes for the region altered, another because of a point mutation near the MLPA probe ligation site, and another that was unexplained. Compared to Southern blot, MLPA has a shorter turn-around time, the analysis is less costly, less time-consuming, and less labor-intensive, and results are generally clear and unambiguous. However, concerns with MLPA include the presence of false-negatives and -positives because of positioning of probes and DNA variants near the probe ligation site. Overall, both Southern blot and MLPA provide important tools for the complete evaluation of patients with Lynch syndrome.
Hereditary nonpolyposis colorectal cancer is an autosomal dominant disorder characterized by the early onset of tumors in the setting of few polyps. The average age of colon cancer diagnosis in individuals with hereditary nonpolyposis colorectal cancer is in the early to middle 40s, although many tumors may occur in the 20s or even in teenage years. In addition to colorectal cancer, several other tumor types, including endometrial, gastric, and ovarian are observed at an increased frequency in families with this disease.1
Approximately two-thirds of patients diagnosed with hereditary nonpolyposis colorectal cancer have germline mutations in any one of several genes involved in DNA mismatch repair (MMR) and have what is now referred to as Lynch syndrome.2,3 DNA MMR is involved in the correction of mutations that occur because of exogenous or endogenous mutagens or misincorporations during DNA replication.4,5,6,7 This DNA repair process involves a complex set of proteins that includes hMLH1, hMLH3, hMSH2, hMSH3, hMSH6, PMS1, and PMS2.7,8,9 The majority of Lynch syndrome cases demonstrate the presence of tumor microsatellite instability and the absence of protein expression within the tumor for one of the genes involved in DNA MMR.8,9 Germline mutations in the MMR genes hMSH2 and hMLH1 account for ∼80 to 90% of the reported mutations in families with Lynch syndrome.7,8 Therefore, screening for germline mutations in hMLH1 and hMSH2 is important for the diagnosis of this syndrome. Both mutation screening techniques (eg, single strand conformational polymorphism, conformation-sensitive gel electrophoresis) and direct sequencing are limited by the fact that they do not detect large deletions, duplications, or other genomic rearrangements that are frequently found in Lynch syndrome kindreds.10,11,12,13 Additionally, Lynch and colleagues14 recently described the identification of an American founder deletion of exons 1 to 6 in hMSH2 in a large outbred US population with a wide geographic distribution. Therefore, due to the presence of large genomic deletions and duplications in hMLH1 and hMSH2, analysis for these rearrangements should be part of a routine mutation detection protocol for Lynch syndrome.
Historically, Southern blot analysis has been used to identify dosage differences in these MMR genes. However, the extent of hMLH1 and hMSH2 rearrangements at the exonic level has sometimes not easily been resolved using traditional cDNA or genomic probes for Southern blot analysis.12,15 In addition to Southern blot analysis, a multiplex polymerase chain reaction (PCR)-based strategy has been suggested to detect deletions of hMLH1 and hMSH2.13 Although this is a rapid, PCR-based method, controlling the end point of linear amplification of genomic DNA is difficult, resulting in issues with dosage interpretation. Other quantitative methods have been suggested but not put into widespread practice.16,17,18,19 Because of these issues, we developed a Southern blot method for the detection of exon deletions/duplications with resolution at the single exon level, using a series of artificially constructed DNA probes. In addition, we have compared this method to a novel PCR-based method: multiplex ligation-dependent probe amplification (MLPA).
Materials and Methods
Patients
Specimens referred for routine clinical testing for Lynch syndrome through the Mayo Clinic Clinical Molecular Genetics Laboratory were used for this study. Patients whose tumors demonstrated the presence of a high level of microsatellite instability (MSI-H), using four mononucleotide and six dinucleotide markers, and loss of protein expression for either hMLH1 or hMSH2 were studied (n = 254). Genomic DNA was isolated from the peripheral blood leukocytes of the patients using Puregene reagents (Gentra Systems, Inc., Minneapolis, MN) according to the manufacturer’s protocol. The study was approved by the Mayo Clinic Institutional Review Board.
Southern Blot Probe Construction
Amplification of Individual Exons
All exons from hMLH1 and hMSH2 were individually amplified by PCR in a 25-μl reaction mixture containing PCR buffer (50 mmol/L KCl, 10mmol/L Tris-HCl, pH 8.3), 200 μmol/L each of dNTP, 1.5 mmol/L MgCl2, 0.2 μmol/L of each primer, and 1.25 U of AmpliTaq Gold (Applied Biosystems, Foster City, CA). PCR was performed using the following conditions: initial denaturation at 95°C for 10 minutes followed by 35 cycles of 95°C for 30 seconds, 55°C for 1 minute, and 72°C for 1 minute. Primers were designed according to the hMLH1 and hMSH2 gene exon sequences (primers available on request) and synthesized by Integrated DNA Technologies (Coralville, IA) with standard desalting purification. The PCR products were separated electrophoretically and stained with ethidium bromide. Each PCR product that corresponded to an individual exon was quantified by densitometric analysis using an α Imager (Alpha Innotech Corp., San Leandro, CA). After quantification, each exon was purified with Microcon YM-100 centrifugal filter devices (Millipore Corp., Bedford, MA) according to the manufacturer’s instructions.
Synthesis of Recombinant Molecules
Ten to fifty ng of each exonic PCR product from above was used for amplification with recombinant PCR primers. Recombinant PCR primers were designed according to the exon groupings for each individual gene (primers available on request). The PCR procedure was the same as described above. After amplification, the PCR product was purified with the Millipore Microcon-YM 100 system. Two or three PCR products were then used as a template for a second PCR amplification. The PCR concentrations and conditions were identical to the first round conditions, with the exception of a final extension at 72°C for 5 minutes. The recombinant PCR products were separated on a 1.5% agarose gel and purified from the gel with a Qiaquick PCR purification kit (Qiagen Inc., Valencia, CA) according to the manufacturer’s procedure.
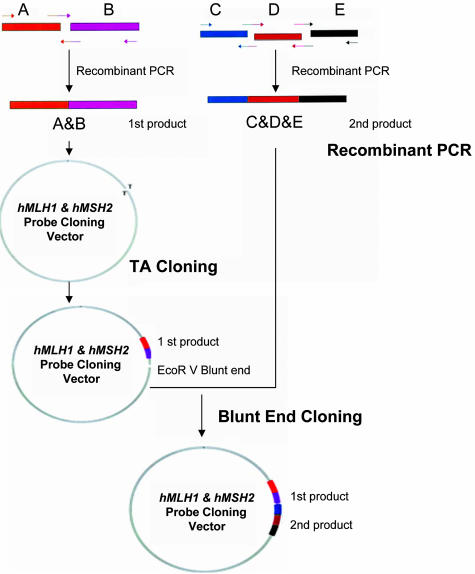
A two-step subcloning procedure was used to link the two recombinant PCR products into a single vector (Figure 1). The first recombinant PCR fragments were cloned by the pGEM-T easy vector system (Promega, Madison, WI) and the pCRII-TOPO vector cloning system (Invitrogen, Carlsbad, CA), as per the manufacturer’s protocols. The insert was sequenced and the subcloning vectors (plasmids) were digested by EcoRV and then treated with calf intestinal alkaline phosphatase. The second recombinant PCR product was treated with T4 kinase, followed by ligation of the recombinant PCR product to the first subcloning vector with T4 DNA ligase. BamHI and NotI were used to remove the insert from the plasmid for each probe. A 1.5% agarose gel was used to separate the vector and insert, and the probe was purified with the Millipore Microcon-YM 100 system.
Figure 1.
hMLH1 and hMSH2 probe construction. Recombinant PCR was used to link two or three exons (A and B; and C, D, and E) into a single molecule. Parallel arrows indicate the recombinant PCR primers. A two-step subcloning method was then used to join two recombinant PCR products together.
Southern Blot Analysis
Genomic DNA from each patient was digested with the use of three restriction endonucleases: EcoRI, BglII, and HindIII. Each individual digested genomic DNA (2.5 μg) was then loaded onto a 0.8% agarose gel for overnight electrophoresis at 55 V. After standard capillary gel transfer to the hybridization membrane, ∼10 ng of each of the purified probes was radioactively labeled with α-32P-dCTP using the High Prime kit (Roche, Basel, Switzerland). The radioactive probes were added to 20 ml of hybridization solution, at a concentration of ∼1 × 106 cpm/ml. Membranes were placed in the probe/hybridization solution, and hybridization took place overnight at 45°C. After hybridization, the membranes were washed three times in 2× standard saline citrate, 0.1% sodium dodecyl sulfate at 60°C for 30 minutes and then once in 0.2× standard saline citrate, 0.1% sodium dodecyl sulfate at 60°C for 30 minutes. The radioactive membranes were then exposed to PhosphorImager (Amersham Biosciences, Piscataway, NJ) screens. After exposure, the PhosphorImager screens were scanned and results were analyzed with ImageQuant 5.0 software (Amersham Biosciences).
Multiplex Ligation-Dependent Probe Amplification
MLPA for gene dosage of hMLH1 and hMSH2 was performed as per the manufacturer’s recommendations (MRC-Holland, Amsterdam, The Netherlands) with minor modifications. All reagents were provided by the manufacturer. Briefly, 400 ng of patient genomic DNA was heated to 95°C for 5 minutes, cooled, then mixed with the P003 probe set and MLPA buffer. Probe hybridization took place at 60°C for 16 hours, followed by probe ligation at 54°C for 10 to 15 minutes. The ligated products were then PCR amplified using 6-FAM-labeled universal primers, and then separated via capillary electrophoresis on the ABI 3100 (Applied Biosystems). Data were collected and analyzed with Genescan and Genotyper (Applied Biosystems) software. The output for each patient displayed a peak for each exon of the hMLH1 and hMSH2 genes, along with several peaks representative of extragenic regions, used for ligation and PCR monitoring. Peak heights for fragments corresponding to specific exons and control regions were binned, appended to a table, and then saved as a text file, all within the Genotyper software. The contents of the tabular text file were then stored in a Microsoft Access database and calculations performed by custom software. Samples were excluded from scoring if two requirements were not met: the 96-bp control peak height/74-bp control peak height ratio ≥5, and identification of all seven extragenic control peaks in the sample. To control for intersample PCR and loading differences, the peak heights from each sample were first normalized by dividing each hMLH1- and hMSH2-associated peak height by the average of the extragenic control peak heights for that sample. Then, each of the sample’s normalized peak heights was compared to the average of at least three normal control’s corresponding normalized peak height, expressed as a percent difference. Any exon with a decrease or increase in peak height of ≥35% was scored as a deletion or duplication, respectively. Both peak height and peak area are suitable for analyses. However, somewhat better results were obtained when peak heights were used.
Results
Southern Blot Probe Design and Construction
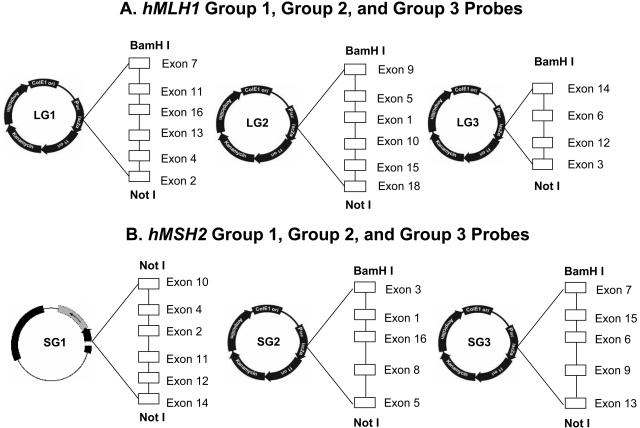
For both hMLH1 (19 exons) and hMSH2 (16 exons), DNA probes were designed and constructed to detect alterations (deletions and duplications) at the individual exon level with multiple probe-enzyme combinations. This was achieved by first using PCR-amplified material from single exons as DNA probes against genomic DNA digested with three different restriction endonucleases (EcoRI, BglII, and HindIII). The size of each migrated band for individual exons from each of the three digests was determined (Table 1). Using this information, exon combinations were strategically selected for groups that did not have overlapping migration patterns. Using EcoRI, BglII, and HindIII, exons 7 and 8 of hMLH1 did not have restriction sites that allowed for separation of the two exons by the three enzymes chosen. Thus, hMLH1 exons 7 and 8 occur on the same fragment, and the exon 7 probe was used to represent both exons 7 and 8. The same situation was true for hMLH1 exons 16 to 19. Here, exons 16 to 19 are all on the same fragment, so the exon 16 and 18 probes were used to represent the four-exon group. Exons for both hMLH1 and hMSH2 were organized into three groups (Figure 2), based on Southern blot patterns that were largely nonoverlapping. Thus, a single probe (probing to multiple exons) was used to determine the rearrangement status of four to six nonoverlapping exons.
Table 1.
hMLH1 and hMSH2 Exon Migration Sizes
| Exon |
hMLH1 migration size (kb)
|
hMSH2 migration size (kb)
|
||||
|---|---|---|---|---|---|---|
| EcoRI | BglII | HindIII | EcoRI | BglII | HindIII | |
| 1 | 11 | 8 | 4.6 | 8.8 | 5.5 | 10 |
| 2 | 11 | 8 | 5.6 | 2.2 | 2.1 | 10 |
| 3 | 4.5 | ND | 6.8 | 2.2 | 2.1 | 2.7 |
| 4 | 4.5 | 3 | 6.8 | 8.8 | 10 | 0.5 |
| 5 | 10 | 15 | 6.8 | 8.8 | 10 | 1.7 |
| 6 | 10 | 15 | 4.5 + 1.4 | 8.8 | 10 | 6.6 |
| 7 | 10 | 15 | 4.5 | 9 | 14 + 0.6 | 1.6 |
| 8 | 10 | 15 | 4.5 | 6.2 | 3.7 | 7/5.3* |
| 9 | 10 | 15 | 1.1 + 0.8 | 3.2 + 2.6 | 5 + 0.7 | 4.4 |
| 10 | 3.8 | 15 | 2.8 | 4.3 | 5 | 1.1 |
| 11 | 2.8 | 15 | 6.2 | 2.6 | 7 | 7.0 |
| 12 | 11/8.2* | 9 | 6.2 + 7.4 | 1.9 | 7 | 1.4 |
| 13 | 11/8.2* | 9 | 7.4 | 1.9 | 3.9 | 10 |
| 14 | 3.6 | ND | 3.6 | 1.2 | 3.9 | 10 |
| 15 | 5 | 15 | 2.3 | 2.8 | 6.5 | 10 |
| 16 | 7 | 15 | 3.3 | 5.3 | 6.5 | 10 |
| 17 | 7 | 15 | 3.3 | |||
| 18 | 7 | 15 | 3.3 | |||
| 19 | 7 | 15 | ND | |||
The number following the/is the size (kb) of an alternative band resulting from a polymorphic restriction site within this fragment. ND means that fragment was too small to detect. If a restriction site is within an exon, 2 fragments may be detected. In such cases, the two fragment sizes are indicated (eg. 4.5 + 1.4).
Figure 2.
hMLH1 and hMSH2 group probes. A: LG1, LG2, and LG3 represent the hMLH1 group 1, group 2, and group 3 probes, respectively. Open blocks represent the exons in the group probe. BamHI and NotI restriction enzymes located at both ends of the inserts were used to release the inserts. B: SG1, SG2, and SG3 are the hMSH2 group 1, group 2, and group 3 probes, respectively. Instead of the pcR2.1-TOPO vector, SG1 was subcloned into the pGEM-T easy vector and the SG1 probe was released by digestion with the NotI restriction enzyme.
To generate multiple-exon probes, recombinant PCR was performed to link noncontiguous (when possible) exons together. First, each exon of hMLH1 and hMSH2 was amplified and purified. In the second-round PCR, two or three first-round PCR products (each individual exon) were used as the templates. During the second-round PCR, it was difficult to obtain a single recombinant PCR product. Thus, a gel purification procedure was used to isolate the band corresponding to the correct size. Then, the recombinant PCR product was ligated into the vector as described above, so that two or three preferably noncontiguous exons were linked into one molecule.
Because the recombinant PCR steps yielded two recombinant PCR products, a two-step subcloning procedure was used to link these two recombinant PCR products into a single vector (Figure 1). The success of the subcloning procedure was confirmed by sequencing the insert. After the initial subcloning procedure, the constructs were digested by EcoRV to create a blunt end. The vector blunt end was treated with calf intestinal alkaline phosphatase to remove phosphate groups and a second recombinant PCR product was treated with T4 kinase to add phosphate groups. T4 DNA ligase was then used to ligate the second recombinant PCR product to the first subcloning vector. Sequencing of the second blunt end subcloning vector was performed to ensure the success of the second step subcloning. The probe was then excised from the insert via restriction enzyme digestion (Figure 1) and routine Southern blot hybridization was performed to evaluate the utility of the probe (Figures 3and 4).
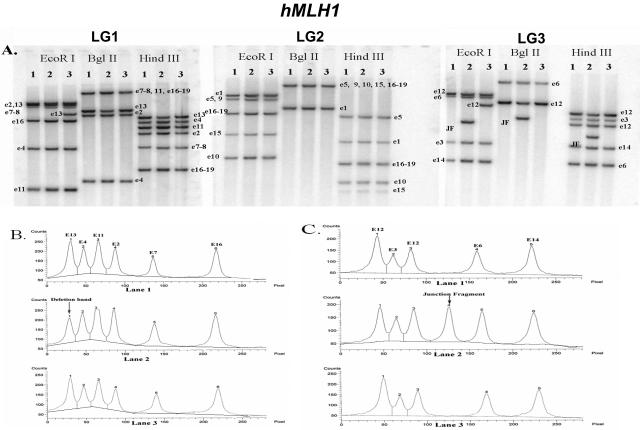
Figure 3.
hMLH1 Southern blot hybridization. A: Southern blot analysis of patients 1 to 3 (lanes 1 to 3) with the LG1, LG2, and LG3 probes. All samples were digested by three restriction enzymes (EcoRI, BglII, and HindIII). Each band is labeled with the exon used in that probe. It is important to note that each fragment may include multiple exons. See Table 1 for overlap of exons within the same fragment. Several polymorphisms can be observed (eg, EcoRI for e13, EcoRI for e12). B: Densitometric analysis of the LG1 HindIII digest of patients 1 to 3 (lanes 1 to 3). Patient 2, peak 1 demonstrates a 50% reduction in density, corresponding to a deletion in exon 13. C: Densitometric analysis of the LG3 HindIII digest of patients 1 to 3. A junction fragment with the exon 12 probe is apparent in patient 2.
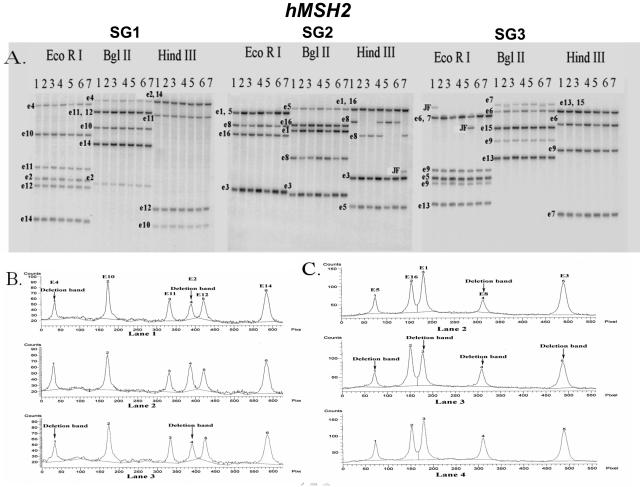
Figure 4.
hMSH2 Southern blot hybridization. A: Southern blot analysis of patients 1 to 7 (lanes 1 to 7) with the SG1, SG2, and SG3 probes. All samples were digested by three restriction enzymes (EcoRI, BglII, and HindIII). Each band is labeled with the exon used in that probe. It is important to note that each fragment may include multiple exons. See Table 1 for overlap of exons within the same fragment. B: Densitometric analysis of the SG1 EcoRI digest of patients 1 to 3 (lanes 1 to 3). Patients 1 and 3 both demonstrate a deletion in exons 2 and 4, whereas patient 2 has no observable deletions for this digest/probe combination. C: Densitometric analysis of the SG3 BglII digest of patients 2 to 4 (lanes 2 to 4). Patient 2 demonstrates a deletion in exon 8; patient 3 demonstrates deletions in exons 1, 3, 5, and 8; and patient 4 has no observable deletions.
Southern Blot Analysis
Examples of three patients tested for hMLH1 large genomic alterations are shown in Figure 3. Neither patient 1 nor patient 3 had a deletion/duplication in hMLH1, whereas patient 2 showed a deletion at exon 13. The deletion at exon 13 could easily be identified based on the following criteria. First, using the LG3 probe, there was a junction fragment that showed that the deletion breakpoint was close to exon 12 with all three digests. According to the densitometry data (Figure 3, B and C), the intensity of the exon 12 band was decreased 50%, suggesting that the junction fragments were from the exon 12 probe. Further evidence to support a deletion in exon 13 came from the LG1 group probe, which demonstrated a 50% reduction in intensity of the exon 13 band without any additional junction fragments. Using the LG3 group probe, it could be demonstrated that exon 14 was intact.
Examples of seven patients analyzed for hMSH2 alterations are shown in Figure 4. Patient 1 had a junction fragment with either the exon 6 or 7 probe, as shown via hybridization with the SG3 probe. Densitometry data for patient 1, from all three multiplex exon groups, demonstrated a 50% decrease in band intensity of exons 1 to 6, but not exon 7. Therefore, patient 1 had a hMSH2 deletion from exons 1 to 6. Using similar logic and analysis as above, patient 2 demonstrated a deletion in exon 8 and patient 3 showed a deletion from exons 1 to 8 of hMSH2. Neither patient 2 nor patient 3 showed any junction fragments. Patient 4 did not demonstrate any hMSH2 exon dosage alterations. Patient 5 had a deletion in exons 1 through 6 and a junction fragment detected with either the exon 6 or 7 probe (SG3 probe). Patient 6 had a deletion in exon 8, with no junction fragments. Although patient 7 had a junction fragment in exon 8 for the HindIII digest (SG2 probe), the intensities of the other exon 8 bands for the other digests (SG2 probe) were not decreased by 50%. Thus, patient 7 had an equivocal alteration in exon 8 or a HindIII polymorphism.
MLPA
As part of the validation process for MLPA, a number of parameters were evaluated. These included the minimum amount of DNA necessary for reproducible results and the various reaction volumes. We performed DNA titrations of 16 samples, using 50, 100, 200, 400, and 800 ng of gDNA. We found that 400 ng of DNA provided the most optimal results, based on reproducibility and minimal sample failures. A failure was defined as a sample that met one or more of these criteria: 1) ABI peak height <500; 2) ratio of the 96-bp to 74-bp control peak heights <5; 3) absence of one or more of the seven extragenic control peaks; and 4) an equivocal percent difference (between 25% and 35%) in peak height.
The volumes of both the ligation and PCR reactions are in excess of what is actually needed to perform fragment analysis. Thus, we sought to determine whether reagent volumes could be reduced 50% while still achieving the same results. We compared 96 samples using full and half reactions and found that full reactions provided the most consistent results, again in terms of reproducibility and minimal sample failures as described above. Some samples failed when using both 400 ng of DNA and full reactions. For these samples (n = 10), because the ligation product was in excess, we repeated the PCR step using the original ligation product to determine whether results could be obtained. This was not the case, indicating that the samples failed either because of inadequacies in the starting genomic DNA quality and/or failure of the ligation step.
Cross-Validation of Southern Blot and MLPA
DNA samples from 254 patients referred for Lynch syndrome testing were analyzed at least once by Southern and at least twice by MLPA for gene dosage alterations in hMLH1 and hMSH2. A total of 118 samples were analyzed for hMLH1 alterations, and 136 samples for hMSH2 alterations. Of the 118 hMLH1 samples, Southern analysis identified 20 samples with large genomic alterations and 98 samples that were negative (Table 2). Of the 20 positive Southern samples, 12 were from probands, whereas the eight remaining positives were relatives of the probands. MLPA analysis identified 19 samples with large genomic hMLH1 alterations and 99 negative samples. Of the positive samples, Southern detected seven and MLPA detected six different hMLH1 deletions/duplications (L.M. Baudhuin and colleagues, submitted for publication), and one major and two minor discrepancies were identified. For the one major discrepancy (Table 2), junction fragments for both exons 12 and 13 of hMLH1 were detected by Southern with the BglII and HindIII restriction endonucleases, but MLPA did not reveal any deletions for these exons in this sample. Exons 12 and 13 of this sample were sequenced but did not show any small alterations that might affect Southern restriction enzyme digestion or MLPA probe binding. Based on the Southern blot results, the deletion was believed to be ∼400 to 500 bp and we hypothesized that the deletion breakpoint regions were downstream of the exon 12 MLPA probes and upstream of the exon 13 MLPA probes, likely in IVS12. Follow-up analyses to characterize the specific region deleted could not be performed because of an insufficient amount of DNA. Minor discrepancies occurred for two cases in which Southern analyses identified larger fragments along with junction fragments for particular exons. Although MLPA detected a deletion in both of these cases, it did not identify the exons with junction fragments as being disrupted.
Table 2.
Intermethod Comparison of Gene Dosage Alterations Identified
| Number of samples
|
||||
|---|---|---|---|---|
|
hMLH1
|
hMSH2
|
|||
| Southern | MLPA | Southern | MLPA | |
| Normal | 98 | 99 | 90 | 94 |
| Equivocal | 0 | 0 | 4 | 0 |
| Deletion | ||||
| Concordant | 15 | 15 | 41 | 41 |
| Discordant* | 1† | 0 | 0 | 1‡ |
| 1§ | 0 | |||
| Duplication | ||||
| Concordant | 4 | 4 | n/a | n/a |
| Discordant | n/a | n/a | n/a | n/a |
| Total | 118 | 118 | 136 | 136 |
Major discrepancies only. See text for further discussion about major/minor discordant and equivocal results.
Southern analysis demonstrated a deletion in hMLH1 exons 12 to 13.
MLPA analysis demonstrated a deletion in hMSH2 exon 16.
Southern analysis demonstrated a hMSH2 promoter deletion.
For hMSH2, a total of 136 samples were analyzed, and both Southern and MLPA analyses identified 42 samples with large genomic hMSH2 alterations, and 94 samples that were negative (Table 2). Southern blot analyses identified 90 negative samples and four samples that were equivocal, whereas MLPA identified all of these same 94 samples as negative for an alteration. One of the equivocal (by Southern) samples had a junction fragment for a particular exon for one digest (Figure 4, patient 7), but did not show a 50% decrease in band intensity of that exon for the other digests. The three other equivocal samples had an ∼30% decrease in band intensity of a particular exon for one of the digests, but the other digest was normal for that exon. Additionally, one of the samples had a slight (20 to 30%) increase in the band intensity of an exon for all three digests. Of the 42 positive samples, 30 were from probands and of these probands, 12 different hMSH2 deletions were characterized by both Southern and MLPA (L.M. Baudhuin and colleagues, submitted for publication). Of the positive results identified between the two methods, there were two major discrepancies (Table 2) and three minor discrepancies. Of the major discrepancies, one sample had a hMSH2 promoter deletion that was detected by Southern, but undetectable by MLPA because of a lack of MLPA probes for that region of the gene (false-negative by MLPA). Another sample was normal by Southern, but showed a deletion in hMSH2 exon 16 by MLPA. Exon 16 was sequenced in this sample, and showed a single base mismatch at nucleotide 2637 (2637C>T), resulting in a nonsense mutation (Gln879Stop). This mutation occurred near the ligation site of the MLPA hMSH2 exon 16 probes and likely led to the decreased peak height observed by MLPA (false-positive by MLPA). For the three minor discrepancies, similar to hMLH1 above, these samples had larger deletions along with junction fragments that were observed by Southern. MLPA detected the deletion but did not detect the exons that were part of the junction fragment as disrupted.
Time and Cost Comparison of Southern Blot versus MLPA
We estimated and compared the cost of reagents and length of time necessary to perform Southern blot and MLPA for gene dosage analysis of hMLH1 and hMSH2. Overall, Southern blot takes ∼6 working days for completion of the test (from set-up to analysis), compared to ∼2 working days for MLPA. For both methods, this estimated time does not include down-time that the technician may have while waiting for transfers, incubations, and so forth. In addition, the estimation for Southern blot does not include weekend (ie, nonworking) days for exposure of the radioactive membrane to the PhosphorImager. Overall, the cost of reagents for one patient for Southern blot is approximately $10 ($5 for hMLH1 and $5 for hMSH2). For MLPA, all reagents for hMLH1 and hMSH2 are provided in a kit, and the cost to run one patient in duplicate (our current protocol) by MLPA is approximately $30. In terms of equipment, both methods require a thermocycler. In addition, the Southern blot method requires miscellaneous equipment for running and transferring gels, hybridization ovens, washing equipment, protective equipment for using radioactive materials, and PhosphorImaging equipment. The MLPA method requires a capillary electrophoresis system, such as the ABI 3100.
Discussion
We evaluated and cross-validated two methods for hMLH1 and hMSH2 gene dosage analysis: a Southern blot method using novel probes for hMLH1 and hMSH2 and MLPA, a commercially available PCR-based approach for gene dosage analysis. The Southern blot method was strategically developed to identify and characterize deletions/duplications in hMLH1 and hMSH2 on an exon-by-exon level. Our design strategy resulted in three probes for each gene, with four to six exons within each probe. The lack of restriction enzyme digestion between a few of the exons required us to combine these exons as a single probe. For example, in hMLH1, intron 7 is very small and there are no EcoRI, BglII, and HindIII restriction digestion sites that allow for separation of exons 7 and 8. Therefore, for the enzyme combination chosen, exons 7 and 8 were combined into a single probe, which was used to represent both exons 7 and 8. The same is true for the hMLH1 exons 16 to 19 probes, in which the probes for exons 16 and 18 represent exons 16 to 19. As a result, this method does not distinguish each individual exon for these regions of hMLH1. However, if necessary, additional digests could be performed with alternative restriction endonucleases to separate these hMLH1 exons out. In the case of hMSH2, there are no exons that are grouped together with all three enzyme digests, so all of the exons can be distinguished individually with at least one restriction enzyme. In addition, the ability to separate all of the remaining hMLH1 exons greatly simplifies the analysis when compared to results obtained with the use of genomic or cDNA probes.
In general, most recombinant PCR protocols are limited by the fact that they can link only two PCR products into one molecule.20,21 However, our goal was to link five to six exons together to minimize the number of probes needed for each gene. After several attempts, we were unable to recombinantly link more than two or three exons successfully. This may have been due to inefficiencies of the DNA polymerase to amplify recombinant molecules comprised of more than three recombinant fragments. Therefore, to obtain four to six exons within one group, we used a two-step subcloning method to link two recombinant molecules into one subcloning vector.
A major advantage of this novel Southern method is that by using multiple exon probes, deletions/duplications can be directly identified and characterized on an exon-by-exon basis. Because the exon probes were subcloned into the vector at equimolar concentrations, the relative densities of each exon’s hybridization band are consistent with each run. We found that hybridization density was associated with the percent GC of each individual exon, with GC percentages lower than 30% resulting in lighter hybridization bands. A likely explanation for this is that the corresponding probe would contain fewer cytosines (α-32P-dCTP) compared to probes targeting exons having a higher GC content. Another explanation is that due to the lower annealing temperature of probes with low GC content, these probes would more easily wash off of the target DNA, resulting in lighter hybridization bands.
After hybridization, densitometry was performed on the resultant hybridization bands followed by quantitative analysis for deletion/duplication status. These results were more reliable than that of a semiquantitative PCR method.13 Because three enzyme digestions were performed, the information was redundant, thereby minimizing false-positive signals. Furthermore, our probe design resulted in exons that were distributed in a noncontiguous manner, so that at least one exon could act as an internal nondeletion control for a given probe/enzyme combination in nearly all cases.
In addition to Southern blot analyses, more recent studies have used PCR-based methods for dosage analyses, including MLPA.22,23,24,25 Several of these studies have indicated that MLPA is a robust assay.22,23 Although for the most part, MLPA performed quite well in our hands, it is a complex assay and requires specific and consistent conditions for it to be robust. We found that MLPA was sensitive to quality and concentration of genomic DNA. Although the manufacturer recommends using 50 to 200 ng of genomic DNA, our titration analyses demonstrated that ∼400 ng of DNA provided more consistent performance. In either case, this is still a very low amount of DNA, which is advantageous, especially when compared to the amount of DNA required for Southern blot and other non-PCR-based gene dosage analysis methods. Quality of DNA was also very important for MLPA analysis. We found that the DNA extraction methodology was a key step in ensuring success of the procedure. Nonetheless, despite the sensitivity of the MLPA assay to DNA quality and quantity, MLPA proved to be a good method for clinical analyses overall.
Manual analysis of MLPA results can be tedious and time-consuming, especially with a large number of samples. To aid the speed of analysis, and to avoid errors that could occur by manual analysis, we developed a software program to analyze the data. As described above, initial analysis of the data were performed using Gene-Scan and Genotyper, and a table was created in Genotyper. The data in the table was then stored in a Microsoft Access database and rapidly analyzed with our in-house software. We validated the software analysis to ensure its performance and found it to be concordant with all aspects of the manual analysis.
As in previous studies,23,24,25 MLPA proved to be an efficient and capable method for gene dosage analysis of hMLH2 and hMSH2. Overall, MLPA and Southern blot results were 98.8% concordant: three major discrepancies were detected. MLPA was able to detect all of the mutations identified by Southern, with the exception of the hMSH2 promoter deletion and the probable hMLH1 IVS12 deletion. The inability to detect the promoter deletion was due to a lack of MLPA probes to that region, rather than to a technical failure of the assay. For the hMLH1 deletion that MLPA did not detect, Southern results suggested that the deletion was ∼400 to 500 bases in length and likely occurred in the IVS12 region, between the MLPA exon 12 and 13 probe binding sites. However, caution should be taken with this interpretation, because there was insufficient DNA to perform confirmatory follow-up analyses that would more clearly identify the deletion in question. For the third major discrepancy, a hMHS2 exon 16 deletion was detected by MLPA but not by Southern. Upon sequencing, a mutation located three nucleotides from the probe ligation site was discovered. This nucleotide change likely led to instability at the ligation site, resulting in a lack of ligation between the exon 16 probes. Thus, when using MLPA for gene dosage analysis, it is important to sequence single exon deletions (and possibly confirm by Southern) to rule out a false-positive result. Other samples that demonstrated single exon deletions by MLPA were concordant with Southern blot, and did not have any detectable alterations by sequencing, indicating that the single exon deletion observed was a true single exon deletion.
Minor discrepancies also occurred between the two methods. Although MLPA accurately identified the presence of a deletion in the majority of cases, MLPA did not correctly define the true extent of the deletion in those cases in which exons were partially deleted. In our sample set, this occurred for five deletions. In these cases, junction fragments were observed by Southern for exons 1, 2, 3, 6, and 9 of hMSH2 and exons 10 and 12 of hMLH1. In all of these cases, although MLPA detected a deletion, it did not identify the exons with junction fragments (as observed by Southern) as being disrupted. Although not verified, this was presumably because the partial exon deletion did not occur in the region that the MLPA probes bound to. Thus, the full extent of the deletion was not clearly defined by the MLPA results. On the other hand, for two other cases with deletions that had junction fragments by Southern, MLPA did identify these exons (hMSH2 exons 8 and 16) as disrupted.
We also encountered four hMSH2 cases that were normal by MLPA but demonstrated equivocal single exon alterations by Southern. As described above, for one of these samples, a junction fragment occurred in exon 8 with a corresponding 50% decrease in the expected band for that exon for one of the digest/probe combinations. However, for the other two-digest/probe combinations, the expected band for that exon was not decreased densitometrically. Thus, it was unclear whether the exon was truly altered, or whether there was a polymorphism at the restriction enzyme recognition site for the digest in which the junction fragment occurred. In the three other equivocal samples, one of the two-probe/digest combinations for exon 4 demonstrated an ∼30% decrease in band density (the other probe/digest combination was normal). Additionally, one of these samples also had an ∼20 to 30% increase in band intensity for all three-probe/digest combinations for exon 3. Thus for these three cases, whether or not the alterations observed in band intensity were real or artifact remains to be determined.
This is the first time that a comprehensive cross-validation has been performed between MLPA and Southern. A previous study of hMLH1 and hMSH2 gene dosage alterations by both MLPA and Southern contained limited information pertaining to the Southern blot methodology and results, and did not demonstrate any discrepancies between the two methods.25 Other studies describing hMLH1 and hMSH2 gene dosage alterations detected by MLPA either did not confirm the results by another method, or used a non-Southern blot method (eg, long-range, inverse, or quantitative PCR) to confirm the results.23,24 Our study demonstrates that there are some discrepancies between results obtained by Southern blot and MLPA, especially in terms of detecting single exon deletions. Thus, it is important to keep in mind that depending on the method used, follow-up analyses may be necessary when encountering these types of gene rearrangements.
Overall, both the MLPA and Southern methods proved to be very useful in the clinical setting for identifying large gene deletions and duplications in hMLH1 and hMSH2. The Southern blot probe design described in this study has several advantages: 1) it allows for the separation of the majority of the exons into single unique fragments with noncontiguous exon analysis; 2) it allows for an internal control for dosage comparison; and 3) it provides built-in redundancy with the three-probe/three-enzyme combinations. MLPA, on the other hand, is useful for samples with limited amounts of DNA, it has a rapid turnaround time, and analysis generally results in an unambiguous interpretation of the deletion/duplication. Because genomic rearrangements of hMLH1 and hMSH2 occur at a relatively high frequency in patients with Lynch syndrome, detection methods such as Southern blot and/or MLPA must be applied when screening for germline mutations in Lynch syndrome patients.
Footnotes
L.M.B and M.M. contributed equally to this study.
References
- Allen BA, Terdiman JP. Hereditary polyposis syndromes and hereditary non-polyposis colorectal cancer. Best Pract Res Clin Gastroenterol. 2003;17:237–258. doi: 10.1016/s1521-6918(02)00149-x. [DOI] [PubMed] [Google Scholar]
- Baudhuin LM, Burgart LJ, Leontovich O, Thibodeau SN: Use of microsatellite instability and immunohistochemistry testing for the identification of individuals at risk for Lynch syndrome. Fam Cancer (in press) [DOI] [PubMed] [Google Scholar]
- Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- Buermeyer AB, Deschenes SM, Baker SM, Liskay RM. Mammalian DNA mismatch repair. Annu Rev Genet. 1999;33:533–564. doi: 10.1146/annurev.genet.33.1.533. [DOI] [PubMed] [Google Scholar]
- Jiricny J, Nystrom-Lahti M. Mismatch repair defects in cancer. Curr Opin Genet Dev. 2000;10:157–161. doi: 10.1016/s0959-437x(00)00066-6. [DOI] [PubMed] [Google Scholar]
- Peltomaki P. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum Mol Genet. 2001;10:735–740. doi: 10.1093/hmg/10.7.735. [DOI] [PubMed] [Google Scholar]
- Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J Med Genet. 1999;36:801–818. [PMC free article] [PubMed] [Google Scholar]
- Cunningham JM, Kim C-Y, Christensen ER, Tester DJ, Parc Y, Burgart LJ, Halling KC, McDonnell SK, Schaid DJ, Walsh-Vockley C, Kubly V, Nelson H, Michels VV, Thibodeau SN. The frequency of hereditary defective mismatch repair in a prospective series of unselected colorectal carcinomas. Am J Hum Genet. 2001;69:780–790. doi: 10.1086/323658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom-Lahti M, Kristo P, Nicolaides NC, Chang SY, Aaltonen LA, Moisio AL, Jarvinen HJ, Mecklin JP, Kinzler KW, Vogelstein B, De La Chapelle A, Peltomäki P. Founding mutations and Alu-mediated recombination in hereditary colon cancer. Nat Med. 1995;1:1203–1206. doi: 10.1038/nm1195-1203. [DOI] [PubMed] [Google Scholar]
- Mauillon JL, Michel P, Limacher JM, Latouche JB, Dechelotte P, Charbonnier F, Martin C, Moreau V, Metayer J, Paillot B, Frebourg T. Identification of novel germline hMLH1 mutations including a 22 kb Alu-mediated deletion in patients with familial colorectal cancer. Cancer Res. 1996;56:5728–5733. [PubMed] [Google Scholar]
- Wijnen J, van der Klift H, Vasen H, Khan PM, Menko F, Tops C, Meijers Heijboer H, Lindhout D, Moller P, Fodde R. MSH2 genomic deletions are a frequent cause of HNPCC. Nat Genet. 1998;20:326–328. doi: 10.1038/3795. [DOI] [PubMed] [Google Scholar]
- Charbonnier F, Raux G, Wang Q, Drouot N, Cordier F, Limacher JM, Saurin JC, Puisieux A, Olschwang S, Frebourg T. Detection of exon deletions and duplications of the mismatch repair genes in hereditary nonpolyposis colorectal cancer families using multiplex polymerase chain reaction of short fluorescent fragments. Cancer Res. 2000;60:2760–2763. [PubMed] [Google Scholar]
- Lynch HT, Coronel SM, Okimoto R, Hampel H, Sweet K, Lynch JF, Barrows A, Wijnen J, van der Klift H, Franken P, Wagner A, Fodde R, de la Chapelle A. A founder mutation of the MSH2 gene and hereditary nonpolyposis colorectal cancer in the United States. JAMA. 2004;291:718–724. doi: 10.1001/jama.291.6.718. [DOI] [PubMed] [Google Scholar]
- Miyaki M, Iijima T, Yamaguchi T, Shirahama S, Ito T, Yasuno M, Mori T. Novel germline hMSH2 genomic deletion and somatic hMSH2 mutations in a hereditary nonpolyposis colorectal cancer family. Mutat Res. 2004;548:19–25. doi: 10.1016/j.mrfmmm.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Casilli F, Di Rocco ZC, Gad S, Tournier I, Stoppa-Lyonnet D, Frebourg T, Tosi M. Rapid detection of novel BRCA1 rearrangements in high-risk breast-ovarian cancer families using multiplex PCR of short fluorescent fragments. Hum Mutat. 2002;20:218–226. doi: 10.1002/humu.10108. [DOI] [PubMed] [Google Scholar]
- Mansfield ES, Robertson JM, Lebo RV, Lucero MY, Mayrand PE, Rappaport E, Parrella T, Sartore M, Surrey S, Fortina P. Duchenne/Becker muscular dystrophy carrier detection using quantitative PCR and fluorescence-based strategies. Am J Med Genet. 1993;48:200–208. doi: 10.1002/ajmg.1320480406. [DOI] [PubMed] [Google Scholar]
- Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time LightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel CT, Kraus C, Rauch A, Ekici AB, Rautenstrauss B, Reis A. A new quantitative PCR multiplex assay for rapid analysis of chromosome 17p11.2-12 duplications and deletions leading to HMSN/HNPP. Eur J Hum Genet. 2003;11:170–178. doi: 10.1038/sj.ejhg.5200920. [DOI] [PubMed] [Google Scholar]
- Innis MA, Gelfand DH, Sninsky JJ, White TJ. Burlington, MA: Academic Press, Inc.; PCR ProtocolsA Guide to Methods and Applications. 1989 [Google Scholar]
- Horton RM, Pease LR. Oxford: IRL Press; Directed MutagenesisA Practical Approach. 1991 [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:57–69. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CF, Charlton RS, Burn J, Sheridan E, Taylor GR. Genomic deletions in MSH2 or MLH1 are a frequent cause of hereditary non-polyposis colorectal cancer: identification of novel and recurrent deletions by MLPA. Hum Mutat. 2003;22:428–433. doi: 10.1002/humu.10291. [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Hampel H, de la Chapelle A. Identification and characterization of genomic rearrangements of MSH2 and MLH1 in Lynch syndrome (HNPCC) by novel techniques. Hum Mutat. 2003;22:258–263. doi: 10.1002/humu.9171. [DOI] [PubMed] [Google Scholar]
- Gille JJ, Hogervorst FB, Pals G, Wijnen JT, van Schooten RJ, Dommering CJ, Meijer GA, Craanen ME, Nederlof PM, de Jong D, McElgunn CJ, Schouten JP, Menko FH. Genomic deletions of MSH2 and MLH1 in colorectal cancer families detected by a novel mutation detection approach. Br J Cancer. 2002;87:892–897. doi: 10.1038/sj.bjc.6600565. [DOI] [PMC free article] [PubMed] [Google Scholar]