Abstract
As part of a national effort to identify biomarkers for the early detection of cancer, we developed a rapid and high-throughput sequencing protocol for the detection of sequence variants in mitochondrial DNA. Here, we describe the development and implementation of this protocol for clinical samples. Heteroplasmic and homoplasmic sequence variants occur in the mitochondrial genome in patient tumors. We identified these changes by sequencing mitochondrial DNA obtained from tumors and blood from the same individual. We confirmed previously identified primary lung tumor changes and extended these findings in a small patient cohort. Eight sequence variants were identified in stage I to stage IV tumor samples. Two of the sequence variants identified (22%) were found in the D-loop region, which accounts for 6.8% of the mitochondrial genome. The other sequence variants were distributed throughout the coding region. In the forensic community, the sequence variations used for identification are localized to the D-loop region because this region appears to have a higher rate of mutation. However, in lung tumors the majority of sequence variation occurred in the coding region. Hence, incomplete mitochondrial genome sequencing, designed to scan discrete portions of the genome, misses potentially important sequence variants associated with cancer or other diseases.
Mitochondrial DNA instability has been reported in degenerative diseases,1,2,3,4 neurodegenerative diseases,5,6 sudden infant death syndrome,7 aging and longevity,3,8,9,10,11,12 and cancer.13,14,15,16,17,18,19,20,21,22,23,24,25 Both solid tumors and hematological diseases including leukemias and lymphomas have been reported to contain changes in the mitochondrial genome.26 The accumulation of sequence variants indicates a possible function as a molecular clock for organ degeneration.27 Mitochondria were first suggested to contribute to carcinogenesis in 197328 when quantitativeand qualitative electron microscopy revealed structural differences in this organelle between cancer patients and normal controls. More detailed analyses of patient specimens revealed mitochondrial microsatellite instability associated with cancer.29,30,31,32,33 Specific point mutations and deletions were subsequently found first by DNA scanning technologies and further identification of specific sequence variants34 were reported.
It is hypothesized that mitochondrial defects are present in tumors due to damaged respiratory systems and ATP production.25 Mitochondria play a fundamental role in energy production and oxidative phosphorylation (OXPHOS).35,36 As a byproduct of this process, toxic reactive oxygen species are generated, resulting in DNA damage. Because mitochondria are the sites of reactive oxygen species production, its DNA is more likely to be damaged than nuclear DNA.37 Mitochondria possess less efficient DNA repair mechanisms than the nucleus,14 therefore, mutations are more likely to persist and be representative of clonal expansion. The evolutionary mutation rate is 17-fold higher compared to single copy genes in the nuclear DNA.14
Advantages of using mitochondria DNA (mtDNA) as a potential biomarker for cancer-specific mutation studies are several. The genome is well characterized, with ∼16,568 bp harboring 37 densely packed genes. Secondly, high copy number (hundreds to thousands of mtDNA copies per cell) is a distinct advantage over nuclear DNA for the detection of sequence variants and translates into less tissue required for analysis. Hence, precious clinical samples are conserved. In addition, the mitochondrial genome is more resistant than nuclear DNA to damage caused by isolation and storage due to the small size and covalently closed, circular structure of mtDNA. Repair is also less efficient in the mitochondrial than nuclear genomes therefore, mutations are more quickly identified. Because mitochondria lack introns, mutations that occur will accumulate in the coding regions and are more likely to have biological consequences.26
Despite the extensive association of mutations with human disease, the role of mitochondrial dysfunction in tumors and the interplay between nuclear and mitochondrial encoded genes remains unclear. Mitochondrial mutations have been observed in colorectal, breast, liver, prostate, pancreatic, and lung cancers. Moreover, sequence variations have been detected in preneoplastic lesions, which suggest mutations occur early in tumor progression.16,19,22 For colorectal cancer, 70% of cell lines examined harbored mitochondrial sequence variations in the coding and noncoding regions.21 DNA alterations were also found in 45% of colorectal cancer specimens. Mitochondrial genome instabilities were found in 48% of breast cancer tissues38 and 42% of breast cancer specimens contained mitochondrial DNA sequence variants in the D-loop.19 The mononucleotide repeat (C-stretch) at D303 to 310 was identified as a candidate hotspot in these breast primary tumors as well as cervical cancer. However, a rapid polymerase chain reaction (PCR) analysis of the C-stretch region from 56 cervical, bladder, and breast tumors, and endometrial neoplasia resulted in the detection of only 13 sequence variants (23%).39
Sequence variants in the D-loop region of the mitochondrial genome are also present in lung cancer. In one study of the noncoding region from 28 cell lines and 55 non-small cell lung cancer samples, sequence alterations were detected in 61% of the cell lines and 20% of the tumors.40 In further studies, lung cancer cell lines and tumor tissues were demonstrated to contain sequence variants at 70% and 43%, respectively.13,14 Sequence variations in mitochondrial DNA may serve as early indicators of lung cancer, thus enabling detection of cancer in early stages when treatment is most effective.
The DNA Technologies Group at National Institute of Standards and Technology (NIST), an Early Detection Research Network Biomarker Validation Laboratory, conducts measurements to validate the utility of genes and gene products as biomarkers after their initial characterization by the Biomarker Discovery Laboratories. The purpose of this study was to evaluate mitochondrial sequence variants as biomarkers of early lung cancer. Lung cancer is the second most common cancer among men and women and is the leading cause of death among the most common cancers.41 The incidence is as great as 117 per 100,000 individuals.42 Reductions in cancer can be expected if improved diagnostics and population screening technologies are implemented. Improvement in lung cancer prognosis can be primarily achieved via the development and validation of accurate early detection assays, which include strategies for noninvasively collected samples coupled with automation and sensitive detection limits. Currently, no clinically applied DNA biomarker exists for lung cancer. Initial pathological diagnosis is based on small bronchoscopic biopsy.43 Thus, reliable, high-throughput assays are needed to determine the role of sequence variation in the mitochondrial genome and the manifestation of cancer in clinical samples and bodily fluids.
In this study, 22 mitochondrial genomes were fully sequenced from blood and tumor DNAs obtained from 11 individuals with lung cancer. Using a nested protocol and M13 primers for sequencing, an automated capillary electrophoresis protocol obtained 97 to 100% (average, 99.8%) sequence coverage for the forward strand and 90 to 100% (average, 98.4%) sequence coverage for the reverse strand. Sequence variants were identified in the tumor samples with respect to patient blood in 5 of 11 individuals (45%). Overall, eight sequence variants were identified. Twenty-two percent of the sequence variants identified were found in the D-loop region, which accounts for only 6.8% of the genome. The other sequence variants were spread throughout the coding region. Despite the reasonable assumption that heteroplasmies should comprise multiple subpopulations of mutated mtDNA molecules, the majority of the tumors were homoplasmic for sequence variants. Our work describes sequence variant detection by fluorescent capillary electrophoresis and compares these results to previous studies using radiolabeled sequencing. Further, limited comparison between these results and those obtained using DNA resequencing microarrays are described. In summary, a robust protocol for the identification of both heteroplasmic and homoplasmic sequence variants in clinical samples (human tissues and bodily fluids) is described.
Materials and Methods
Nucleic Acid Isolation
DNA was extracted from microdissected tissue obtained from cryostat-embedded snap-frozen sections. DNA from primary tumors and bodily fluids was evaluated. Paired normal and tumor specimens were collected after surgical resection with prior consent from patients in the Johns Hopkins University Hospital. DNA from tumor sections was digested with 1% sodium dodecyl sulfate/Proteinase K, extracted by phenol chloroform, and ethanol precipitated. Tumor samples were obtained from males and females and represented broncholoalveolar, squamous cell, and adenocarcinomas. Samples were collected from stage I and IV tumors (Table 1).
Table 1.
Pathology and Staging of Clinical Samples
| Sample | Age | Pathology | Stage | JHU sequence | Sequence variants |
|---|---|---|---|---|---|
| 1 | 59 | Bronchioloalveolar | I | Complete | 1 |
| 2 | 74 | Adenocarcinoma | I | D-loop | 0 |
| 3 | 62 | Squamous cell | I | D-loop | 0 |
| 4 | 84 | Adenocarcinoma | I | D-loop | 0 |
| 5 | 47 | Adenocarcinoma | IV | Complete | 1 |
| 6 | 78 | Adenocarcinoma | I | Complete | 0 |
| 7 | 67 | Adenocarcinoma | I | D-loop | 0 |
| 8 | 64 | Bronchioloalveolar | IV | D-loop | 1 |
| 9 | 82 | Squamous cell | I | Complete | 4 |
| 10 | 41 | Adenocarcinoma | I | D-loop | 1 |
| 11 | 66 | Large cell undifferentiated | IV | D-loop | 0 |
PCR Amplification
PCR amplification primers selected were reported previously44 to specifically amplify mitochondrial encoded DNA sequences. PCR amplification conditions were modified to optimize these primers and to use PCR-ready amplification microtiter plates. Briefly, primers were synthesized by Operon/Qiagen (Alameda, CA) and were shipped frozen at 100 μmol/L. Amplification was performed in two steps: a primary amplification followed by a nested, secondary amplification. For the primary amplification, nine overlapping primer sets were used to amplify the entire mtDNA genome from 20 ng of DNA template (Table 2). These PCR products ranged in size from 1886 to 2075 bp. The DNA, primers (0.6 μmol/L of each), and deionized H2O were added to a Clontech BD Sprint Advantage 96-well plate (Clontech Laboratories, Inc., Palo Alto, CA) containing lyophilized BD Sprint Advantage polymerase mix, dNTPs, optimized PCR buffer, and a mix of cryoprotectants for a total reaction volume of 25 μl. Thermal cycling conditions were as follows: preamplification denaturation: (1 cycle), 95°C for 1 minute; amplification (30 cycles): 95°C for 30 seconds; annealing, 58°C for 1 minute; elongation, 68°C for 3 minutes; final elongation (1 cycle), 68°C for 3 minutes; 4°C hold.
Table 2.
Primary and Nested Primers for Full Mitochondrial Genome Sequence Analysis
| Primer Set 1 | NT Position | Product Length | Primer Set 2 | NT Position | Product Length | Primer Set 3 | NT Position | Product Length |
|---|---|---|---|---|---|---|---|---|
| Tay-1 | 503–2484 | 1982 | Tay-2 | 2384–4249 | 1886 | Tay-3 | 4155–5220 | 2066 |
| tay2-1* | 516–1190 | 711 | tay2-4 | 2395–3074 | 716 | tay2-7 | 4184–4869 | 722 |
| tay2-2 | 1138–1801 | 700 | tay2-5 | 2995–3648 | 687 | tay2-8 | 4832–775 | 775 |
| tay2-3
|
1754–2444
|
725
|
tay2-6
|
3536–4239
|
740
|
tay2-9
|
5526–6188
|
699
|
| Primer Set 4
|
NT Position
|
Product Length
|
Primer Set 5
|
NT Position
|
Product Length
|
Primer Set 6
|
NT Position
|
Product Length
|
| Tay-4 | 6113–8017 | 1905 | Tay-5 | 7925–9884 | 1980 | Tay-6 | 9167–11748 | 1982 |
| tay2-10 | 6115–6781 | 703 | tay2-13 | 7960–8641 | 718 | tay2-16 | 9821–10516 | 732 |
| tay2-11 | 6730–7398 | 705 | tay2-14 | 8563–9231 | 705 | tay2-17 | 10394–11032 | 675 |
| tay2-12
|
7349–8012
|
697
|
tay2-15
|
9181–9867
|
723
|
tay2-18
|
10985–11708
|
760
|
| Primer Set 7
|
NT Position
|
Product Length
|
Primer Set 8
|
NT Position
|
Product Length
|
Primer Set 9
|
NT Position
|
Product Length
|
| Tay-7 | 11614–13638 | 2025 | Tay-8 | 13539–15431 | 1893 | Tay-9 | 15331–836 | 2075 |
| tay2-19 | 11634–12361 | 765 | tay2-22 | 13568–14275 | 745 | tay2-25 | 15372–16067 | 732 |
| tay2-20 | 12284–13007 | 758 | tay2-23 | 142527–14928 | 738 | tay2-D1 | 15879–16545 | 703 |
| tay2-21 | 12951–13614 | 700 | tay2-24 | 14732–15420 | 724 | tay2-D2 | 16495–390 | 482 |
| tay2-D3 | 213–803 | 525 |
Nested primers contain M13 sequence (forward or reverse) for fluorescent sequencing.44
PCR amplification products were subsequently analyzed for both quality and quantity of DNA using the Caliper AMS 90 SE electrophoresis system (Caliper Technologies Corp., Mountain View, CA). To facilitate sequencing, the nine amplicons were reamplified using M13-tagged primers. The secondary amplification uses 28 primer pairs to amplify smaller fragments of the primary amplicons (482 bp and 775 bp, Table 2). Secondary amplification reactions were conducted using 2 μl (0.6 to 45 ng) of the primary PCR product and the Clontech 96-well plates and reagents used in the primary PCR step. Thermal cycling conditions were as follows: preamplification denaturation: (1 cycle), 95°C for 1 minute; amplification (30 cycles): 95°C for 30 seconds; annealing, 58°C for 1 minute; elongation, 68°C for 1 minute; final elongation (1 cycle), 68°C for 3 minutes; 4°C hold.
PCR Clean-Up
PCR clean-up protocols were optimized for high-throughput sequencing using both Exo l-SAP and filtration. The first method, Exo I-SAP, was used for partial plates and individual reactions. The following reagents were added to a final 25-μl reaction volume: 1.0 unit of SAP (USB Corp., Cleveland, OH), 5.0 units of Exo I (USB), and 4.85 μl of deionized H2O. For multiple reactions, a master mix of these reagents was made and dispensed in 6.25-μl aliquots. Samples were then placed on the 9700 thermocycler (Applied Biosystems, Foster City, CA) for 15 minutes at 37°C, then 30 minutes at 72°C. The samples were then diluted with 40 μl of deionized H2O for analysis on the AMS 90 SE.
The second method used for PCR clean-up was the Montage PCRμ96 plate (Millipore Corp., Billerica, MA). This method was used for full 96-well plates containing the secondary PCR reactions. Seventy-five μl of deionized H2O were added to each well of the secondary amplification plate to a final volume of 100 μl/well. The contents of each well were then transferred to the Millipore filtration plate and placed on the vacuum manifold for 30 to 45 minutes or until all of the wells were dry. Once dry, 65 μl of deionized H2O were added to each well. The plate was then placed on a shaker at high speed for 10 minutes. The samples were then transferred to a 96-well plate compatible with the Caliper AMS 90 SE for quantification.
DNA Quantification
After clean-up, samples were separated on the AMS 90 SE to size and quantify the DNA. Fifty ng of each sample were then transferred to a new plate and dried in a vacuum centrifuge. The samples were resuspended in 50 μl of deionized water to a final concentration of 1 ng/μL for sequencing.
DNA Sequencing
Secondary products tagged with M13 forward and reverse primers, (tgtaaaacgacggccagt and ggaaacagctatgaccat, respectively) were sequenced using the Big Dye Terminator (BDT) version 3.1 cycle sequencing kit (Applied Biosystems). A one-eighth cycle-sequencing reaction was used for all sequencing. The 28 secondary PCR amplicons for each sample were sequenced on both strands with the M13 primers, for a total of 56 sequencing reactions for each mitochondrial genome. Each reaction contained 1 μl of each of the following reagents: BigDye Terminator, DNA (1 ng/μl), M13 primer (forward or reverse; 5 pmol/μl), 5× dilution buffer (Applied Biosystems), and dH2O to a final volume of 5 μl. Cycling sequencing conditions were as follows: (40 cycles): 96°C for 10 seconds; annealing, 50°C for 5 seconds; elongation, 60°C for 4 minutes; 4°C hold. All separations were performed using the ABI 3100 genetic analyzer using an 80-cm capillary and POP4 polymer system. Samples were electrokinetically injected (30 seconds, 1 kV) and separated at 14.6 kV. Sequences were aligned using the DNA Star SeqMan II (5.05) program (DNASTAR, Inc, Madison, WI) and scanned for polymorphisms and sequence variants. Both homoplasmic (identical mitochondrial genomes) and heteroplasmic (differences in mitochondrial DNA genomes present) sequence variants were expected.13,14 The sensitivity of sequence variation detection methods is a critical element to determining the degree of heteroplasmy.
Sequencing Clean-Up
The Montage SEQ96 plate (Millipore) was used for clean-up after cycle sequencing. Thirty μl of wash solution (Millipore) was added to each well of the cycle-sequencing plate. The samples were transferred to the clean-up plate and placed on the vacuum manifold for 15 to 20 minutes or until the wells were dry. A second wash of 30 μl of wash solution was added and vacuumed dry for an additional 25 to 30 minutes. Once dry, 20 μl of injection solution (Millipore) were added to each well and the plate was mixed vigorously on a plate shaker for 10 minutes. Resuspended samples were transferred to a 3100 optical plate (Applied Biosystems) and diluted with 15 μl of HI-DI formamide (Applied Biosystems).
Results
DNA Amplification
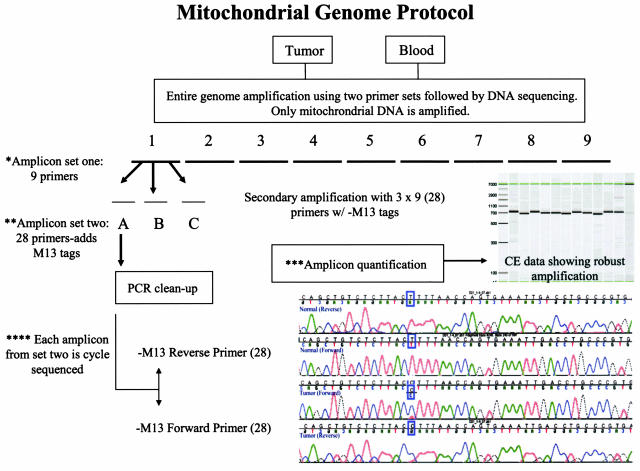
Minimum quantities of DNA required to fully sequence a mitochondrial genome were determined for the amplification of DNA from both tumor and blood. Reproducible results were obtained with 10 to 20 ng of DNA obtained from the clinical laboratory. The best results were obtained with a previously described44 nested PCR protocol, in which nine overlapping amplicons ranging in size from ∼1.9 to 2.1 kb were first generated spanning the entire mitochondrial genome (Table 2). These PCR products were then reamplified to generate 28 secondary PCR products with M13 primers attached to facilitate sequencing (Table 2). Because core sequencing laboratories request that common primer sequences, such as the M13 forward and reverse primers, be used in their facilities, our protocol allows research and clinical laboratories to send their amplified clinical samples to core laboratories for sequencing. An illustration of the amplification and sequencing methodology is shown in Figure 1.
Figure 1.
Validation scheme. Strategy for amplifying and sequencing entire mitochondrial genome. Each sample is individually amplified and sequenced. Mitochondrial genomes isolated from primary lung tumors and blood were initially amplified with nine overlapping primer sets that cover the entire genome. They were divided for reamplification and incorporation of M13 tags (Table 2). The quantity of every amplicon is determined and normalized. The 28 secondary PCR products are sequenced with M13 forward and reverse primers.
Primer Compatibility for Haplotyping
Further, the primers used in this study were carefully examined to ensure their ability to be used for determining the haplogroup status of the three major ethnic groups in humans, which are represented by seven single nucleotide polymorphisms (Table 3). Although single nucleotide polymorphism T14783C is contained within primer 17F, primer set 16 has overlapping coverage for this single nucleotide polymorphism. The other six single nucleotide polymorphisms defining the three major ethnic groups were not present in either the primary or secondary primer sets, therefore could easily be detected in our study design. Our 11 patients were haplotyped in both blood and tumor DNA samples to ensure that the samples were matched, thereby assuring the traceability of the sequence data to the patient specimens. In all cases, the haplogroup association of blood and tumor sequences matched (data not shown).
Table 3.
Coding Region SNPs that Identify Major Human Haplogroups
| European/Asian (haplogroup N) | Asian (haplogroup M) | African (haplogroup L) |
|---|---|---|
| A8701 | A8701G | |
| T9540 | T9540C | |
| A10398 | A10398G | |
| T10873 | T10873C | |
| C10400 | C10400T | |
| T14783 | T14783C | |
| G15043 | G15043A |
PCR Product Quality Control
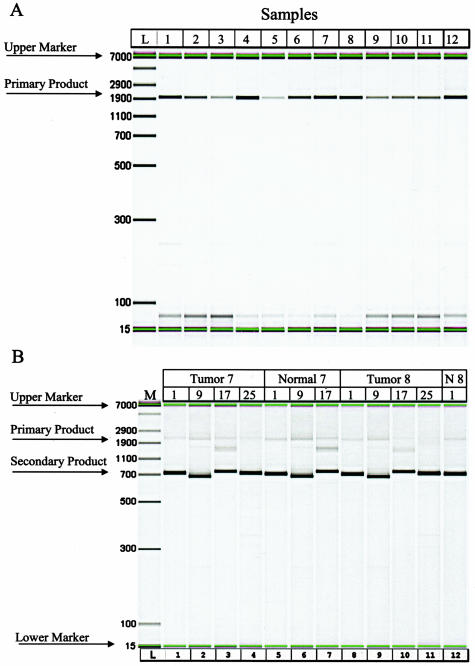
The quality and quantity of amplicons from both primary and secondary amplification reactions were determined with a Caliper AMS-90 SE instrument. Representative results for both primary and secondary amplifications are in Figure 2, A and B, respectively. In general, 40 μl of primary amplicons varied from 11.6 to 892 ng generated. In some cases, low quantities of nonspecific products were also observed. The samples were reamplified with the nested primers if the specific target amplicons represented the majority product. Secondary PCR products varied between 72 ng and 2.7 μg. These products were examined by AMS 90 analysis, and high-quality products, those in which the expected amplicon was the majority product, were used for sequencing.
Figure 2.
A: Gel images of primary and secondary amplification primers. Nine primer sets were used to amplify clinical samples. Primary amplicon products are shown as 1905-bp migration products. One of the nine primer pairs (primer pair no. 4 shown here) was used to amplify the mtDNA from clinical samples. B: Twenty-eight primer sets were used to add tags to facilitate DNA sequencing. Secondary amplicon products are shown as 711, 699, 675 and 732 for primer pairs 1, 9, 17, and 25, respectively.
DNA Sequencing
By using M13-tagged primers, the cycle sequencing conditions were reduced to two reactions (forward and reverse), thus streamlining the assay and reducing costs for clinical laboratories requiring full genome sequencing. The Applied Biosystems (80 cm × 50 μm) capillary for long sequence reads (800 to 1000 nucleotides) was used for sequencing the 482- to 775-bp secondary amplicons. The minimum required quantity of DNA and cycle sequencing reagents were determined for these long reads. Overall, sequencing coverage for DNAs obtained from both tumor and blood approached 100%, with forward strand coverage generally higher than reverse (Table 4). In only one genome was sequence coverage of more than one strand lower than 98%. In those samples with gaps in sequence coverage on both strands (sample 10, blood; sample 11, blood and tumor), the gaps occurred in the same positions. Hence, complete coverage was not achieved for these samples; the forward gap in all three samples represents the number of nucleotides missing to attain full coverage on at least one strand.
Table 4.
Sequencing Coverage for Clinical Samples
| Normal (blood)
|
Tumor
|
|||
|---|---|---|---|---|
| % Forward coverage | % Reverse coverage | % Forward coverage | % Reverse coverage | |
| Sample 1 | 100 | 99.4 | 100 | 99.7 |
| No. nt* missing | 99 | 60 | ||
| Sample 2 | 100 | 99.91 | 100 | 100 |
| No. nt missing | 29 | |||
| Sample 3 | 100 | 100 | 100 | 99.96 |
| No. nt missing | 24 | |||
| Sample 4 | 100 | 99.7 | 100 | 99.71 |
| No. nt missing | 49 | 48 | ||
| Sample 5 | 100 | 99.81 | 100 | 100 |
| No. nt missing | 31 | |||
| Sample 6 | 100 | 95.35 | 100 | 97.13 |
| No. nt missing | 772 | 476 | ||
| Sample 7 | 100 | 98.25 | 100 | 93.49 |
| No. nt missing | 290 | 1079 | ||
| Sample 8 | 100 | 100 | 100 | 100 |
| No. nt missing | ||||
| Sample 9 | 100 | 99.11 | 100 | 99.53 |
| No. nt missing | 168 | 79 | ||
| Sample 10 | 99.98 | 98.09 | 100 | 97.54 |
| No. nt missing | 26 | 329 | 392 | |
| Sample 11 | 98.48 | 97.98 | 97.33 | 90.42 |
| No. nt missing | 289 | 335 | 458 | 1588 |
Nucleotides.
Analysis of Variants
In this study, a robust fluorescent sequencing protocol was developed for the identification of both heteroplasmic and homoplasmic mitochondrial sequence variants in small quantities of human tissues and bodily fluids. To determine whether changes in the human mitochondrial genome are associated with lung tumors, we compared the sequences of primary lung tumor samples with matched blood samples from the same individual. Tumor mtDNA sequences were further compared to the revised human mitochondrial DNA Cambridge sequence45,46 found at Mitomap (http://www.mitomap.org). Twenty-two mitochondrial genomes were fully sequenced at NIST. High-quality sequence data were obtained from as little as 1.0 ng of amplified DNA.
Sequence variants were identified in the tumor samples from 5 of 11 individuals, An overall summary of the sequencing data are shown in Table 5. Sample 5 contained a single sequence change within the D-loop region, and samples 1, 8, and 10 contained single sequence changes within the coding region. Sample 9 contained four sequence changes, both in coding and noncoding regions.
Table 5.
Summary of Sequence Variants Detected
| Sample number | Nucleotide position | Reference* | Blood | Tumor | Consensus |
|---|---|---|---|---|---|
| 1 | 2664 | T | T | Y | Agree |
| 2 | No sequence variants | Agree | |||
| 3 | 16519 | T | T | C | JHU only† |
| 4 | No sequence variants | Agree | |||
| 5 | 302 | 7 C’S | 8 C’S | 7 C’S | Agree |
| 6 | No sequence variants | Agree | |||
| 7 | No sequence variants | Agree | |||
| 8 | 15229 | T | Y | Y | NIST only |
| 9 | 6 | 2 NIST only | |||
| Variants‡ | 2 JHU only | ||||
| 10 | 7734 | T | T | C | NIST only |
| 11 | No sequence variants | Agree |
Reference sequence is the revised Cambridge human mitochondrial sequence (http://www.mitomap.org).
Consensus sequence is the consensus between the JHU-radiolabeled protocol and the NIST fluorescent-sequencing protocol.
Four variants reported for radiolabeled sequencing,14 four reported for fluorescent sequencing, two overlap for both methods.
Comparison to Previous Results
Samples provided to NIST for this study had been previously sequenced, in part, at The Johns Hopkins University (JHU) using a radiolabeled sequencing protocol.13,14 After full mitochondrial sequencing was completed at NIST, the presence and positions of sequence variants between tumor and matched blood were compared to previous JHU results.
Complete mitochondrial genome sequence data from JHU was available for 4 of the 11 paired samples. Incomplete sequence was obtained for the remaining 7. As shown in Table 5, the sequence data from JHU agreed fully with the sequence data from NIST for 7 of the 11 paired samples; 5 of these contained no sequence variants within the entire genome. The other two samples in full agreement both had single point mutations within the mitochondrial genome; one within the hypervariable 2 region of the D-loop in the C stretch at D303 to 310, the second within the 16S ribosomal RNA gene at Mitomap position 2664.
For the four paired samples (samples 3 and 8 to 10) in which differences were identified between JHU and NIST, samples 3, 8, and 10 harbored single sequence variants within the coding region. JHU reported a homoplasmic mutation in sample 3 at nucleotide position 16,519 (within the D-loop) that was not confirmed at NIST. In samples 8 and 10, single heteroplasmic mutations were detected within the coding region of the mitochondrial genome using the fluorescent sequencing protocol described herein. In these samples, JHU sequenced only the hypervariable region, and would therefore not have observed these mutations.
The results obtained for sample 9, the fourth sample in which discrepancies were noted between NIST and JHU, are shown in Table 6. Sample 9 was fully sequenced at both JHU and NIST. Four mutations were observed at JHU, two within the coding region at positions 5521 and 12,345, and two within the D-loop region at nucleotide positions 16,183 and 16,187. Analysis at NIST confirmed the two coding region variants, but failed to identify the two D-loop variants. In this sample, NIST detected two new sequence variants at nucleotide positions 1463 and 16,274.
Table 6.
Sequence Variants Detected in Sample 9
| Position | Reference | NIST
|
JHU
|
||
|---|---|---|---|---|---|
| Blood | Tumor | Blood | Tumor | ||
| 1463 | C | C | T | C | C |
| 5521 | G | G | A | G | A |
| 12,345 | G | A | R | G | A |
| 16,183 | A | A | A | C | A |
| 16,187 | C | T | T | C | T |
| 16,274 | G | G | A | G | G |
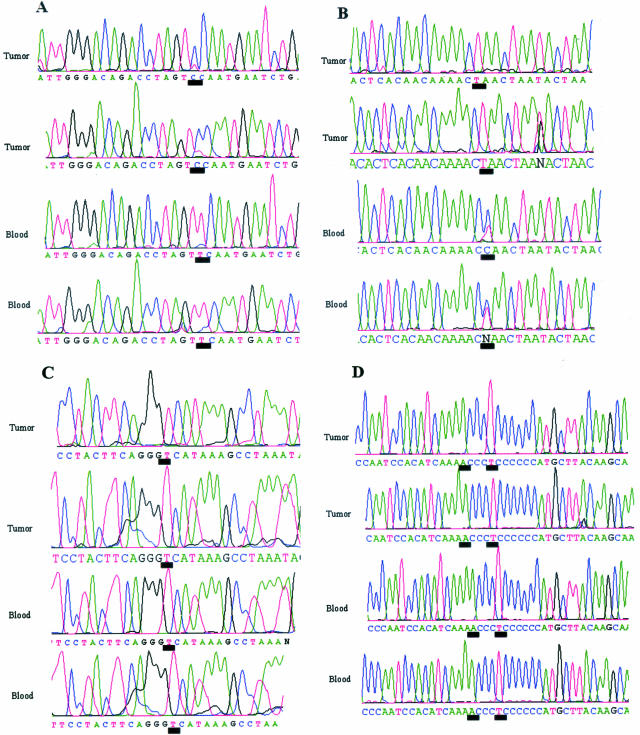
The electropherograms illustrating the discrepant sequence variants discussed above are shown in Figure 3. Sequence variants detected by fluorescent sequencing in samples 8 and 10 are shown in Figure 3, A and B, respectively. Note that both of these sequence variants appear to be heteroplasmic for the tumor sample and that the variant in sample 8 is heteroplasmic in both tumor and blood. The electropherograms for the sequence positions in samples 3 and 9 reported by JHU to contain sequence variants are shown in Figure 3, C and D, respectively. No sequence variants were observed by fluorescent sequencing.
Figure 3.
Discrepant sequence variants. A: Sequence data from mitochondrial sample no. 8 illustrating low-level heteroplasmy between DNAs obtained from tumor and blood (control) samples. Sequence data are shown for both forward (top) and reverse (bottom) sequence reactions. The black line underscores the nucleotide position 15,229. B: Sequence data from mitochondrial sample no. 10 illustrating heteroplasmic sequence variation between DNAs obtained from tumor and blood (control) samples. Sequence data are shown for both forward (top) and reverse (bottom) sequence reactions. The black line underscores the nucleotide position 7734. C: Sequence data from mitochondrial sample no. 3 illustrating no sequence variation between DNAs obtained from tumor and blood (control) samples, previously reported.14 Sequence data are shown for both forward (top) and reverse (bottom) sequence reactions. The black line underscores the nucleotide position 16,519. D: Sequence data from mitochondrial sample no. 9 illustrating no sequence variation between DNAs obtained from tumor and blood (control) samples using the described fluorescent sequencing method. Sequence data are shown for both forward (top) and reverse (bottom) sequence reactions. The black lines underscore the nucleotide positions 16,183 and 16,187, previously reported to contain sequence variants.14
Discussion
NIST has focused on metrics for the identification of mutations for the early detection of cancer. Currently, there is no universal standard method for detecting disease-specific mitochondrial sequence variations. Our goal was to develop a standard capillary electrophoresis method and assist the Early Detection Research Network to clinically evaluate whether mtDNA sequencing could serve as a biomarker for lung cancer. Our high-throughput capillary electrophoresis analysis of mitochondrial DNA from lung cancer patients detected sequence variants in stage I and IV tumors, suggesting that mitochondrial mutation(s) could serve as a biomarker for early detection. The capillary electrophoresis protocol can be reliably used by other laboratories to detect cancer in asymptomatic patients, make a diagnosis once symptoms appear, or monitor cancer patients for recurrence or individuals known to be at high risk. Further, in addition to clinical applications, the full mitochondrial genome-sequencing protocol was able to determine the haplogroups of the patients in this study, suggesting that this protocol would be useful for human identification.
The analysis of the entire mitochondrial genomes from 22 patient samples resulted in the detection of eight sequence variants identified in 5 of 11 (45%) matched lung tumor samples. Our observed 45% is consistent with other lung tumor studies that report a detection range of 20 to 43% in lung cancer.13,14,40 Of the eight sequence variants identified in this study, two (22%) were detected in the D-loop region, which accounts for 6.8% of the mitochondrial genome. Six variants were found in the coding region; three within RNA-encoding regions and two of the remaining three were silent mutations.
Although not every tumor possessed sequence variants, one sample contained a number of variants, sample 9. The presence of multiple variants in tumors is in agreement with previous observations. In Fliss and colleagues,14 two lung tumors were reported with more than one sequence variant (nos. 2 and 4, respectively). Further, multiple variants were found in head and neck and bladder cancers as well (three to five variants per tumor). Sample 9, obtained from a stage 1 squamous cell carcinoma, contains four to six sequence variants, as reported by the two laboratories using different sequencing technologies.14 Although fluorescent sequencing detected four variants in this sample, only two of these variants, found within the coding region, were previously reported.14 Two additional variants were found by fluorescent sequencing at nucleotide positions 1463 and 16,274 within the human mitochondrial genome. Two other variants previously reported14 at positions 16,183 and 16,187 were not detected in this study.
During this study, a redesigned Affymetrix mitochondrial DNA array was used to sequence a subset of the same lung cancer samples reported here.47 The results obtained using this chip were very promising; it detected the majority of those sequence variants detected at NIST, as well as others present as heteroplasmies at low levels (under the 10 to 20% detection limit of the fluorescent sequencing protocol). The MitoChip results confirmed our results for samples 3 and 8, in which no sequence variants were found. Further, the MitoChip confirmed all of the coding region variants detected at NIST for sample 9, and confirmed that the sequence variant at position 12,345 was heteroplasmic (G > R), previously reported as a G > A transition by radiolabeled sequencing.14 Moreover, because of the greater sensitivity, the MitoChip detected a fourth variant at position 12,308. Although we detected a nearby heteroplasmic variant at 12,345, fluorescent sequencing was not able to detect this additional heteroplasmy. As reported in the MitoChip study, mixed populations (ie, heteroplasmies) were detected at a 49:1 mixture.47 This suggests that the mutation at position 12,308 was a heteroplasmy present at a level lower than the detection limits of fluorescent sequencing (10 to 20%). The limit of detecting heteroplasmic variants with the MitoChip is unknown and further studies to address this question are ongoing. The remaining three variants reported by NIST and JHU are found within the D-loop region, which is not tiled on the MitoChip.
It is a reasonable assumption that heteroplasmies should comprise multiple subpopulations of mutated mtDNA molecules. Consistent with this hypothesis, heteroplasmy was detected in three of the eight sequence variants (37.5%) in this study. The fluorescent sequence technology used in this study is only capable of detecting heteroplasmies (or mixed samples) present at 10 to 20% of the population. As reported, the MitoChip was capable of detecting a sequence variant present at the 2% level (1 in 49),47 and detected an additional heteroplasmic variant in sample 9. Because heteroplasmy was found in three of eight variants, the MitoChip may confer a real advantage in cancer diagnostics. Further studies are needed to determine the overall sensitivity of the MitoChip for heteroplasmy detection.
As the mitochondrial genome becomes fully characterized across healthy and diseased human populations,48 the differences between normal and disease-linked variations should become clear. The relative mutation rates for each mitochondrial nucleotide have been studied49 and recent haplogroup data suggest that mitochondrial sequence variations are a nonrandom process.50 Little is known about the aging effects on lung mitochondria, although sequence variants have been found to accumulate with aging in other tissues.2 Although the sample set is small, we note that no correlation could be made with respect to presence of sequence variants and patient age from which the tumor sample was derived. For example, one individual, age 82, contained four to seven sequence variants whereas two others, ages 78 and 84, contained no sequence variation (Table 1). Similarly, no correlation was found with respect to the presence of sequence variants and either tumor classification or stage of tumor progression.
In summary, the comparison of sequencing results from three platforms showed that there was a general agreement in the presence or absence of sequence variations in matched lung primary cancer samples (8 of 11). As NIST fully sequenced all 22 mitochondrial genomes, we were able to detect more coding region variants than previously reported for lung cancer.14 Our study extends the findings that lung cancer, as well as other diseases, contain sequence variants spanning the entire mitochondrial genome and therefore full genome sequencing provides the cancer diagnostic community with a useful biomarker assay.51
Footnotes
Supported in part by the National Cancer Institutes-Early Detection Research Network (interagency agreements Y1CN010309 and Y1CN2020).
Certain commercial equipment, instruments, materials, or companies are identified in this article to specify adequately the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are the best available for the purpose.
References
- Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm for degenerative diseases and aging. Ageing vulnerability: causes and interventions. Novartis Found Symp. 2001;235:247–263. doi: 10.1002/0470868694.ch20. [DOI] [PubMed] [Google Scholar]
- Melov S, Shoffner JM, Kaufman S, Wallace DC. Marked increase in the number and variety of mitochondrial DNA rearrangements in aging human skeletal muscle. Nucleic Acid Res. 1995;23:4122–4126. doi: 10.1093/nar/23.20.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melov S, Hinerfeld D, Esposito L, Wallace DC. Multi-organ characterization of mitochondrial genomic rearrangements in ad libitum and caloric restricted mice show striking somatic mitochondrial DNA rearrangements with age. Nucleic Acids Res. 1997;25:974–982. doi: 10.1093/nar/25.5.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Murdock DG. Mitochondria and dystonia: the movement disorder connection? Proc Natl Acad Sci USA. 1999;96:1817–1819. doi: 10.1073/pnas.96.5.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun AS, Brown MD, Wallace DC. A mitochondrial DNA mutation at nucleotide pair 14459 of NADH dehydrogenase subunit 6 gene associated with maternally inherited Leber hereditary optic neuropathy and dystonia. Proc Natl Acad Sci USA. 1994;91:6206–6210. doi: 10.1073/pnas.91.13.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdal SH, Vege A, Egeland T, Musse MA, Rognu TO. Possible role of mtDNA mutations in sudden infant death. Pediatr Neurol. 2002;27:23–29. doi: 10.1016/s0887-8994(02)00384-3. [DOI] [PubMed] [Google Scholar]
- Chomyn A, Attardi G. MtDNA mutations in aging and apoptosis. Biochem Biophys Res Commun. 2003;304:519–529. doi: 10.1016/s0006-291x(03)00625-9. [DOI] [PubMed] [Google Scholar]
- Melov S, Wallace DC. Mitochondrial DNA rearrangements in aging human brain and in-situ PCR of mtDNA. Neurobiol Aging. 1999;20:565–571. doi: 10.1016/s0197-4580(99)00092-5. [DOI] [PubMed] [Google Scholar]
- Jazwinski SM. Metabolic control and ageing. Trends Genet. 2000;16:506–511. doi: 10.1016/s0168-9525(00)02119-3. [DOI] [PubMed] [Google Scholar]
- Murdock DG, Christacos ND, Wallace DC. The age related accumulation of a mitochondrial DNA control region mutation in muscle, but not brain, detected by a sensitive PNA directed PCR clamping based method. Nucleic Acids Res. 2000;28:4350–4355. doi: 10.1093/nar/28.21.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardi G. Role of mitochondrial DNA in human aging. Mitochondrion. 2002;2:27–37. doi: 10.1016/s1567-7249(02)00032-6. [DOI] [PubMed] [Google Scholar]
- Polyak K, Li Y, Zhu H, Lengauer C, Willson JKV, Markowitz SD, Trush MA, Kinzler KW, Vogelstein B. Somatic mutations of the mitochondrial genome in human colorectal tumors. Nat Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, Jen J, Sidransky D. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;28:2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- Nomoto S, Yamashita K, Koshikawa K, Nakao A, Sidransky D. Mitochondrial D-loop mutations as clonal markers in multicentric hepatocellular carcinoma and plasma. Clin Cancer Res. 2002;8:481–487. [PubMed] [Google Scholar]
- Ha PK, Tong BC, Westra WH, Sanchez-Cespedes M, Parrella P, Zahurak M, Sidransky D, Califano JA. Mitochondrial C-tract alteration in premalignant lesions of the head and neck: a marker for progression and clonal proliferation. Clin Cancer Res. 2002;8:2260–2265. [PubMed] [Google Scholar]
- Sanchez-Cespedes M, Parrella P, Nomoto S, Cohen D, Xiao Y, Esteller M, Jernomino C, Jordan RC, Nicol T, Koch WM, Schoenberg M, Mazzarelli P, Fazio VM, Sidransky D. Identification of a mononucleotide repeat as a major target for mitochondrial DNA alterations in human tumors. Cancer Res. 2001;61:7015–7019. [PubMed] [Google Scholar]
- Jones JB, Song JJ, Hempen PM, Parmigiani G, Hruban RH, Kern SE. Detection of mitochondrial DNA mutations in pancreatic cancer offers a “mass”-ive advantage over detection of nuclear DNA mutations. Cancer Res. 2001;61:299–304. [PubMed] [Google Scholar]
- Parrella P, Xiao Y, Fliss M, Sanchez-Cespedes M, Mazzarelli P, Rinaldi M, Nicol T, Gabrielson E, Cuomo C, Cohen D, Pandit S, Spencer M, Rabitti C, Fazio VM, Sidransky D. Detection of mitochondrial DNA mutations in primary breast cancer and fine-needle aspirates. Cancer Res. 2001;61:7623–7626. [PubMed] [Google Scholar]
- Chen JZ, Gokden N, Greene GF, Mukunyadzi P, Kadlubar FF. Extensive somatic mitochondrial mutations in primary prostate cancer using laser capture microdissection. Cancer Res. 2002;62:6470–6474. [PubMed] [Google Scholar]
- Copeland WC, Wachsman JT, Johnson FM, Penta JS. Mitochondrial DNA alterations in cancer. Cancer Invest. 2002;20:557–569. doi: 10.1081/cnv-120002155. [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Nomoto S, Caballero OL, Usadel H, Henrique R, Varzim G, Oliveira J, Lopes C, Fliss MS, Sidransky D. Mitochondrial mutations in early stage prostate cancer and bodily fluids. Oncogene. 2001;20:5195–5198. doi: 10.1038/sj.onc.1204646. [DOI] [PubMed] [Google Scholar]
- Okochi O, Hibi K, Uemura T, Inoue S, Takeda S, Kaneko T, Nakao A. Detection of mitochondrial DNA alterations in the serum of hepatocellular carcinoma patients. Clin Cancer Res. 2002;9:2875–2878. [PubMed] [Google Scholar]
- Hibi K, Nakayama H, Yamazaki T, Takase T, Taguchi M, Kasai Y, Ito K, Akiyama S, Nakao A. Detection of mitochondrial DNA alterations in primary tumors and corresponding serum of colorectal cancer patients. Int J Cancer. 2001;94:429–431. doi: 10.1002/ijc.1480. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Penta JS, Johnson RM, Wachsman JT, Copeland WC. Mitochondrial DNA in human malignancy. Mutat Res. 2001;488:119–133. doi: 10.1016/s1383-5742(01)00053-9. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial DNA sequence variation in human evolution and disease. Proc Natl Acad Sci USA. 1994;91:8739–8746. doi: 10.1073/pnas.91.19.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumacher HR, Szelkely IE, Patel SB, Fisher DR. Mitochondrial, a clue to oncogenesis. Lancet. 1973;2:327. doi: 10.1016/s0140-6736(73)90836-2. [DOI] [PubMed] [Google Scholar]
- Neubert D, Hopfenmuller W, Fuchs G. Manifestation of carcinogenesis as a stochastic process on the basis of altered mitochondrial genome. Arch Toxicol. 1981;48:89–125. doi: 10.1007/BF00310481. [DOI] [PubMed] [Google Scholar]
- Cavalli IR, Liang BC. Mutagenesis, tumorigenicity, and apoptosis, are the mitochondrial involved? Mutat Res. 1998;398:19–26. doi: 10.1016/s0027-5107(97)00223-6. [DOI] [PubMed] [Google Scholar]
- Shay JW, Werbin H. New evidence for the insertion of mitochondrial DNA into the human genome. Mutat Res. 1992;275:227–235. doi: 10.1016/0921-8734(92)90026-l. [DOI] [PubMed] [Google Scholar]
- Baggetto LG. Role of mitochondria in carcinogenesis. Eur J Cancer. 1992;291:156–159. doi: 10.1016/0959-8049(93)90598-a. [DOI] [PubMed] [Google Scholar]
- Bandy B, Davison AJ. Mitochondrial mutations may increase oxidative stress. Free Radic Biol Med. 1990;10:515–519. doi: 10.1016/0891-5849(90)90152-9. [DOI] [PubMed] [Google Scholar]
- Holt I, Harding AE, Morgan-Hughes JA. Deletion of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- Toescu EC, Myronova N, Verkhratsky A. Age-related structural and functional changes of brain mitochondria. Cell Calcium. 2000;28:329–338. doi: 10.1054/ceca.2000.0167. [DOI] [PubMed] [Google Scholar]
- Toyokuni S, Okamoto K, Yodoi J, Hiai H. Persistent oxidative stress in cancer. FEBS Lett. 1995;358:1–3. doi: 10.1016/0014-5793(94)01368-b. [DOI] [PubMed] [Google Scholar]
- Yakes RM, Van Housten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci USA. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard SM, Bailliet G, Paez GL, Bianchi MS, Peltomaki P, Bianchi NO. Nuclear and mitochondrial genome instability in human breast cancer. Cancer Res. 2000;60:4231–4237. [PubMed] [Google Scholar]
- Parrella P, Seripa D, Matera MG, Rabitti C, Rinaldi M, Mazzarelli P, Gravina C, Gallucci M, Altomare V, Flammia G. Mutations of the D310 mitochondrial mononucleotide repeat in primary tumors and cytological specimens. Cancer Lett. 2003;190:73–77. doi: 10.1016/s0304-3835(02)00578-5. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Toyooka S, Miyajima K, Iizasa T, Fujisawa T, Bekele NB, Gazdar AF. Alterations in the mitochondrial displacement loop in lung cancers. Clin Cancer Res. 2003;9:5636–5641. [PubMed] [Google Scholar]
- Hirsch FR, Franklin WA, Gazdar AF, Bunn PA., Jr Early detection of lung cancer; clinical perspectives of recent advances in biology and radiology. Clin Cancer Res. 2001;7:5–22. [PubMed] [Google Scholar]
- Miller BA, Kolonel LN, Bernstein L, et al (eds): Racial/Ethnic Patterns of Cancer in the United States. 1988–1992, National Cancer Institute: US Cancer Patterns. Bethesda, National Institutes of Health pub 96–4104 [Google Scholar]
- Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn Matt, Rosen GD, Perou CM, Whyte RI. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RW, Taylor GA, Durham SE, Turnbull DM. The determination of complete mitochondrial DNA sequences in single cells: implications for the study of somatic mitochondrial DNA point mutations. Nucleic Acids Res. 1999;29:e74–e84. doi: 10.1093/nar/29.15.e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Maitra A, Cohen Y, Gillespie SE, Mambo E, Fukushima N, Hoque MO, Shah N, Goggins M, Califano J, Sidransky D, Chakravarti A. The human MitoChip: a high-throughput sequencing microarray for mitochondrial mutation detection. Genome Res. 2004;14:812–819. doi: 10.1101/gr.2228504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LJ, Liang MH, Kwon H, Park J, Bai RK, Tan DJ. Comprehensive scanning of the entire mitochondrial genome for mutations. Clin Chem. 2002;48:1901–1912. [PubMed] [Google Scholar]
- Meyer S, von Haeseler A. Identifying site-specific substitution rates. Mol Biol Evol. 2003;20:182–189. doi: 10.1093/molbev/msg019. [DOI] [PubMed] [Google Scholar]
- Torroni A, Rengo C, Guida V, Cruciani F, Sellitto D, Coppa A, Calderon F, Simionati B, Valle G, Richards M, Macaulay V, Scozzari R. Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am J Hum Genet. 2001;69:348–356. doi: 10.1086/324511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker PE. Cancer biomarker validation: roles for the National Institute of Standards and Technology (NIST). Ann NY Acad Sci. 2003;983:142–150. doi: 10.1111/j.1749-6632.2003.tb05969.x. [DOI] [PubMed] [Google Scholar]