Abstract
A multiplex polymerase chain reaction assay was developed for the rapid simultaneous detection of category A select bacterial agents (Bacillus anthracis and Yersinia pestis) and parasitic pathogens (Leishmania species) in blood using the Cepheid Smart Cycler platform. B. anthracis (Sterne) and Yersinia. pseudotuberculosis were used in the assay for optimization for B. anthracis and Y. pestis, respectively. The specificity of the target amplicons [protective antigen gene of B. anthracis and rRNA genes of other pathogens or human (internal control)] was evaluated by staining the amplicons with SYBR Green I and determining their individual melting temperatures (Tm). As a novel approach for pathogen semiquantitation, the Tm peak height of the amplicon was correlated with a known standard curve of pathogen-spiked samples. This assay was able to detect DNA in blood spiked with less than 50 target cells/ml for all of the pathogens. The sensitivity of this assay in blood was 100% for the detection of Leishmania donovani from leishmaniasis patients and B. anthracis (Sterne) from symptomatic mice. The time necessary for performing this assay including sample preparation was less than 1.5 hours, making this a potentially useful method for rapidly diagnosing and monitoring the efficacy of drugs or vaccines in infected individuals.
Blood transfusion saves millions of lives each year. Eighty percent of the world’s population lives in developing countries with an access to only 20% of the world’s safe blood supply (World Health Organization, press releases, 2000, http://www.who.int/inf-pr-2000/en/state2000-09.html). Blood has the potential to be contaminated by numerous pathogens.1 Contamination of blood or blood products could be accidental, mainly due to donation from infected asymptomatic individuals or a deliberate bioterror attempt to spread the infectious agents. The organisms with maximum potential to be used as biothreats are usually highly virulent and often difficult to diagnose.
Several bacterial and viral pathogens are listed by the Centers for Disease Control and Prevention as category A agents, which may appear in blood during infection. Some of them are gram-positive spore-forming bacteria, Bacillus anthracis and Clostridium botulinum that cause anthrax and botulism, respectively,2 gram-negative bacterium Yersinia pestis that causes plague,3 and viruses responsible for various hemorrhagic fevers.4,5 Death due to anthrax occurs when the bacteremia reaches 107 to 108 bacilli/ml of blood.6 Y. enterocolitica, a common contaminant of packed red blood cells, was responsible for 50% of all clinical sepsis episodes associated with the transfusion of red blood cells, of which 61% were fatal.7 Packed red blood cells or whole blood, usually stored at 4°C, can allow the growth of these gram-negative bacteria.8 Among the bacteria mentioned above, B. anthracis and Y. pestis disseminate easily in the environment and result in high mortality rates.9,10 Concern about the usage of category A select organisms as bioweapons has increased because of recent intentional dissemination of B. anthracis spores in the United States.11 Although the average incubation period for such bacteria in the body before the onset of symptoms is only a few days (2 to 10 days),2 asymptomatic individuals could potentially donate infected blood in this window period.
There are several protozoan parasites that are blood borne, each of which causes a unique severe disease and that are of concern to the blood supply. For example, parasites belonging to the trypanosomatid family such as Leishmania donovani and Leishmania major, cause the fatal visceral (kala-azar) and cutaneous leishmaniasis, respectively.12 Similarly, Trypanosoma brucei and Trypanosoma cruzi are the causative agents for African sleeping sickness and Chagas disease, respectively.13 Thousands of immigrants to the United States from Chagas disease endemic regions, carrying asymptomatic chronic T. cruzi infections, represent a reservoir population for potential transfusion transmission of T. cruzi.14 At least six T. cruzi transfusion-transmitted cases have been reported so far in the United States and Canada.14 Leishmaniasis currently threatens 1.5 to 2.0 million people annually with an estimated death toll of 50,000 persons/year in 88 countries around the world.12 The intracellular parasite Leishmania is present in blood for an undefined period of time without showing any clinical symptoms during the initial phase of the disease.15 Such individuals can potentially donate infected blood, because Leishmania not only survives blood-banking storage conditions, but also retains its infectivity as demonstrated by studies in animal models, such as hamsters and dogs.16,17 Leishmania species such as L. donovani causing visceral disease have been shown to be transmitted by blood transfusion in several human cases.15 US army personnel stationed in endemic areas are exposed to Leishmania infection. In a recent Centers for Disease Control and Prevention/Department of Defense Morbidity and Mortality Weekly Report, ∼500 cases of cutaneous leishmaniasis and 2 visceral cases have been reported from US soldiers deployed in both Iraq and Afghanistan.18 Hence the Department of Defense and the American Association of Blood Banks implemented a 1-year deferral period for soldiers returning from Iraq after deployment and permanent deferral for a diagnosed case of leishmaniasis. This deferral does not extend to US soldiers in Afghanistan since they have been deferred because of Malaria. The Food and Drug Administration in the United States is in agreement with such a deferral policy as a measure to ensure the safety of the nation’s blood supply from transmission of Leishmania (Blood Products Advisory Committee Meeting, December, 2003).
Under the circumstances described above rapid and accurate detection of contamination or diagnosis of infection due to either bacterial or parasitic agents in blood is needed. Various methodologies have been reported to diagnose B. anthracis, Y. pestis, and Leishmania species with a wide range of specificity and sensitivity, mostly for an individual organism.3,13,19,20,21,22,23,24,25,26 The present study describes the development and evaluation of a single-tube rapid multiplex polymerase chain reaction (PCR) for the simultaneous detection of B. anthracis, Y. pestis, and Trypanosomatida in blood. In the current study the sensitivity and specificity of a multiplex PCR assay for the detection of the three types of blood borne pathogens are assessed.
Materials and Methods
Bacterial and Parasitic Species and Culture Conditions
Vaccine strain of B. anthracis (Sterne 34F2) (Colorado Serum Co., Denver, CO) was cultured in SG sporulation medium.27 Y. pseudotuberculosis (Pfeiffer) Smith and Thal [American Type Culture Collection (ATCC) no. 6905] and Y. enterocolitica strain 8081 were cultured in Luria-Bertani liquid medium.28 The bacterial cells were stored at −80°C according to standard procedure.28 To calculate cell concentration of the stored cultures, an aliquot of cultures was used to count cells by serial dilution and plating on Luria-Bertani agar plates. Different species of Leishmania and T. brucei from our laboratory stocks were cultured using published protocols29,30 and cells were counted using a Coulter particle counter (Beckman Coulter, Miami, FL). In vitro grown L. donovani axenic amastigote parasites (strain 1S, clone 2D, World Health Organization designation MHOM/SD/62/1S-CL2D)31 that closely resemble the amastigote stage of Leishmania found in human macrophages were used to spike Leishmania into blood samples.12
Target Genes and Design of Primers
The amplicons that were chosen are from the genes that have multiple copies in both selected pathogens and humans. The B. anthracis detection target is in the protective antigen gene located on plasmid pXO1, present also in the vaccine strain. The Y. pestis target is in the 16S rRNA gene region conserved in all of the Yersinia species. Similarly, to detect Leishmania species the target is in the conserved region of the 18S rRNA gene. To increase the confidence of the assay and to recognize the false-negatives that may arise because of a reaction failure, an internal control was used. The internal control target is in the human 18S rRNA gene.
To establish a multiplex assay, we identified primer sets specific for these pathogen targets and the human ribosomal RNA control that have similar annealing temperatures and an optimal Mg2+ concentration using the Primer 3′ program.32 The length of all of the primers was between 18 to 23 bases with an optimum (50%) GC content. Optimum melting temperature (Tm) of the oligos was 58°C at a salt concentration of 50 mmol/L. The primers (sequences not provided for security reasons) of B. anthracis, Yersinia, and Leishmania were screened against the human RepBase library [a database of repetitive DNA sequence elements found in a variety of eukaryotic organisms (http://www.hgmp.mrc.ac.uk/Software/EMBOSS/Apps/eprimer3.html)] to avoid repetitive regions. The specificity of the primers and avoidance of cross-hybridization with human sequences was assured by testing in BLAST searches (http://www.ncbi.nlm.nih.gov/blast). To avoid formation of primer homodimers and heterodimers, the primer sequences were tested in the Amplify 1.2′ program (http://engels.genetics.wisc.edu/amplify). To discriminate the amplified products, care was taken while designing the oligos to select different sized amplicons that have different melting temperature values. The lengths of the four amplicons were 99 bp for B. anthracis, 121 bp for Yersinia, 163 bp Leishmania, and 141 bp for the internal control. The amplicon sequences were tested in the Mfold program to avoid secondary structures on DNA where the primers anneal.33
DNA Extraction from Bacteria, Parasite, and Whole Blood
Total DNA from cultured B. anthracis (Sterne), Y. pseudotuberculosis, Leishmania species, and T. brucei was extracted, purified, and measured using the Genome DNA kit (BIO 101, Carlsbad, CA) and manufacturer’s protocol. To determine the limit of detection, 200 μl of healthy anonymous donor human whole blood (National Institutes of Health blood bank) containing heparin as anti-coagulant was seeded with serial dilution of each pathogen and the DNA was extracted with the QIAamp DNA blood mini kit (Qiagen, Valencia, CA). To extract DNA from the blood of visceral leishmaniasis patients, 200 μl of blood was directly used for the extraction. However, to extract DNA from the mice infected with B. anthracis (Sterne) 10 μl of the blood was mixed with 190 μl of healthy human blood and used for extraction. The total processing time for a DNA extraction was less than 30 minutes. The DNA at the final step was dissolved in 50 μl of Qiagen elution buffer and 5 μl of this was used in each multiplex PCR reaction.
Multiplex PCR Assay for Detection and Semiquantitation of Pathogen Cells
A master mix was prepared (for each 25-μl reaction volume) containing 1× buffer (TaKaRa), 6 mmol/L MgCl2, 0.25 U Ex Taq (TaKaRa), 0.25 mmol/L dNTP, 0.3 μmol/L each primer, 1× SYBR Green I (vendor stock 10,000X; Molecular Probes, Eugene, OR), and water to 20 μl and aliquoted into Smart Cycler PCR reaction tubes (Cepheid, Sunnyvale, CA). Then 5 μl of template DNA was added to each tube. Cycling parameters were: preheat at 94°C for 150 seconds and then 45 two-step cycles of 94°C for 15 seconds and 64°C for 30 seconds. After the last amplification cycle, PCR products were analyzed by melting curve analysis in the Smart Cycler by slowly increasing the temperature to 95°C. Total reaction and melting curve analysis time for a sample was 55 minutes. The reactions were run in triplicate with appropriate controls. The fidelity of the amplified products of all of the organisms was confirmed by individually cloning the amplified products into pCRII-TOPO plasmid (Invitrogen, Carlsbad, CA) and sequencing.
The lowest level of detection of each pathogen was determined by using the extracted DNA obtained from 200 μl of blood spiked with serial dilutions of cells from each individual pathogen (ie, 10,000, 1000, 100, 10, and 5 cells). To calculate the pathogen load in the blood, a standard curve based on the fluorescence peak height of the amplified products and pathogen cell number in blood was generated.
Detection of Pathogens from Blood Samples of B. anthracis-Infected Mice and Leishmaniasis Patients
The sensitivity of the multiplex assay was tested on the blood obtained from mice infected with B. anthracis (Sterne). The mice were inoculated intraperitoneally with a target dose (lethal dose 90%) of 2 to 2.5 × 107 spores of bacteria essentially as described previously.34 Blood samples were collected in the presence of anti-coagulant from these infected mice at different time points after challenge. Samples from noninoculated mice were included in the assay as negative controls.
Blood samples were collected from 11 kala azar (visceral leishmaniasis) patients from Bihar, India, and reporting to Safdarjung Hospital, New Delhi, India, at the pretreatment stage. The patients exhibited characteristic disease symptoms such as fever, hepatosplenomegaly, anemia, and leukopenia. Only patients that were clinically diagnosed for kala azar and found positive for the presence of the parasite in bone marrow aspirates at the pretreatment stage26 were used in this study. In addition, blood samples obtained from three patients after treatment with anti-leishmanial drugs were also included in the study. All of the collected blood samples were heparinized, their identity was blinded and stored at 4°C. The coding of the clinical status of the patient samples were broken after the assay was performed and results were then analyzed. All of the animal and human experiments were done under National Institutes of Health guidelines for animal and human protection (http://oacu.od.nih.gov/ARAC).
Results
Evaluation of the Multiplex PCR Assay
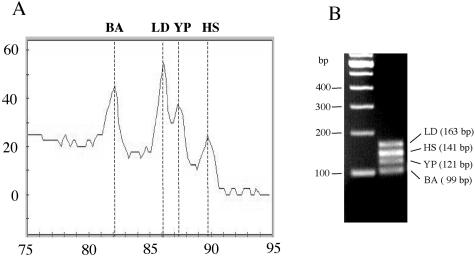
Because the virulent strain of B. anthracis is a select agent, we used the animal vaccine (Sterne) strain for the assay standardization. Because both Y. pestis and Y. enterocolitica are infectious, the closely related species Y. pseudotuberculosis, which is not a human pathogen was used as a surrogate. L. donovani cells were used to test the primer set that has a target in all of the Leishmania and Trypanosoma species. To test the multiplex assay, the DNA of B. anthracis (Sterne) (100 fg), Y. pseudotuberculosis (100 fg), and L. donovani (50 pg) was mixed with human DNA [a 10th of the DNA (∼300 ng) obtained from 200 μl of blood] and used in a multiplex PCR. Amplification of all of the four amplicons was observed by the appearance of their expected fluorescence Tm peaks (Figure 1A). The Tm peak for B. anthracis (Sterne) was 82.2°C, for Y. pseudotuberculosis was 87.3°C, for L. donovani was 86.1°C, and for the human control was 88.8°C. The agarose gel picture of the PCR reaction shows the amplified products of all of the organisms with expected size DNA fragments (Figure 1B). The fidelity of the sequence of the amplified products was further confirmed by sequence analysis.
Figure 1.
A: Melt derivative analysis of the multiplex fluorescence PCR showing the amplification of specific products of B. anthracis (Sterne) (BA) with Tm 82.2°C, L. donovani (LD) with Tm 86.1°C, Y. pseudotuberculosis (YP) with TM 87.3°C, and the internal human control (HS) with Tm 88.8°C. B: Agarose gel confirmation of the products of the multiplex PCR stained with ethidium bromide from A showing the amplified products of: LD, L. donovani; HS, human; YP, Y. pseudotuberculosis; and BA, B. anthracis (Sterne).
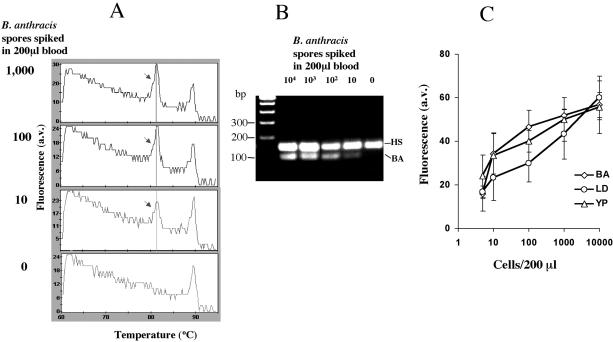
After confirming the feasibility of the multiplex assay conducted using the DNA of the pathogens directly, the lower level of detection (LOD) of the pathogens in the blood was determined by doing the assay on DNA obtained from 200 μl of whole blood spiked with a known number of B. anthracis (Sterne) spores, 10,000, 1000, 100, 10, 5, and 0. A Tm peak corresponding to the amplified product of B. anthracis (Sterne) was observed at 82°C at all of the concentrations (Figure 2A). We also observed a gradual reduction in the height of the B. anthracis Tm peaks (Figure 2A) and the intensity of the amplified DNA bands of B. anthracis on the gel (Figure 2B) with the decreasing number of pathogen spores in the blood. However, in all of the reactions, the height of the internal control (human rRNA gene fragment amplification) Tm peak remained constant. We have used the height of the fluorescence peak above baseline of B. anthracis (Sterne) of different spore concentrations in plotting a standard graph for quantification (Figure 2C). All of the negative samples (nine) had baseline fluorescence. Similar studies on the determination of LOD were performed for the other two organisms, Y. pseudotuberculosis and L. donovani, and shown in Figure 2C. The analysis showed that the assay system can detect <10 cells per 200 μl of blood for all of the three pathogens.
Figure 2.
A: Melt derivative analysis of the multiplex PCR assay with human blood carrying a known number of spiked B. anthracis (Sterne) spores. B: Agarose gel confirmation of the products of the multiplex PCR showing the linear relationship between the logarithm of the number of spores spiked and the amount of amplified band intensity of B. anthracis (Sterne). C: Quantitation graph showing the linear relationship between the height of the fluorescence peak above baseline of the specific amplicons (y axis) and the logarithm of the number of bacterial or parasitic pathogen cells individually spiked in 200 μl of blood (x axis). BA, B. anthracis (Sterne); YP, Y. pseudotuberculosis; LD, L. donovani; a.v., arbitrary value. The data represent the means ± SD of three independent experiments.
Specificity and Sensitivity of PCR
Each of the primer sets added in the multiplex reaction amplified its product only in the presence of the DNA from its corresponding organism. When the primer sets were tested individually on the templates of other organisms such as Escherichia coli and Bacillus cereus they did not amplify any DNA fragment (data not included). The primers neither cross-hybridized with each other nor did they react with any amplified products of other pathogens in the assay. They did not amplify any nonspecific fragments other than the four expected amplicons. However, Yersinia primers also amplified DNA from Y. enterocolitica (with Tm peak at 87°C) when tested in a separate reaction (data not shown). Similarly, the primers designed for Leishmania recognized, in addition to L. donovani, the DNA extracted from other closely related Leishmania and Trypanosoma species that cause different diseases, namely L. chagasi, L. infantum, and L. mexicana that cause visceral leishmaniasis; L. major and L. tropica that cause cutaneous leishmaniasis; L. braziliensis that causes mucocutaneous leishmaniasis; and Trypanosoma brucei that causes African sleeping sickness (data not shown).
Detection of Anthrax in Blood Samples from in Vivo-Infected Mice Using Multiplex PCR
Multiplex PCR analysis was conducted with blood samples collected during a time course after infection from laboratory mice inoculated with B. anthracis (Sterne). The sample type and PCR results are shown in Table 1. Multiplex PCR conducted on the blood samples at 0 (prebleed) and 12 hours after the intraperitoneal injection of the spores of B. anthracis (Sterne) did not show the fluorescence peak for B. anthracis (Table 1). However, blood samples collected at 24-, 36-, and 48-hour time points after the inoculation were positive for the bacteria at an increasing rate with time after infection (Table 1). The bacteremia in the animal that tested PCR-positive after 24 hours of exposure was estimated to be 200 to 1000 cells/ml (cell concentration range obtained from the Tm peak standard curve) (Figure 2C). Similarly, four of five animals that were positive after 36 to 48 hours of exposure had bacteremia in their blood between 104 to 105 cells/ml. All these bacteremic mice were lethargic with scruffy hair and died within 24 hours.
Table 1.
Multiplex Fluorescence PCR with Blood Samples from Mice Infected with the Spores of B. anthracis (Sterne)
| Blood collection time (hours after challenge) | Total number of samples tested | Number of PCR-positive samples (% positives in parentheses) | Approximate B. anthracis (Sterne) cell number in blood per ml |
|---|---|---|---|
| 0 | 6 | 0 | ND |
| 12 | 6 | 0 | ND |
| 24 | 4 | 1 (25) | 200 to 1000 |
| 36 | 3 | 2 (66) | 1 × 104 to 105 |
| 48 | 2 | 2 (100) | 1 × 104 to 105 |
ND, not detected.
Detection of L. donovani in the Blood Samples from Visceral Patients Using Multiplex PCR
The multiplex PCR assay was evaluated with the blood samples collected from 11 leishmaniasis patients who were identified as positive for the presence of the parasite (Table 2). The approximate number of parasites calculated with the help of the standard graph ranged from 50 to 1000 cells per ml of blood in these patients. Whereas samples obtained from three patients who had undergone drug treatment showed no amplification for the presence of the parasite (Table 2).
Table 2.
Multiplex Fluorescence PCR with Blood Samples from Visceral Leishmaniasis Patients Infected with L. donovani
| Source of blood samples | Total number of samples tested | Number of PCR-positive samples | Approximate Leishmania cell number in blood per ml |
|---|---|---|---|
| Patients with VL | 11 | 11 | 50 to 1000 |
| VL patients after treatment | 3 | 0 | ND |
ND, not detected.
Discussion
Individual detection of either B. anthracis,19,20,24 Y. pestis,22,23 or Leishmania26,35 through conventional or real-time PCR has been reported. With an increased concern for blood safety due to emerging pathogens and the threat of bioterror agents, which can be transmissible through blood, the need for rapid pathogen detection becomes essential. The cost and the logistical burden of testing for many phylogenetically diverse pathogens call for a multiplex assay. However, little is known on the simultaneous detection of pathogens of diverse genera through multiplex PCR. Technological hurdles to detection in the unique matrix of whole blood and the increase in complexity of the PCR reaction that occurs when run in a multiplex format have slowed development of such an assay. There is a need to develop tests that could be used for both donor screening and diagnosis ideally based on multiplexing for such pathogens. In the present study, we have developed an assay that can simultaneously identify the presence of both bacterial (B. anthracis and Y. pestis) and parasitic (Trypanosomatid species) pathogens in blood. We selected target genes that are present in multiple copies in these organisms to achieve enhanced assay sensitivity.
Specific primer sets were designed to detect B. anthracis, Yersinia, and Leishmania to work uniformly in a multiplex PCR reaction using SYBR Green I in a Smart Cycler platform. The primers were generated such that each of the primer sets of an organism amplify a distinct length of amplicon so that the amplified products have distinct melting temperatures and could be resolved on agarose gel if needed. We performed melt-derivative analysis of the PCR products and did not rely on the cycle threshold (CT) value, because the CT value would be a collective fluorescence of all of the amplified products, stained with SYBR Green I in this multiplex reaction. Melt derivatives obtained from the melting curve analysis conducted at the end of the last PCR cycle resolve the amplified products into individual peaks depending on their Tm values. Tm value differentiates the amplified products based on their length, sequence, and GC content. Such an analysis in this report resolved all of the four amplicons into four distinctive Tm peaks.
We are unaware of any reports with a method to quantitate the pathogen cells in biological samples in a multiplex PCR using SYBR Green I-based melting curve analysis. Hence, we have used the Tm peak heights of the products that correspond proportionally to the approximate pathogen cell number per unit volume of blood as a novel way to achieve semiquantitative measure of each pathogen.
Because SYBR Green I can also bind nonspecifically to any amplified DNA, steps were taken to avoid nonspecific amplifications. The enzyme, TaKaRa EX Taq polymerase (TaKaRa Shuzo Co., Ltd., Shiga, Japan) premixed with neutralizing monoclonal antibody to the polymerase, that requires hot start for the release of the antibody that activates the polymerase and reduces nonspecific amplification was used in the assay. The reaction samples were hot started at 95°C for 150 seconds before the first denaturation step of thermocycling. A combined PCR annealing and extension of the product was accomplished at a relatively high temperature, ie, 64°C for 30 seconds in each cycle. These features probably are the reason that we did not observe any nonspecific DNA amplification in the reaction. In earlier studies by others in the blood samples spiked with either B. anthracis (Sterne),25 Y. enterocolitica,36 or Leishmania infantum,37,38 the lowest limits of detection observed were 400, 8, and 10 to 100 cells/ml, respectively. Whereas in this study, the lower detection level achieved for three pathogens was <50 cells/ml. This suggests that the assay presented here has a higher sensitivity for B. anthracis and Leishmania detection in blood samples. This assay is rapid and takes less than 1.5 hours to detect any of these organisms either individually or simultaneously if present in blood. There is no such rapid method designed for identifying Yersinia sp. in blood. However, an assay developed by Mayo-Roche to detect B. anthracis individually in blood takes <1.5 hours.39 Previously tests developed to detect Leishmania in blood using fluorescence PCR have shown to take >2 hours.40,41
In addition to the high specificity of the primers selected for all of the three pathogens, primers for Yersinia and Leishmania could also detect related Yersinia and Leishmania species. For example, the primers of Y. pestis also recognized the DNA of Y. enterocolitica, an another blood borne species of Yersinia that causes septicemia disorder. Similarly the primers of Leishmania recognized DNA of several species of Leishmania and T. brucei confirming that these primers can identify other blood-borne Trypanosomatid parasites as well. Thus the assay has the ability to detect a limited group of organisms that are all important to detect for blood safety. Primer sets to identify the individual pathogens are available and could be adapted to the Smart Cycler platform. We are in the process of testing the primers described in this study for virulent forms of B. anthracis and Y. pestis to ensure their ability to detect the corresponding virulent agents.
The test was also validated successfully using peripheral blood samples obtained from mice infected with B. anthracis (Sterne) and from visceral leishmaniasis patients. The sensitivity of B. anthracis (Sterne) detection with either regular or real-time PCR using tissue materials other than blood reported earlier was between 89 to 100%.21,42 Similarly, reports on detection of Leishmania species from blood samples using a PCR assay, have shown 45 to 96% sensitivity with different degree of sample sizes.26,43,44,45,46,47,48,49 The sensitivity of the current multiplex PCR assay to detect Leishmania or B. anthracis (Sterne) in blood is 100% from either patients or symptomatic mice, respectively. Infected mice that failed to give a positive PCR test were not symptomatic at the time of collection of the blood sample implying a very low level of bacteremia.
In conclusion, unique primer sets, careful optimization of the conditions, extensive development, and testing with laboratory cultured pathogens spiked into human blood have resulted in an assay that proved capable of detecting Leishmania and B. anthracis (Sterne) from infected human and mouse blood samples, respectively. This assay meets the requirement of simultaneous, specific, sensitive, and rapid detection of more than one divergent blood-borne pathogen in a single reaction tube. This semiquantitative assay has the potential to be automated and incorporated into high-throughput screening. Optimization toward increasing the number of pathogens detected in a multiplex format and high-throughput screening is under exploration. This assay also has the potential to be used for other body fluid samples. Similar multiplex assays could be designed for other biological agents that pose a bioterror threat.
Acknowledgments
We thank James E. Rogers (Edgewood Chemical Biological Center, U.S. Army Soldier and Biological Chemical Command, U.S. Army Edgewood, MD) for providing Y. pseudotuberculosis; Karen Meysick (Division of Bacterial, Parasitic, and Allergenic Products, CBER, Food and Drug Administration, Bethesda, MD) for providing Y. enterocolitica; Mike Klutch (Division of Viral Products, CBER, Food and Drug Administration, Bethesda, MD) for sequencing the amplicons; and Indira Hewlett and Alain Debrabant (Division of Emerging and Transfusion Transmitted Diseases, Center for Biologics Evaluation and Research, Food and Drug Administration, Bethesda, MD) for critical review of this manuscript.
Footnotes
Supported by the Competitive Grant for Counter Terrorism Special Needs Projects, Center for Biologics Evaluation and Research, Bethesda MD, and Cooperative Research and Development Agreement, Hematech, Westport, CT.
References
- Chamberland ME, Epstein J, Dodd RY, Persing D, Will RG, DeMaria A, Jr, Emmanuel JC, Pierce B, Khabbaz R. Blood safety. Emerg Infect Dis. 1998;4:410–411. doi: 10.3201/eid0403.980317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan EH. Detecting bioterror attacks by screening blood donors: a best-case analysis. Emerg Infect Dis. 2003;9:909–914. doi: 10.3201/eid0908.030079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelthaler DM, Hinnebusch BJ, Rittner CM, Gage KL. Quantitative competitive PCR as a technique for exploring flea-Yersina pestis dynamics. Am J Trop Med Hyg. 2000;62:552–560. doi: 10.4269/ajtmh.2000.62.552. [DOI] [PubMed] [Google Scholar]
- Leroy EM, Baize S, Volchkov VE, Fisher-Hoch SP, Georges-Courbot MC, Lansoud-Soukate J, Capron M, Debre P, McCormick JB, Georges AJ. Human asymptomatic Ebola infection and strong inflammatory response. Lancet. 2000;355:2210–2215. doi: 10.1016/s0140-6736(00)02405-3. [DOI] [PubMed] [Google Scholar]
- Oliveira De Paula S, Malta Lima D, Clotteau M, Pires Neto Rd Rda J, Lopes da Fonseca BA. Improved detection of dengue-1 virus from IgM-positive serum samples using C6/36 cell cultures in association with RT-PCR. Intervirology. 2003;46:227–231. doi: 10.1159/000072432. [DOI] [PubMed] [Google Scholar]
- Swartz MN. Recognition and management of anthrax—an update. N Engl J Med. 2001;345:1621–1626. doi: 10.1056/NEJMra012892. [DOI] [PubMed] [Google Scholar]
- Klein HG, Dodd RY, Ness PM, Fratantoni JA, Nemo GJ. Current status of microbial contamination of blood components: summary of a conference. Transfusion. 1997;37:95–101. doi: 10.1046/j.1537-2995.1997.37197176958.x. [DOI] [PubMed] [Google Scholar]
- Wagner SJ, Friedman LI, Dodd RY. Transfusion-associated bacterial sepsis. Clin Microbiol Rev. 1994;7:290–302. doi: 10.1128/cmr.7.3.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan PJ, Rosenstein NE, Gillen M, Meyer RF, Kiefer MJ, Deitchman S, Besser RE, Ehrenberg RL, Edwards KM, Martinez KF. Responding to detection of aerosolized Bacillus anthracis by autonomous detection systems in the workplace. MMWR Recomm Rep. 2004;53:1–12. [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MT, Prentice MB, Sebaihia M, James KD, Churcher C, Mungall KL, Baker S, Basham D, Bentley SD, Brooks K, Cerdeno-Tarraga AM, Chillingworth T, Cronin A, Davies RM, Davis P, Dougan G, Feltwell T, Hamlin N, Holroyd S, Jagels K, Karlyshev AV, Leather S, Moule S, Oyston PC, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- Sobel J, Khan AS, Swerdlow DL. Threat of a biological terrorist attack on the US food supply: the CDC perspective. Lancet. 2002;359:874–880. doi: 10.1016/S0140-6736(02)07947-3. [DOI] [PubMed] [Google Scholar]
- Handman E. Leishmaniasis: current status of vaccine development. Clin Microbiol Rev. 2001;14:229–243. doi: 10.1128/CMR.14.2.229-243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desquesnes M, Davila AM. Applications of PCR-based tools for detection and identification of animal trypanosomes: a review and perspectives. Vet Parasitol. 2002;109:213–231. doi: 10.1016/s0304-4017(02)00270-4. [DOI] [PubMed] [Google Scholar]
- Leiby DA, Herron RM, Jr, Read EJ, Lenes BA, Stumpf RJ. Trypanosoma cruzi in Los Angeles and Miami blood donors: impact of evolving donor demographics on seroprevalence and implications for transfusion transmission. Transfusion. 2002;42:549–555. doi: 10.1046/j.1537-2995.2002.00077.x. [DOI] [PubMed] [Google Scholar]
- Luz KG, da Silva VO, Gomes EM, Machado FC, Araujo MA, Fonseca HE, Freire TC, d’Almeida JB, Palatnik M, Palatnik-de Sousa CB. Prevalence of anti-Leishmania donovani antibody among Brazilian blood donors and multiply transfused hemodialysis patients. Am J Trop Med Hyg. 1997;57:168–171. doi: 10.4269/ajtmh.1997.57.168. [DOI] [PubMed] [Google Scholar]
- Palatnik-de-Sousa CB, Paraguai-de-Souza E, Gomes EM, Soares-Machado FC, Luz KG, Borojevic R. Transmission of visceral leishmaniasis by blood transfusion in hamsters. Braz J Med Biol Res. 1996;29:1311–1315. [PubMed] [Google Scholar]
- Giger U, Oakley DA, Owens SD, Schantz P. Leishmania donovani transmission by packed RBC transfusion to anemic dogs in the United States. Transfusion. 2002;42:381–383. doi: 10.1046/j.1537-2995.2002.00061.x. [DOI] [PubMed] [Google Scholar]
- Aronson N, Ananthakrishnan M, Bernstein W, Hochberg L, Marovich M, Ockenhouse C, Yoon I, Weina P, Benson P, Fischer J, Hack D, Hawkes C, Polhemus M, Wortmann G, McEvoy P, Neafie R, Defraites R, Herwaldt BL. Update: Cutaneous leishmaniasis in U.S. military personnel—Southwest/Central Asia, 2002–2004. Morb Mort Wkly Rep. 2004;53:264–265. [Google Scholar]
- Cheun HI, Makino SI, Watarai M, Shirahata T, Uchida I, Takeshi K. A simple and sensitive detection system for Bacillus anthracis in meat and tissue. J Appl Microbiol. 2001;91:421–426. doi: 10.1046/j.1365-2672.2001.01395.x. [DOI] [PubMed] [Google Scholar]
- Fasanella A, Losito S, Trotta T, Adone R, Massa S, Ciuchini F, Chiocco D. Detection of anthrax vaccine virulence factors by polymerase chain reaction. Vaccine. 2001;19:4214–4218. doi: 10.1016/s0264-410x(01)00159-1. [DOI] [PubMed] [Google Scholar]
- Hoffmaster AR, Meyer RF, Bowen MD, Marston CK, Weyant RS, Thurman K, Messenger SL, Minor EE, Winchell JM, Rassmussen MV, Newton BR, Parker JT, Morrill WE, McKinney N, Barnett GA, Sejvar JJ, Jernigan JA, Perkins BA, Popovic T. Evaluation and validation of a real-time polymerase chain reaction assay for rapid identification of Bacillus anthracis. Emerg Infect Dis. 2002;8:1178–1182. doi: 10.3201/eid0810.020393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo AC, Almeida AM, Leal NC. Retrospective study of a plague outbreak by multiplex-PCR. Lett Appl Microbiol. 2003;37:361–364. doi: 10.1046/j.1472-765x.2003.01377.x. [DOI] [PubMed] [Google Scholar]
- Neubauer H, Meyer H, Prior J, Aleksic S, Hensel A, Splettstosser W. A combination of different polymerase chain reaction (PCR) assays for the presumptive identification of Yersinia pestis. J Vet Med B Infect Dis Vet Public Health. 2000;47:573–580. doi: 10.1046/j.1439-0450.2000.00384.x. [DOI] [PubMed] [Google Scholar]
- Ramisse V, Patra G, Garrigue H, Guesdon JL, Mock M. Identification and characterization of Bacillus anthracis by multiplex PCR analysis of sequences on plasmids pXO1 and pXO2 and chromosomal DNA. FEMS Microbiol Lett. 1996;145:9–16. doi: 10.1111/j.1574-6968.1996.tb08548.x. [DOI] [PubMed] [Google Scholar]
- Rantakokko-Jalava K, Viljanen MK. Application of Bacillus anthracis PCR to simulated clinical samples. Clin Microbiol Infect. 2003;9:1051–1056. doi: 10.1046/j.1469-0691.2003.00736.x. [DOI] [PubMed] [Google Scholar]
- Salotra P, Sreenivas G, Pogue GP, Lee N, Nakhasi HL, Ramesh V, Negi NS. Development of a species-specific PCR assay for detection of Leishmania donovani in clinical samples from patients with kala-azar and post-kala-azar dermal leishmaniasis. J Clin Microbiol. 2001;39:849–854. doi: 10.1128/JCM.39.3.849-854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caipo ML, Duffy S, Zhao L, Schaffner DW. Bacillus megaterium spore germination is influenced by inoculum size. J Appl Microbiol. 2002;92:879–884. doi: 10.1046/j.1365-2672.2002.01597.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Cold Spring Harbor: Cold Spring Harbor Laboratory; Molecular CloningA Laboratory Manual. 1989 [Google Scholar]
- Joshi M, Dwyer DM, Nakhasi HL. Cloning and characterization of differentially expressed genes from in vitro-grown ‘amastigotes’ of Leishmania donovani. Mol Biochem Parasitol. 1993;58:345–354. doi: 10.1016/0166-6851(93)90057-5. [DOI] [PubMed] [Google Scholar]
- Wang Z, Morris JC, Drew ME, Englund PT. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- Debrabant A, Joshi MB, Pimenta PF, Dwyer DM. Generation of Leishmania donovani axenic amastigotes: their growth and biological characteristics. Int J Parasitol. 2004;34:205–217. doi: 10.1016/j.ijpara.2003.10.011. [DOI] [PubMed] [Google Scholar]
- van Baren MJ, Heutink P. The PCR suite. Bioinformatics. 2004;20:591–593. doi: 10.1093/bioinformatics/btg473. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov VA, Robinson TM, Riemenschneider J, Golding B, Kennedy M, Shiloach J, Alibek K. Treatment of anthrax infection with combination of ciprofloxacin and antibodies to protective antigen of Bacillus anthracis. FEMS Immunol Med Microbiol. 2004;40:71–74. doi: 10.1016/S0928-8244(03)00302-X. [DOI] [PubMed] [Google Scholar]
- Bell AS, Ranford-Cartwright LC. Real-time quantitative PCR in parasitology. Trends Parasitol. 2002;18:337–342. [PubMed] [Google Scholar]
- Sen K, Asher DM. Multiplex PCR for detection of Enterobacteriaceae in blood. Transfusion. 2001;41:1356–1364. doi: 10.1046/j.1537-2995.2001.41111356.x. [DOI] [PubMed] [Google Scholar]
- Lachaud L, Dereure J, Chabbert E, Reynes J, Mauboussin JM, Oziol E, Dedet JP, Bastien P. Optimized PCR using patient blood samples for diagnosis and follow-up of visceral Leishmaniasis, with special reference to AIDS patients. J Clin Microbiol. 2000;38:236–240. doi: 10.1128/jcm.38.1.236-240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanakos G, Patsoula E, Kremastinou T, Saroglou G, Vakalis N. Development of a PCR-based method for diagnosis of Leishmania in blood samples. Mol Cell Probes. 2002;16:415–420. doi: 10.1006/mcpr.2002.0436. [DOI] [PubMed] [Google Scholar]
- Uhl JR, Bell CA, Sloan LM, Espy MJ, Smith TF, Rosenblatt JE, Cockerill FR., III Application of rapid-cycle real-time polymerase chain reaction for the detection of microbial pathogens: the Mayo-Roche rapid anthrax test. Mayo Clin Proc. 2002;77:673–680. doi: 10.4065/77.7.673. [DOI] [PubMed] [Google Scholar]
- Schulz A, Mellenthin K, Schonian G, Fleischer B, Drosten C. Detection, differentiation, and quantitation of pathogenic leishmania organisms by a fluorescence resonance energy transfer-based real-time PCR assay. J Clin Microbiol. 2003;41:1529–1535. doi: 10.1128/JCM.41.4.1529-1535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossolasco S, Gaiera G, Olchini D, Gulletta M, Martello L, Bestetti A, Bossi L, Germagnoli L, Lazzarin A, Uberti-Foppa C, Cinque P. Real-time PCR assay for clinical management of human immunodeficiency virus-infected patients with visceral leishmaniasis. J Clin Microbiol. 2003;41:5080–5084. doi: 10.1128/JCM.41.11.5080-5084.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry KS, Tuteja U, Santhosh PK, Lalitha MK, Batra HV. Identification of Bacillus anthracis by a simple protective antigen-specific mAb dot-ELISA. J Med Microbiol. 2003;52:47–49. doi: 10.1099/jmm.0.05027-0. [DOI] [PubMed] [Google Scholar]
- Adhya S, Chatterjee M, Hassan MQ, Mukherjee S, Sen S. Detection of Leishmania in the blood of early kala-azar patients with the aid of the polymerase chain reaction. Trans R Soc Trop Med Hyg. 1995;89:622–624. doi: 10.1016/0035-9203(95)90416-6. [DOI] [PubMed] [Google Scholar]
- Andresen K, Gasim S, Elhassan AM, Khalil EA, Barker DC, Theander TG, Kharazmi A. Diagnosis of visceral leishmaniasis by the polymerase chain reaction using blood, bone marrow and lymph node samples from patients from the Sudan. Trop Med Int Health. 1997;2:440–444. [PubMed] [Google Scholar]
- Katakura K, Kawazu S, Naya T, Nagakura K, Ito M, Aikawa M, Qu JQ, Guan LR, Zuo XP, Chai JJ, Chang KP, Matsumoto Y. Diagnosis of kala-azar by nested PCR based on amplification of the Leishmania mini-exon gene. J Clin Microbiol. 1998;36:2173–2177. doi: 10.1128/jcm.36.8.2173-2177.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzum E, White F, III, Thakur C, Dietze R, Wages J, Grogl M, Berman J. Diagnosis of symptomatic visceral leishmaniasis by use of the polymerase chain reaction on patient blood. J Infect Dis. 1995;171:751–754. doi: 10.1093/infdis/171.3.751. [DOI] [PubMed] [Google Scholar]
- Osman OF, Oskam L, Zijlstra EE, Kroon NC, Schoone GJ, Khalil ET, El-Hassan AM, Kager PA. Evaluation of PCR for diagnosis of visceral leishmaniasis. J Clin Microbiol. 1997;35:2454–2457. doi: 10.1128/jcm.35.10.2454-2457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Curran MD, Rastogil AK, Middleton D, Sundar S. Diagnostic PCR with Leishmania donovani specificity using sequences from the variable region of kinetoplast minicircle DNA. Trop Med Int Health. 1999;4:448–453. doi: 10.1046/j.1365-3156.1999.00416.x. [DOI] [PubMed] [Google Scholar]
- Smyth AJ, Ghosh A, Hassan MQ, Basu D, De Bruijn MH, Adhya S, Mallik KK, Barker DC. Rapid and sensitive detection of Leishmania kinetoplast DNA from spleen and blood samples of kala-azar patients. Parasitology. 1992;105:183–192. doi: 10.1017/s0031182000074096. [DOI] [PubMed] [Google Scholar]