Abstract
Although microsatellite instability (MSI) testing is a useful tool for molecular screening of hereditary nonpolyposis colorectal cancer (HNPCC) carcinomas, conflicting results have been obtained in colorectal adenomas. This might result from different techniques of tissue sampling and MSI analysis. Alternatively, some HNPCC-associated adenomas may follow a molecular route that differs from the MSI pathway. In the present study we examined the MSI status of 18 adenomas from 17 HNPCC patients by comparing manual adenoma dissection under gross visual control with laser microdissection of single adenoma crypts. After manual gross dissection, 50% (9 of 18) and 11.1% (2 of 18) of the adenomas displayed high-level (MSI-H) and low-level (MSI-L) MSI, respectively. The same set of adenomas split into 83.3% (15 of 18) MSI-H and 5.6% (1 of 18) MSI-L after laser microdissection. The expression pattern of mismatch repair (MMR) proteins showed a higher concordance rate with the MSI status in laser-dissected (94%) than gross-dissected (47%) adenomas. Whereas two adenomas remained microsatellite stable (MSS) and MMR proficient even after laser-assisted dissection, two MSI-H cases showed either rare instabilities at coding microsatellites or intratumoral heterogeneity of MSI with and without MSH2 expression. This suggests that in some adenomas development of MMR dysfunction occurs stepwise with MSI, arising before complete loss of MMR gene expression, whereas other HNPCC-associated adenomas might develop independently of MMR deficiency.
Hereditary nonpolyposis colorectal cancer (HNPCC) is an autosomal dominant syndrome representing the most common form of hereditary colorectal cancer.1,2 Approximately 5 to 8% of all colorectal cancers have been classified as HNPCC1 by using the criteria proposed by the International Collaborative Group on HNPCC.2 Most cases of HNPCC are caused by germline mutations in one allele of mismatch repair (MMR) genes MSH2 or MLH1, or less frequently in the genes MSH6 and PMS2.3,4,5,6,7,8,9,10,11 For PMS1 or MLH3 a pathogenic germline mutations in a HNPCC-family is not reported yet, however as a partner of MLH1 in case of PMS1 or functional (MLH3) it might also be important.
Mutations in these genes impair the function of MMR proteins, their role being to recognize and repair mismatches and insertion/deletion loops caused by slippage of DNA polymerase.12,13 MMR deficiency leads to microsatellite instability (MSI), characterized by widespread insertions and deletions of one or few bases within short tandem repeats in tumor DNA, compared to matching normal DNA.14,15,16 It has been shown that more than 90% of colorectal cancers in HNPCC patients are MSI, whereas this is only the case in 15% of sporadic colorectal cancers.17 Microsatellite analysis has therefore been proposed as a useful diagnostic tool in screening for HNPCC syndrome.18,19,20 Moreover, it is possible to identify tumors with a defective MMR system by immunohistochemistry because inactivation of MMR genes may cause loss of expression of the corresponding protein.21,22,23,24,25,26
In sporadic colorectal adenomas a very low frequency of MSI, ranging from 0 to 3%, has been found,27,28,29 whereas a higher but variable MSI frequency ranging from 10 to 90% has been documented in HNPCC-associated adenomas.27,30,31,32,33,34,35 It can be hypothesized that the variable MSI detection rate observed in HNPCC adenomas might be related to the tissue sampling procedure.27,28,30,31,32,33,34 The significance of intralesional molecular heterogeneity in relation to the degree of dysplasia as a potential source of false-negative MSI tests has not yet been studied specifically. On the other hand, although MSI has been suggested as an early event in HNPCC-associated carcinogenesis, MMR deficiency might not be the only cause for the development of early adenomas in HNPCC patients.27,33 The aim of this study was to investigate early steps of tumorigenesis in a series of HNPCC-related adenomas using laser-assisted microdissection and to compare the results with the immunohistochemical profile.
Materials and Methods
Patients and Tumors
Our retrospective study is based on 17 HNPCC patients (nine males, eight females; age range, 24 to 78 years; mean age, 45.9 years) forming part of a prospective multicentric study throughout Germany supported by the Deutsche Krebshilfe (German Cancer Aid). Family history of these patients fulfilled the Amsterdam I or II criteria. These patients were selected because they had developed synchronous adenomas in 15 cases and metachronous adenomas in 2 cases, one of the latter showing two synchronous adenomas next to each other (Table 1, case 17). One patient (case 3) suffered from endometrial cancer and developed a metachronous adenoma and was the mother of case 13. For each patient, paraffin blocks of carcinoma, adenoma, and normal colonic mucosa were available from the archives of the Institute of Pathology, Klinikum Kassel, Kassel, Germany, where the tumor tissue was studied on referral basis within the German HNPCC-consortium that currently serves for more than 2000 registered families meeting the Bethesda criteria (for details see: http//www.krebshilfe.de). Eight patients with a sporadic colorectal carcinoma and synchronous or metachronous adenomas served as a control group.
Table 1.
Patient Characteristics
| Patients
|
Carcinoma and Adenoma
|
MS-status manually dissected | MMR proteins
|
Laser dissected lesion | Microsatellite loci
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | F/M | YS | Type* | Location** | CA-stage AD-size | CA-grade AD-dyspl. | MSH2 | MLH1 | MSH6 | BAT40 | BAT25 | BAT26 | APC | D2S123 | Mfd15 | ||
| 1 | F | 30 | CA | T | T3N1MX | 2 | MSI-H | + | − | + | nd | + | + | − | + | − | |
| tAD | 2 cm | 0.5 cm | D3 | MSI-H | + | − | + | nd | + | + | + | + | − | ||||
| MSI-H | D1 | + | + | + | + | + | − | ||||||||||
| MSI-H | D2 | + | + | + | − | − | + | ||||||||||
| MSI-H | D3 | + | + | + | + | + | − | ||||||||||
| 2 | F | 49 | CA | T | T3N0M0 | 2 | MSI-H | − | + | (+) | nd | + | + | + | + | − | |
| tvAD | 5 cm | 1.5 cm | D3 | MSI-H | − | + | (+) | nd | + | + | + | − | + | ||||
| MSI-H | D1 | + | + | + | − | − | + | ||||||||||
| MSI-H | D2 | + | + | + | + | − | + | ||||||||||
| MSI-H | D3 | + | + | + | + | − | + | ||||||||||
| 3 | F | 56 | CA | END | T1N0M0 | 2 | MSI-H | − | + | (+) | nd | − | + | + | − | + | |
| tADm | S | 1 cm | D3 | MSI-H | − | + | (+) | nd | + | + | + | + | + | ||||
| MSI-H | D1 | + | + | + | + | + | + | ||||||||||
| MSI-H | D2 | + | + | + | + | + | + | ||||||||||
| MSI-H | D3 | + | + | + | + | + | + | ||||||||||
| 4 | M | 46 | CA | T | T3N0M0 | 2 | MSI-H | + | − | + | nd | + | + | + | + | + | |
| tvAD | 2 cm | 1.5 cm | D3 | MSI-H | + | − | + | nd | + | + | + | nd | + | ||||
| MSI-H | D2 | + | + | + | + | + | + | ||||||||||
| MSI-H | D3 | + | + | + | + | + | + | ||||||||||
| 5 | F | 78 | CA | S | T3N1MX | 3 | MSI-H | − | + | − | nd | + | + | + | + | + | |
| tAD | 2 cm | 0.8 cm | D3 | MSI-H | − | + | − | nd | + | + | + | + | + | ||||
| MSI-H | D2 | + | + | + | + | + | + | ||||||||||
| MSI-H | D3 | + | + | + | + | + | + | ||||||||||
| 6 | F | 63 | CA | T | T2N0MX | 2 | MSI-H | + | − | + | nd | + | + | nd | + | + | |
| tAD | adj. | 0.8 cm | D3 | MSI-H | + | − | + | nd | + | + | − | + | + | ||||
| MSI-H | D2 | − | + | + | + | + | + | ||||||||||
| MSI-H | D3 | − | + | + | + | − | + | ||||||||||
| 7 | M | 43 | CA | A | T3N2MX | 3 | MSI-H | − | + | (+) | nd | + | + | + | nd | + | |
| tAD | 4 cm | 0.5 cm | D1 | MSI-H | − | + | (+) | nd | + | + | + | − | + | ||||
| MSI-H | D1 | − | + | + | + | − | + | ||||||||||
| 8 | M | 40 | CA in | R | T1N1M0 | 2 | MSI-H | − | + | − | pT1 | + | + | + | + | + | + |
| tvAD | 2.5 cm | D2 | MSI-H | (+) | + | (+) | nd | + | + | − | + | + | |||||
| MSI-H | + | D1 | − | + | + | − | + | − | |||||||||
| MSI-H | + | D2 | + | + | + | − | + | − | |||||||||
| MSI-H | − | D1 | − | + | + | + | + | + | |||||||||
| MSI-H | − | D2 | − | + | + | + | + | + | |||||||||
| 9 | F | 39 | CA | T | T4N2MX | 2 | MSI-H | + | − | + | nd | + | + | − | + | + | |
| tAD | 6 cm | 1.5 cm | D2 | MSI-L | + | − | + | nd | − | + | − | − | − | ||||
| MSI-H | D1 | − | − | + | + | + | − | ||||||||||
| MSI-H | D2 | − | + | + | + | − | + | ||||||||||
| 10 | F | 35 | CA | R | T2N0M0 | 2 | MSI-H | + | − | + | nd | + | + | + | + | + | |
| tAD | 1 cm | 0.4 cm | D1 | MSI-L | + | − | + | nd | − | − | − | − | + | ||||
| MSI-H | D1 | + | − | + | + | − | − | ||||||||||
| MSI-H | D2 | + | − | + | + | − | + | ||||||||||
| 11 | M | 44 | CA | A | T2N0MX | 2 | MSI-H | + | − | + | nd | + | + | + | + | + | |
| tvAD | 6 cm | 1.5 cm | D2 | MSS | + | − | + | nd | − | − | − | − | − | ||||
| MSI-H | D1 | + | + | − | − | − | + | ||||||||||
| MSI-H | D2 | + | − | − | + | − | − | ||||||||||
| 12 | M | 50 | CA | S | T3N0M0 | 2 | MSI-H | − | + | − | nd | + | + | − | + | nd | |
| tAD | 8 cm | 0.5 cm | D2 | MSS | − | + | − | nd | − | − | − | − | − | ||||
| MSI-L | D1 | + | − | − | − | − | − | ||||||||||
| MSI-H | D2 | + | − | − | − | − | + | ||||||||||
| 13 | M | 24 | CA in | R | T1N1M0 | 2–3 | MSI-H | − | + | (+) | pT1 | + | + | + | + | − | + |
| fAD | 3.5 cm | D3 | MSS | − | + | (+) | − | (+) | − | − | − | − | |||||
| MSI-L | D1 | − | − | + | − | − | − | ||||||||||
| MSI-L | D2 | − | + | − | + | − | − | ||||||||||
| MSI-H | D3 | + | + | − | + | − | − | ||||||||||
| 14 | F | 44 | CA in | T | T1N0M0 | 3 | MSI-H | + | − | + | pT1 | + | + | + | + | + | + |
| fAD | 4 cm | D3 | MSS | + | − | + | nd | − | − | − | − | − | |||||
| MSI-H | D3 | + | + | + | + | + | + | ||||||||||
| 15 | M | 41 | CA in | R | T1N0MX | 2 | MSI-H | + | + | + | pT1 | + | − | − | + | − | + |
| fAD | 1.0 cm | D3 | MSS | + | + | + | nd | − | − | − | − | − | |||||
| MSS | D1 | − | − | − | − | − | − | ||||||||||
| MSS | D2 | − | − | − | − | − | − | ||||||||||
| MSI-L | D3 | − | − | − | − | − | + | ||||||||||
| 16 | M | 55 | CA | A | T3N0M0 | 2 | MSI-H | − | + | − | nd | + | + | + | + | + | |
| tAD | 3 cm | 0.7 cm | D1 | MSS | + | + | + | nd | − | − | − | − | − | ||||
| MSS | D1 | − | − | − | − | − | − | ||||||||||
| 17 | M | 42 | CA | A | T3N0M0 | 3 | MSI-H | + | − | + | nd | + | + | + | + | + | |
| tvADm | R | 2 cm | D3 | MSI-H | + | − | + | nd | + | + | + | + | + | ||||
| MSI-H | + | − | + | nd | + | + | + | + | + | ||||||||
| tADm | R | 0.8 cm | D2 | MSS | + | + | + | nd | − | − | − | − | − | ||||
| MSS | D1 | − | − | − | − | − | − | ||||||||||
| MSS | D2 | − | − | − | − | − | − | ||||||||||
The patients identification number (No.), sex (F/M), age at diagnosis of the cancer (YS) are listed in the first column (patients). In the second column (carcinoma and adenoma) data of tumor type (AC, adenocarcinoma; AD, adenoma), localization of the tumor (A; ascending colon; T; transverse colon; S; sigmoid colon; R; rectum; END; endometrium; adj.; adjacent to carcinoma) tumor stage, and grade are presented. In the second row the data are given for the syn- or metachronous adenomas including adenoma type (t; tubulous; tv; tubulovillous; f; flat), the distance to a synchronous cancer (if applicable [cm]) and size of the lesion (maximum diameter, [cm]) as well as the grading (D1; mild dysplasia; D2; moderate dysplasia; D3; severe dysplasia). In the third column (MS status) results of the MSI analysis are listed with MS status after manual dissection being underlined. In the fourth column (MMR proteins) staining results of MSH2, MLH1, and MSH6 are shown. In the fifth column (laser) the different grades of dysplasia which were collected by laser microdissection are presented. The last column (microsatellite loci) lists data of the MSI analysis for each single locus.
Tumor Histology
For each adenoma and carcinoma, two representative cross sections were subjected to hematoxylin and eosin (H&E) and periodic acid-Schiff staining to evaluate the histology. Typing and grading of carcinomas as well as typing of adenomas followed the recommendations of the World Health Organization classification of tumors.36 Accordingly, adenomas were grouped into elevated and flat with either a tubular, villous, or tubulovillous growth pattern. Two cases of the flat adenomas have been reported previously (Table 1, cases 13 and 14).37,38 Various degrees of dysplasia in different areas of the same adenoma were identified and graded as mild (D1), moderate (D2), or severe (D3), with D1 and D2 corresponding to low-grade and D3 to high-grade intraepithelial neoplasia. Finally, each adenoma was classified in relation to the most advanced focus of dysplasia. The assessment of dysplasia was performed independently by two pathologists with good correlation and reproducibility. Gross tumor parameters (site, size and stage) were recorded according to the UICC.39 In four cases, adenocarcinoma developed from a tiny early focus either from flat adenoma (3×) or from an elevated tubulovillous adenoma (so-called carcinoma in adenoma). All tumor blocks and blood samples were studied after obtaining the patients informed consent under an approved protocol by the ethics committee of each participating center.
Microdissection and DNA Extraction
Manual Dissection
Paraffin-embedded carcinoma, adenoma and corresponding normal tissue sections (5 μm thick) were mounted on glass slides and stained with H&E. Cross sections were prepared for each tumoral lesion and additionally stained with periodic acid-Schiff reagent. Areas of interest of each adenoma and carcinoma in every individual patient were marked using a microscope on the H&E-stained slide by a pathologist. These marked areas, which sometimes comprised the whole adenoma, were scraped off additional unstained cross sections. The tissue underwent proteinase K digestion at a final concentration of 2 μg/ml (Qiagen, Valencia, CA). DNA was extracted with the QIAamp DNA mini kit (Qiagen) according to the manufacturer’s recommendations and purified with the PCR Template Preparation kit (Roche, Mannheim, Germany).
Laser Microdissection
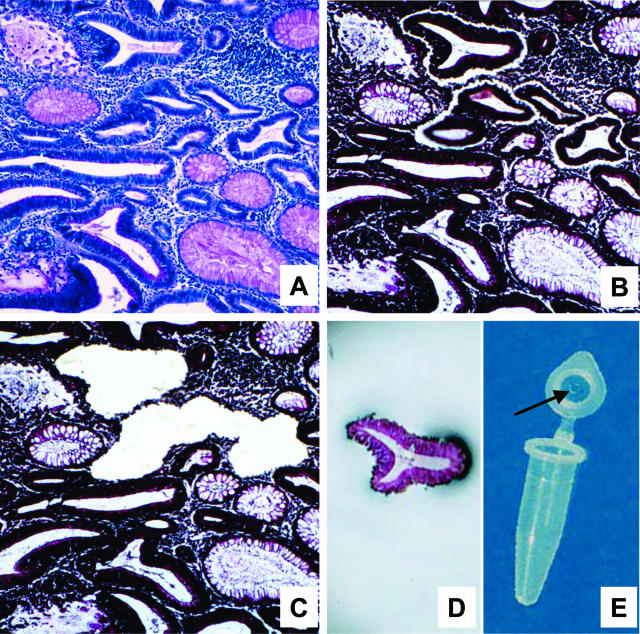
Additional sections of the same paraffin blocks containing normal and adenomatous tissue were prepared for laser microdissection. Briefly, the sections were mounted onto a polyethylene membrane, previously attached to a glass slide with rubber cement (Fixogum; Marabuwerke, Tamm, Germany). Slides were deparaffinized in xylene, rehydrated in descending alcohols, and H&E stained. Laser microdissection was performed using a Leica AS LMD system (Leica Microsystems GmbH, Wetzlar, Germany). Approximately 150 to 500 epithelial cells (mean, 375 epithelial cells) were harvested for each analysis, the cells being harvested from areas within the adenoma showing different degrees of dysplasia (D1 to D3) as well as from small foci of carcinoma in adenoma. Usually two independent samples were taken for each degree of dysplasia (Figure 1). Cell lysis was performed by dispensing 20 μl of lysis buffer into each cap. The buffer consisted of 10% proteinase K (2 mg/ml; Roche), 0.5% Tween 20 (Merck, Darmstadt, Germany), and 1× TaqPCR Buffer (Roche). After digestion for 18 hours at 50°C, proteinase K was inactivated by incubation at 94°C for 10 minutes.
Figure 1.
Laser microdissection. A: A periodic acid-Schiff-stained section used to identify the degree of dysplasia. B, C, E: The laser cut line surrounds the dysplastic cells (B); these cells were harvested in the cap of a microtube (C, E). D: The tissue section after laser microdissection: DNA contamination by stromal and inflammatory cells surrounding the desired cells was avoided.
Analysis of MSI
In accordance to the recommendations by the National Cancer Institute and the ICG-HNPCC15,40 five microsatellite loci were used in all tumor lesions both for manual and laser-based dissection to detect MSI: two loci with mononucleotide runs (BAT25, BAT26) and three loci with CA dinucleotide repeats (D2S123, D5S346, D17S250). In addition, samples obtained by laser microdissection were also tested for instability at BAT40. The primer sequences of these microsatellite loci have been reported elsewhere.41 Eight adenomas of seven patients (Table 1, cases 3, 6, 12 to 14, 16, and 17) were investigated by an extended primer panel demonstrating instabilities at mononucleotide repeats within the coding regions of 26 different genes that are essential for tumorigenesis, such as transforming growth factor-β receptor type II, BAX, CASP5, and others (for details see Woerner et al42,43).
Polymerase chain reaction (PCR) amplifications were performed in an Eppendorf Mastercycler Gradient (Eppendorf, Hamburg, Germany). Primers were 5′-labeled with HEX, FAM, or TET (PE Applied Biosystems, Foster City, CA). DNA amplification was performed in a standard reaction mix reported elsewhere with cycling conditions using Ampli Taq Gold (PE Applied Biosystems).4,41 The PCR products were run on an ABI Prism 310 genetic analyzer (PE Applied Biosystems) according to the manufacturer’s instructions and microsatellite status was analyzed using GeneScan fragment analysis software (GeneScan, PE Applied Biosystems). Only cases showing unequivocally distinct additional peaks or shifts in tumor tissue DNA in comparison to normal tissue DNA were recorded and classified as MSI.
The microsatellite status of each sample was determined based on the percentage of unstable loci. The status was defined as MSI-high (MSI-H) when more than 30% of markers displayed instability and as MSI-low (MSI-L) with less than 30% of markers exhibiting instability.15 In cases in which only dinucleotides of the original five National Institutes of Health marker panel were mutated shifts at BAT40 excluded MSI-L according to updated Bethesda (II) recommendations.40 A sample was classified as microsatellite stable (MSS) when no MSI was found.
Immunohistochemistry
Loss of MMR protein expression (MSH2, MLH1, MSH6) was determined by immunohistochemical analysis both in carcinomas and adenomas. Briefly, sections were dewaxed in xylene and rehydrated in graded ethanols. After washing in distilled water, the sections were placed in citrate buffer (10 mmol/L, pH 6.0) in a microwave oven (800 W, 20 minutes) to allow antigen retrieval. Endogenous peroxidase was blocked with 3% H2O2 for 5 minutes, while nonspecific binding sites were blocked by incubating sections with goat serum for 30 minutes. The slides were incubated overnight at 4°C with primary antibody against MSH2 (clone FE11, dilution 1:100; Oncogene Sciences, Cambridge, MA), MLH1 (clone G168-728, dilution 1:200; PharMingen, San Diego, CA), or MSH6 (clone 44, dilution 1:600; Becton Dickinson Transduction Laboratories, San Diego, CA) proteins. Subsequently, the sections were washed with phosphate-buffered saline (PBS) and incubated with a secondary biotinylated antibody (Vector Laboratories, Burlingame, CA). After rinsing with PBS, the sections were incubated with streptavidin-conjugated horseradish peroxidase (Vector Laboratories). For detection, the chromogen 3-amino-9-ethylcarbazole (Sigma, St. Louis, MO) was used according to the manufacturer’s recommendations and finally the slides were lightly counterstained with hematoxylin. Only slides with distinct positive nuclear staining in the basal crypt cells of normal mucosa, stromal as well as inflammatory cells were evaluated. Loss of MMR protein expression within the neoplastic lesions was defined as complete when less than 10% of tumor cells were stained. Incomplete loss (10 to 50% stained cells) or only focal staining (cohesive nests of stained tumor cells) was recorded as (+) (Table 1).
Statistical Analysis
Statistical analysis was performed using χ2 and Fisher’s exact test. A value less than P = 0.05 was considered as statistically significant.
Results
MSI and MMR Expression in Carcinomas
All cancer tissues from HNPCC-patients were classified as MSI-H and all but one exhibited loss of MMR protein expression. In particular, eight (47.1%) carcinomas showed complete loss of MLH1 expression and normal immunoreactivity for MSH2 and MSH6, whereas eight (47.1%) exhibited complete loss of MSH2 expression; in addition, four (23.5%) cases of this latter group showed also a complete loss of MSH6 expression. The only MMR protein-positive carcinoma showed MSI exclusively at dinucleotide loci with stable mononucleotides when the original five National Cancer Institute marker panel was used (Table 1, case 15).
MSI after Manual and Laser-Based Dissection in Adenomas
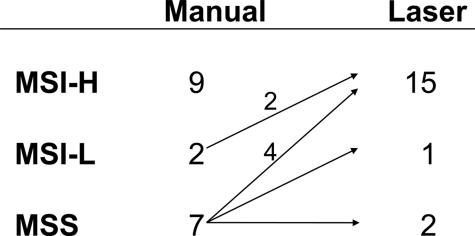
After manual dissection, MSI analysis of representative cross sections of the 18 adenomas from the 17 HNPCC patients demonstrated the presence of nine (50%) MSI-H, two (11.1%) MSI-L, and seven (38.9%) MSS tumors. However, after laser microdissection 15 of 18 adenomas (83.3%) were MSI-H, 1 of 18 (5.6%) was MSI-L, and 2 of 18 (11.1%) remained MSS. Six adenomas were reclassified as MSI-H, two of which had been classified as MSI-L and four of which had been categorized as MSS by manual dissection (Figure 2). To rule out the possibility that instability may have resulted from analysis of too few cells for laser microdissection showing a mobility shift, all eight adenomas from the control group were analyzed with the same amount of cells and none of them displayed MSI (data not shown). Interestingly, testing of the two MSS adenomas by an extended primer panel did not reveal any instabilities of coding mononucleotide repeats within 26 target genes. Six MSI-H adenomas, however, showed instabilities at more than 40% of the tested coding repeats (data not shown).
Figure 2.
Microsatellite status after manual and laser-guided dissection indicating the cases with a changed MSI level.
MSI, Location and Type of Adenoma
Thirteen adenomas occurred separately from the carcinoma, only six of these were classified as MSI-H after manual dissection, while two remained MSS even after laser microdissection. The remaining five adenomas were located in close vicinity to the carcinoma. Four of these were large adenomas with only small foci of (micro-)invasive adenocarcinomas (pT1); in three of these tumors MSI status could correctly be classified only after laser microdissection (Table 1, cases 13 to 15).
There was a significant increase in the number of affected MS loci with increasing degree of dysplasia. Instability affecting at least 83% (5 of 6) of microsatellite markers occurred in most of the microdissected high-grade intraepithelial neoplasias (70%, 7 of 10) but was significantly less common in low-grade intraepithelial neoplasias with medium grade (40%, 6 of 15) and low-grade dysplasia (14.3%, 2 of 14; P = 0.0375). Gain of instability with tumor progression could also be shown in the carcinoma-in-adenoma cases (Table 1, cases 8, and 13 to 15). The invasive part of these neoplasias showed the highest number of unstable microsatellite loci (≥5 markers) in 3 of 4 (microinvasive) carcinomas but only in 3 of 11 microdissected dysplastic adenomatous lesions (75% versus 27%, P = 0.092). No correlation was found between MSI status and growth pattern of adenomas.
MMR Protein Immunoexpression in Adenomas
Loss of MMR protein expression was detected in 14 of 18 (77.7%) adenomas. In particular, 8 of 18 (44.4%) cases showed complete loss of MLH1 expression and normal positive nuclear staining for MSH2 and MSH6, while 6 of 18 (33.3%) cases exhibited a complete loss of MSH2 expression; 2 of 6 adenomas of this latter group showed a complete loss of MSH6 expression in addition to the loss of MSH2. The remaining four adenomas stained positive for all three MMR proteins. In the control group all adenomas revealed a positive nuclear staining for MLH1, MSH2 and MSH6.
Relationship between MMR Protein Expression and Microsatellite Status
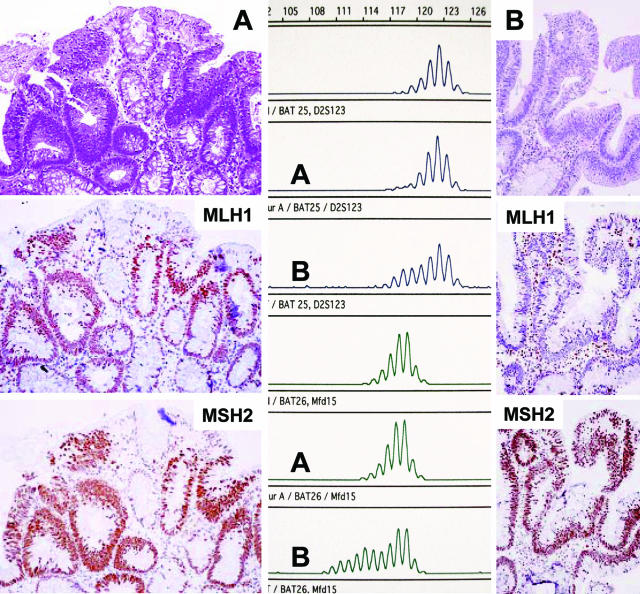
MMR protein expression pattern showed the highest concordance rate with the MSI status in laser-dissected (94%) rather than gross-dissected (47%) adenomas. After laser microdissection of the 15 MSI-H adenomas 14 displayed loss of MMR protein expression (ie, 93.3% concordance rate). One of the MSI-H adenomas showed intratumoral heterogeneity with loss of MSH2 staining in approximately two-thirds of the adenoma but retained expression in the remaining part. In this case (Table 1, case 8) we analyzed these areas of different MMR expression for MSI by laser microdissection and we were able to demonstrate a correlation between the loss of protein expression and the extent of the mobility shift (Figure 3).
Figure 3.
Carcinoma arising in an adenoma in a case with partial loss of MSH2 in the adenoma. A: Focus of MSH2 expression in the adenoma (->, retained MSH2 immunostaining; AD, adenoma; CA, carcinoma; N, normal mucosa; * , lymphoid follicle as internal positive staining control). B: Enlarged inset from A with focal MSH2 staining and C shows the expression of MLH1 within the same area. D and E: MSI at BAT40 (D) and BAT25 (E) of MSH2-negative cells (D: lane 1, normal; lanes 2 and 3, adenomatous tissue without MSH2; lanes 4 and 5, with preserved MSH2 expression; E: lane 1, normal; lane 2, adenomatous tissue with MSH2 staining; and lanes 3 and 4, without MSH2 staining).
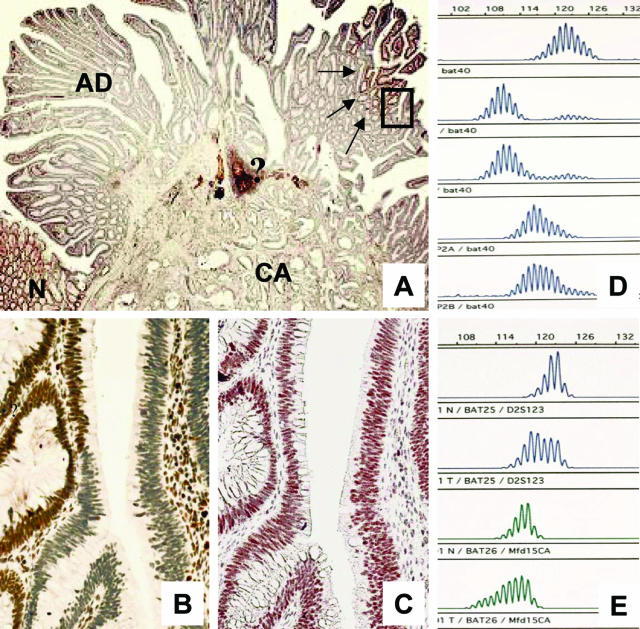
Of the four cases with preserved MMR gene expression, laser microdissection disclosed already BAT40 mutations (Table 1, case 12). This adenoma was the smallest in this series (∼3 mm in diameter) and mobility shifts at coding MS loci were restricted to 1 of 22 evaluable target genes. Two other adenomas that stained positive for all three MMR proteins were MSS even though the corresponding cancers showed MSI-H and loss of one MMR protein. Interestingly, in one of these two cases (Table 1, case 17) the patient suffered from two synchronous adenomas that occurred 2 years after cancer resection in the rectal stump, one of which was MSI-H and had lost MLH1 expression, whereas the other adenoma was MSS and expressed all MMR proteins (Figure 4). The fourth adenoma with regular MMR protein expression exhibited low-level MSI even after laser microdissection (Table 1, case 15). In this case the corresponding carcinoma showed only dinucleotide instabilities and no immunohistochemical loss of MMR proteins.
Figure 4.
A known HNPCC patient with synchronous adenomas. One adenoma showed nuclear staining for MLH1 and MSH2 (A) whereas the other revealed a loss of MLH1 (B). Corresponding MSI analysis disclosed instabilities at BAT25 (top set) and BAT 26 (bottom set) for the one (B) and no instabilities for the other (A) adenoma (lanes 1 and 4, normal control).
Discussion
In the present study we obtained different results for MSI analysis of adenomas of known HNPCC patients, depending on the method of microdissection used. These adenomas arose either synchronously or metachronously with carcinomas that had been shown to be MSI-H and had complete loss of MMR proteins in all but one case. Using manual dissection of marked areas, which were scraped off additional unstained cross sections, we found MSI in 11 of 18 adenomas (61%) (MSI-H, 9 of 18 cases; MSI-L, 2 of 18 cases), whereas 7 of 18 adenomas (38.9%) were classified as MSS. After laser microdissection, six adenomas had to be reclassified as MSI-H, two of which had been classified as MSI-L and four of which had been classified as MSS by manual dissection. Overall the results with laser microdissection showed MSI in 16 of 18 adenomas (88.9%), 15 of which were MSI-H and one of which was MSI-L. Only two cases remained in the MSS category. These findings clearly demonstrate an increased sensitivity of MSI detection by laser microdissection. This might explain why the rate of MSI in adenomas of patients with MMR deficiencies varies widely between 10% and 90%.27,28,33
One of the reasons for the higher sensitivity of MSI screening using laser-microdissected tissues is that adenomatous cells were harvested selectively, excluding stromal and inflammatory cells. Accordingly we found an increased number of unstable MSI loci with a degree of dysplasia, which is also confirmed by our observation that in early carcinomas within adenomas usually most MSI loci are affected. Another explanation might be the heterogeneity in the precursor lesion where some areas are more affected by MSI than others. This problem can be approached by microdissecting multiple areas of the lesion and then comparing the relative patterns. Therefore, we dissected at least two areas with equal grades of dysplasia in each adenoma for MSI analysis. MSI concordance rate between these two areas ranged from 45% in low-grade dysplastic lesions to more than 90% in high-grade dysplastic lesions, indicating that MSI may be missed, particularly in low-grade intraepithelial neoplasia (data not shown). To avoid the number of cells obtained using laser microdissection falling below a necessary minimum, which might cause other problems as shown previously,38 we amplified at least 150 cells. In our opinion the optimum number of template cells ranges between 200 to 300 cells.
Another issue is the possibility that different MSI results may be attributed to various methodologies applied in the literature for evaluating MSI. In fact, only few studies33,35 have used the guidelines for the evaluation of MSI recommended by the National Cancer Institute workshop, which we used in the present study.15,41 Additionally we recommend immunohistochemical analysis for the correct interpretation of the MSI data. In five of six cases in our study, which were classified as MSI-H only after laser microdissection, the result was confirmed by the immunohistological data, with loss of expression in one of the responsible MMR-proteins. In general we found that the shift of the microsatellite peaks is more marked in the areas that have lost expression of the MMR proteins but interestingly, the instability is present in areas of the adenoma that are still expressing the protein. The observation in one adenoma (case 12) that MSI can be an earlier event than immunohistochemically detectable loss of MMR expression together with technical limitations of immunohistochemistry, namely sensitivity to overfixation and other false-negatives, emphasizes that MSI studies should be done in conjunction with immunohistochemistry and cannot be replaced by immunohistochemistry alone. Taking these aspects into consideration, MSI analysis in adenomas could be regarded as being relatively specific in analyzing patients at risk for having HNPCC syndrome. This was also hypothesized by Loukola and colleagues28 in cases in which clinical or family history suggested a hereditary predisposition.
One adenoma revealed a regular protein expression of the tested MMR proteins and was classified as MSI-L. In this case the carcinoma was also MMR protein-positive (case 15) and instability was restricted to dinucleotide loci with stable mononucleotides. Because the family history fulfilled the Amsterdam II criteria and no germline mutation could be demonstrated in either of the three tested MMR genes, this patient most likely belongs to a non-MMR related form of HNPCC as proposed by the most recent Bethesda workshop.41
It is well accepted that in HNPCC patients the progression time to carcinoma is much reduced when compared with patients without genetic predisposition.35,44,45,46 One explanation may be the higher frequency of high-grade intraepithelial neoplasias.35,46 In our cohort there was also an increased number of flat adenomas in HNPCC patients. These lesions show a much shorter progression time to carcinoma than elevated or pedunculated forms.47,48 Whether the formation of flat adenomas is due to genetic or mechanical factors is, however, a matter of debate.
In our own experience MSI analysis is highly prone to give false-negative results, particularly in such flat adenomas. Using manual dissection, contamination of adenomatous tissue with adjacent normal mucosa and no specific selection of high-grade dysplasia or even the tiny early adenocarcinoma foci in the so-called carcinoma in adenoma neoplasias turned out to be the most important factors.37,38 Also the role of hyperplastic polyps is not resolved yet. Whereas Rijcken and colleagues49 describe the role of the MMR defect as unlikely for the development of hyperplastic polyps, Jass and colleagues50 recently discussed a role of these polyps for malignant transformation.57
As published by Sparks and colleagues52 MSI occurs very early and may represent the first genetic alteration in HNPCC-related tumors. This is in concordance with our data demonstrating MSI in adenomas of different degrees of dysplasia and even in D1 adenomas. Our data are also in accordance with the work of Iino and colleagues35 who found that adenomas displaying high-grade dysplasia revealed MSI-H more frequently than those with low-grade dysplasia.
The gain of instability with tumor progression could be shown in cases in which large adenomas showed only small microinvasive carcinomas (carcinoma in adenoma). Whereas in 3 of 4 (microinvasive) carcinomas at least 5 of 6 microsatellites were affected this was only the case in 3 of 11 preinvasive dysplastic lesions. Although not laser microdissected, 8 of 17 (47%) carcinomas displayed instabilities at all five tested microsatellite loci that could only be detected in 3 of 18 (17%) adenomas after manual dissection.
However, even after laser microdissection two adenomas appear not to follow the mutator pathway, showing MSS and expression of all MMR proteins, even though the carcinoma in those patients revealed a loss of MMR proteins (one MSH2, one MLH1). It is unclear whether these adenomas have a different pathway of molecular carcinogenesis or whether they represent very early stages of the lesion before the second MMR allele is deleted. This hypothesis is supported by the MSI-H adenoma with only partial loss of MSH2 expression. Low-level MSI may thereby form a transitional stage between MSS and MSI-H status as shown by Iino and colleagues.35 This is in accordance to our case (case 12) consistently showing BAT40 instabilities at different adenoma sites but only one dinucleotide mutation. Most importantly in this case just 1 of 22 evaluable coding MS loci showed a mobility shift demonstrating early intralesional heterogeneity with stepwise development of MSI. According to the original Bethesda guidelines this case would have been classified as MSI-L.15 However, as outlined by the revised Bethesda (II) recommendations mononucleotide instabilities (eg, at BAT40 or MYCL1) indicate a MMR-related MSI-H lesion particularly in those cases in which dinucleotide repeats of the original five-NCI primer panel are affected.40 In contrast, the one above-mentioned adenoma that turned out to be MSI-L even after laser microdissection is most likely not related to the known MMR related forms of HNPCC showing selective dinucleotide instabilities and no loss of MMR genes. Presumably this case represents a true MSI-L lesion that will most probably not progress to MSI-H and thus belongs to the third molecular pathway of non-MIN and non-CIN type.50,52,53
In summary, MSI analysis of tumors, especially adenomas, is challenging because of the lesional heterogeneity that can be reduced by laser microdissection. Immunohistochemical staining can be used to confirm the MSI data and to avoid false-negative results, which could lead to a failure to identify a patient with a MMR deficiency, thereby excluding him/her from the more intense screening and surveillance.
Appendix
The German HNPCC-Consortium consists of the following centers (in alphabetic order): clinical centers in Bochum (in addition to author: Frank Brasch, Jörg T. Epplen, Stefan Hahn, Erdmute Kunstmann, Christian Pox Jörg Willert), Bonn (in addition to authors: Constanze Pagenstecher, Waltraut Friedl, Holger Lauschke, Andreas Hirner, Christof Lamberti, Peter Propping, Tilman Sauerbruch), Düsseldorf (in addition to author: Gabriela Möslein), Dresden (in addition to authors: Daniela E. Aust, Friedrich Balck, Ruth Höhl, Friedmar R. Kreuz, Stefan Krüger, Steffen R. Pistorius, Jens Plaschke), Heidelberg (in addition to authors; Peter Kienle, Hanns-Peter Knaebel, Miriam Tariverdian, Uta Mazitschek, Friedrich Cremer, Monika Keller, Magnus von Knebel-Doeberitz), München-Regensburg (in addition to author: Manfred Gross, Reinhard Kopp, Peter Lohse, Michael Muders, Yvonne Müller-Koch, Holger Vogelsang), the center for reference pathology Kassel (in addition to author: Ernst Heinmöller), and the center for documentation and biometry in Leipzig (in addition to author: Jochen Forberg, Marlies Herold, Markus Löffler).
Footnotes
Supported by the German Cancer Aid (Deutsche Krebshilfe) (grant 70-240I-Rüto to J.R. and R.B.), the Deutsche Forschungsgemeinschaft (Bu 672/10-1, 10-2 to R.B.), the German Ministry for Education and Research (BMBF) (grant AN 020113007601 to J.R.), the Ministero dell’Istruzione, dell′Universita′ e della Ricerca (MIUR) (to G.T.), and the Consiglio Nazionale delle Ricerche (CNR) (Legge 449/97, Progetto Diagnostica Molecolare in Oncologia, contribution no. CU02.00343.ST97 to G.T.).
G.G. and A.M. share first authorship.
References
- Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J Med Genet. 1999;36:801–818. [PMC free article] [PubMed] [Google Scholar]
- Vasen HFA, Watson P, Mecklin J-P, Lynch HT, the ICG-HNPCC New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the international collaborative group on HNPCC. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- Akiyama Y, Sato H, Yamada T, Nagasaki H, Tsuchiya A, Abe R, Yuasa Y. Germline mutation of the hMSH6/GTBP in an atypical hereditary nonpolyposis colorectal cancer kindred. Cancer Res. 1997;57:3920–3923. [PubMed] [Google Scholar]
- Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A, Tannergard P, Bollag RJ, Godwin AR, Ward DC, Nordenskjold M, Fishel R, Kolodner R, Liskay RM. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonan P, Aaltonen LA, Nystrom L, Guan YY, Zhang J, Meltzer PS, Yu JW, Kao FT, Chen DJ, Cerosaletti KM, Fournier REK, Todd S, Lewis T, Leach RJ, Naylor SR, Green J, Jass J, Watson P, Lynch HT, Trent JM, de la Chapelle A, Kinzler KW, Vogelstein B. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- Miyaki M, Konishi M, Tanaka K, Kikuchi-Yanoshita R, Muraoka M, Yasuno M, Igari T, Koike M, Chiba M, Mori T. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet. 1997;17:271–272. doi: 10.1038/ng1197-271. [DOI] [PubMed] [Google Scholar]
- Nicoloaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, Rosen CA, Haseltine WA, Fleishmann RD, Fraser CM, Adams MD, Venter JC, Dunlop MG, Hamilton SR, Petersen GM, de la Chapelle A, Vogelstein B, Kinzler KW. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD, Venter JC, Hamilton SR, Peterson GM, Watson P, Lynch HT, Peltomäki P, Mecklin JP, de la Chapelle A, Kinzler KW, Vogelstein B. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- Peltomäki P, Vasen HFA, the International Collaborative Group on HNPCC Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. Gastroenterology. 1997;113:1146–1158. doi: 10.1053/gast.1997.v113.pm9322509. [DOI] [PubMed] [Google Scholar]
- Wu Y, Berends MJW, Mensink RGJ, Verlind E, Sijmons RH, van der Zee AGJ, Hollema H, Kleibeuker JH, Buys CHCM, Hofstra RMW. Germline hMLH3 mutations in patients with suspected HNPCC. Am J Hum Genet. 2000;67:17–25. [Google Scholar]
- Buermeyer AB, Deschênes SM, Baker SM, Liskay RM. Mammalian DNA mismatch repair. Annu Rev Genet. 1999;33:533–564. doi: 10.1146/annurev.genet.33.1.533. [DOI] [PubMed] [Google Scholar]
- Jiricny J, Nyström-Lahti M. Mismatch repair defects in cancer. Curr Opin Genet Dev. 2000;10:157–161. doi: 10.1016/s0959-437x(00)00066-6. [DOI] [PubMed] [Google Scholar]
- Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- Peltomäki P. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum Mol Genet. 2001;10:735–740. doi: 10.1093/hmg/10.7.735. [DOI] [PubMed] [Google Scholar]
- Rüschoff J, Bocker T, Schlegel J, Stamm G, Hofstaedter F. Microsatellite instability: new aspects in the carcinogenesis of colorectal carcinoma. Virchows Arch. 1995;426:215–222. doi: 10.1007/BF00191357. [DOI] [PubMed] [Google Scholar]
- Lamberti C, Kruse R, Ruelfs C, Caspari R, Wang Y, Jungck M, Mathiak M, Malayeri HRH, Friedl W, Sauerbruch T, Propping P. Microsatellite instability—a useful diagnostic tool to select patients at high risk for hereditary non-polyposis colorectal cancer: a study in different groups of patients with colorectal cancer. Gut. 1999;44:839–843. doi: 10.1136/gut.44.6.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukola A, Eklin K, Laiho P, Salovaara R, Kristo P, Järvinen H, Mecklin J-P, Launonen V, Aaltonen LA. Microsatellite marker analysis in screening for hereditary nonpolyposis colorectal cancer (HNPCC). Cancer Res. 2001;61:4545–4549. [PubMed] [Google Scholar]
- Thibodeau SN, French AJ, Roche PC, Cunningham JM, Tester DJ, Lindor NM, Moslein G, Baker SM, Liskay RM, Burgart LJ, Honchel R, Halling KC. Altered expression of hMSH2 and hMLH1 in tumors with microsatellite instability and genetic alterations in mismatch repair genes. Cancer Res. 1996;56:4836–4840. [PubMed] [Google Scholar]
- Jass JR. hMLH1 and hMSH2 immunostaining in colorectal cancer. Gut. 2000;47:315–316. doi: 10.1136/gut.47.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W, Burgart LJ, Krause-Paulus R, Thibodeau SN, Almeida M, Bocker Edmonston T, Boland CR, Sutter C, Jass JR, Lindblom A, Lubinski J, MacDermot K, Sanders DSA, Morreau H, Müller A, Oliani C, Orntoft T, Ponz De Leon M, Rosty C, Rodriguez-Bigas M, Rüschoff J, Ruszkiewicz A, Sabourin J, Salovaara R, Möslein G, the ICG-HNPCC The reliability of immunohistochemistry as a prescreening method for the diagnosis of hereditary nonpolyposis colorectal cancer (HNPCC)—results of an international collaborative study. Familial Cancer. 2001;1:87–92. doi: 10.1023/a:1013840907881. [DOI] [PubMed] [Google Scholar]
- Plaschke J, Krüger St, Pistorius St, Theissig F, Saeger HD, Schackert HK. Involvement of hMSH6 in the development of hereditary and sporadic colorectal cancer revealed by immunostaining is based on germline mutations, but rarely on somatic inactivation. Int J Cancer. 2002;97:643–648. doi: 10.1002/ijc.10097. [DOI] [PubMed] [Google Scholar]
- Lanza G, Gafà R, Maestri I, Santini A, Matteuzzi M, Cavazzini L. Immunohistochemical pattern of MLH1/MSH2 expression is related to clinical and pathological features in colorectal adenocarcinomas with microsatellite instability. Mod Pathol. 2002;15:741–749. doi: 10.1097/01.MP.0000018979.68686.B2. [DOI] [PubMed] [Google Scholar]
- Rigau V, Sebbagh N, Olschwang S, Paraf F, Mourra N, Parc Y, Flejou JF. Microsatellite instability in colorectal carcinoma. The comparison of immunohistochemistry and molecular biology suggests a role for hMSH6 immunostaining. Arch Pathol Lab Med. 2003;127:694–700. doi: 10.5858/2003-127-694-MIICC. [DOI] [PubMed] [Google Scholar]
- Aaltonen LA, Peltomäki P, Mecklin JP, Jarvinen H, Jass JR, Green JS, Lynch HT, Watson P, Tallqvist G, Juhola M. Replication errors in benign and malignant tumors from hereditary nonpolyposis colorectal patients. Cancer Res. 1994;54:1645–1648. [PubMed] [Google Scholar]
- Loukola A, Salovaara R, Kristo P, Moisio A-L, Kääriäinen H, Ahtola H, Eskelinen M, Härkönen N, Julkunen R, Kangas E, Ojala S, Tulikoura J, Valkamo E, Järvinen H, Mecklin J-P, de la Chapelle A, Aaltonen LA. Microsatellite instability in adenomas as a marker for hereditary nonpolyposis colorectal cancer. Am J Pathol. 1999;155:1849–1853. doi: 10.1016/S0002-9440(10)65503-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J, Leggett B, Gustafson C, Ward M, Searle J, Thomas L, Buttenshaw R, Chenevix-Trench G. Genomic instability occurs in colorectal carcinomas but not in adenomas. Hum Mutat. 1993;2:351–354. doi: 10.1002/humu.1380020505. [DOI] [PubMed] [Google Scholar]
- Jacoby RF, Marshall DJ, Kailas S, Schlack S, Harms B, Love R. Genetic instability associated with adenoma to carcinoma progression in hereditary nonpolyposis colon cancer. Gastroenterology. 1995;109:73–82. doi: 10.1016/0016-5085(95)90270-8. [DOI] [PubMed] [Google Scholar]
- Konishi M, Kikuchi-Yanoshita R, Tanaka K, Muraoka M, Onda A, Okumura Y, Kishi N, Iwama T, Mori T, Koike M, Ushio K, Chiba M, Nomizu S, Konishi F, Utsunomiya J, Miyaki M. Molecular nature of colon tumors in hereditary nonpolyposis colon cancer, familial polyposis, and sporadic colon cancer. Gastroenterology. 1996;111:307–317. doi: 10.1053/gast.1996.v111.pm8690195. [DOI] [PubMed] [Google Scholar]
- Akiyama Y, Iwanaga R, Saitoh K, Shiba K, Ushio K, Ikeda E, Iwama T, Nomizu T, Yuasa Y. Transforming growth factor β type II receptor gene mutations in adenomas from hereditary nonpolyposis colorectal cancer. Gastroenterology. 1997;112:33–39. doi: 10.1016/s0016-5085(97)70216-6. [DOI] [PubMed] [Google Scholar]
- Pedroni M, Sal E, Scarselli A, Borghi F, Menigatti M, Benatti P, Percesepe A, Rossi G, Foroni M, Losi L, DiGregorio C, De Pol A, Nascimbeni R, Di Betta E, Salerni B, Ponz de Leon M, Roncucci L. Microsatellite instability and mismatch-repair protein expression in hereditary and sporadic colorectal carcinogenesis. Cancer Res. 2001;61:896–899. [PubMed] [Google Scholar]
- Rashid A, Zahurak M, Goodman SN, Hamilton SR. Genetic epidemiology of mutated K-ras proto-oncogene, altered suppressor genes, and microsatellite instability in colorectal adenomas. Gut. 1999;44:826–833. doi: 10.1136/gut.44.6.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino H, Simms L, Young J, Arnold J, Winship IM, Webb SI, Furlong KL, Leggett B, Jass JR. DNA microsatellite instability and mismatch repair protein loss in adenomas presenting in hereditary non-polyposis colorectal cancer. Gut. 2000;47:37–42. doi: 10.1136/gut.47.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton SR, Aaltonen LA. Pathology and genetics of tumours of the digestive system. Lyon: IARC Press; World Health Organization Classification of Tumours. 2000 [Google Scholar]
- Heinmöller E, Renke B, Beyser K, Dietmaier W, Langner C, Rüschoff J. Pitfalls in diagnostic molecular pathology—significance of sampling error. Virchows Arch. 2001;439:504–511. doi: 10.1007/s004280100450. [DOI] [PubMed] [Google Scholar]
- Müller A, Giuffre G, Bocker Edmonston T, Mathiak M, Roggendorf B, Heinmöller E, Brodegger T, Tuccari G, Mangold E, Buettner R, Rüschoff J, the German HNPCC Consortium, German Cancer Aid (Deutsche Krebshilfe) Challenges and pitfalls in HNPCC screening by microsatellite analysis and immunohistochemistry. J Mol Diagn. 2004;6:308–315. doi: 10.1016/S1525-1578(10)60526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittekind CH, Meyer HJ, Bootz F. Sobin LH, Wittekind C, editors. Berlin: UICC,; TNMKlassifikation Maligner Tumoren. 2002:66–70. [Google Scholar]
- Umar A, Boland CR, Terdiman JP, Syngal S, Chapelle de la A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, Hamilton SR, Hiatt RA, Jass J, Lindblom A, Lynch HT, Peltomaki P, Ramsey SD, Rodriguez-Bigas MA, Vasen HFA, Hawk ET, Barett JC, Freedman AN, Srivastava S. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietmaier W, Wallinger S, Bocker T, Kullmann F, Fishel R, Rüschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–4756. [PubMed] [Google Scholar]
- Woerner SM, Benner A, Sutter C, Schiller M, Yuan YP, Keller G, Bork P, von Knebel Doeberitz M, Gebert JF. Pathogenesis of DNA repair-deficient cancers: a statistical meta-analysis of putative real common target genes. Oncogene. 2003;22:2226–2235. doi: 10.1038/sj.onc.1206421. [DOI] [PubMed] [Google Scholar]
- Woerner SM, Kloor M, Müeller A, Rüeschoff J, Friedrichs N, Buettner R, Buzello M, Kienle P, Knaebel HP, Kunstmann E, Pagenstecher C, Schackert HK, Moslein G, Vogelsang H, von Knebel Doeberitz M, Gebert JF. Microsatellite instability of selective target genes in HNPCC-associated colon adenomas. Oncogene. 2005 doi: 10.1038/sj.onc.1208456. Feb. 7 (epub) [DOI] [PubMed] [Google Scholar]
- Lindgren G, Liljegren A, Jaramillo E, Rubio C, Linblom A. Adenoma prevalence and cancer risk in familial non-polyposis colorectal cancer. Gut. 2002;50:228–234. doi: 10.1136/gut.50.2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong AE, Morreau H, van Puijenbroek M, Eilers PH, Wijnen J, Griffioen G, Cats A, Menko FH, Kleibeuker JH, Vasen HFA. The role of mismatch repair gene defects in the development of adenomas in patients with HNPCC. Gastroenterology. 2004;12:42–48. doi: 10.1053/j.gastro.2003.10.043. [DOI] [PubMed] [Google Scholar]
- Rijcken FE, Hollema H, Kleibeuker JH. Proximal adenomas in hereditary non-polyposis colorectal cancer prone to rapid malignant transformation. Gut. 2002;50:382–386. doi: 10.1136/gut.50.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togashi K, Konishi F, Koinuma K, Ishitsuka T, Kojima M, Okada MH. Flat and depressed lesions of the colon and rectum: pathogenesis and clinical management. Ann Acad Med Singapore. 2003;32:152–158. [PubMed] [Google Scholar]
- Sakashita M, Aoyama N, Maekawa S, Kuroda K, Shirasaka D, Ichihara T, Kuroda Y, Minami R, Maeda S, Kasuga M. Flat-elevated and depressed, subtypes of flat early colorectal cancers, should be distinguished by their pathological features. Int J Colorectal Dis. 2000;15:275–281. doi: 10.1007/s003840000244. [DOI] [PubMed] [Google Scholar]
- Rijcken FE, Tineke van der Suis I, Hollema H, Kleibeuker JH. Hyperplastic polyps in hereditary nonpolyposis colorectal cancer. Am J Gastroenterol. 2003;10:2306–2316. doi: 10.1111/j.1572-0241.2003.07629.x. [DOI] [PubMed] [Google Scholar]
- Jass JR, Iino H, Ruszkiewicz A, Painter D, Solomon MJ, Koorey DJ, Cohn D, Furlong KL, Walsh MD, Palazzo J, Bocker Edmonston T, Fishel R, Young J, Leggett BA. Neoplastic progression occurs through mutator pathways in hyperplastic polyposis of the colorectum. Gut. 2000;47:43–49. doi: 10.1136/gut.47.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jass JR. Hyperplastic polyps and colorectal cancer: is there a link? Clin Gastroenterol Hepatol. 2004;22:1–8. doi: 10.1016/s1542-3565(03)00284-2. [DOI] [PubMed] [Google Scholar]
- Sparks AB, Morin PJ, Vogelstein B, Kinzler KW. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res. 1998;58:1130–1134. [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;10:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]