Abstract
Somatic mutations in the tyrosine kinase domain of the epidermal growth factor receptor (EGFR) gene are present in lung adenocarcinomas that respond to the EGFR inhibitors gefitinib and erlotinib. Two types of mutations account for ∼90% of mutated cases: short in-frame deletions in exon 19 and a specific point mutation in exon 21 at codon 858 (L858R). Screening for these mutations has been based mainly on direct sequencing. We report here the development and validation of polymerase chain reaction-based assays for these two predominant types of EGFR mutations. The assay for exon 19 mutations is based on length analysis of fluorescently labeled polymerase chain reaction products, and the assay for the exon 21 L858R mutation is based on a new Sau96I restriction site created by this mutation. Using serial dilutions of DNAs from lung cancer cell lines harboring either exon 19 or 21 mutations, we detected these mutations in the presence of up to ∼90% normal DNA. In a test set of 39 lung cancer samples, direct sequencing detected mutations in 25 cases whereas our assays were positive in 29 cases, including 4 cases in which mutations were not apparent by sequencing. These assays offer higher sensitivity and ease of scoring and eliminate the need for sequencing, providing a robust and accessible approach to the rapid identification of most lung cancer patients likely to respond to EGFR inhibitors.
A recent finding that is having a major impact on adult solid tumor oncology is that of somatic mutations in the tyrosine kinase domain of the epidermal growth factor receptor (EGFR) gene in lung adenocarcinomas that respond to the EGFR inhibitor gefitinib (Iressa).1,2,3 Tumors sensitive to erlotinib (Tarceva), another EGFR inhibitor with a mechanism of action similar to that of gefitinib, also contain the same types of EGFR mutations.3 Given the annual incidence of lung adenocarcinomas, eg, ∼70,000 in the US, the volume of molecular diagnostic assays for EGFR mutations could rival that of ERBB2 amplification assays in breast cancers, if EGFR testing becomes part of standard lung cancer management.
Two mutations account for ∼90% of EGFR mutations reported to date in lung adenocarcinomas (Table 1). The most common mutation type, seen in ∼46% of cases with EGFR mutations, is a short in-frame deletion of 9, 12, 15, 18, or 24 nucleotides in exon 19. The second most common mutation, seen in ∼43% of cases with EGFR mutations, is a point mutation (CTG to CGG) in exon 21 at nucleotide 2573, that results in substitution of leucine by arginine at codon 858 (L858R). Other much less common mutations have been described in exons 18, 20, and 21 (Table 1). Combining data from four studies,1,2,3,4 it appears that ∼80% of tumors that respond to gefitinib or erlotinib contain missense mutations or in-frame deletions in the EGFR tyrosine kinase domain, compared to none of 36 drug-refractory tumors (P < 0.05). These studies show that these EGFR mutations correlate strongly with sensitivity to specific EGFR inhibitors and that their detection could be used to predict which patients will respond to these drugs.
Table 1.
Summary of Published Data on EGFR Mutation Types in Lung Adenocarcinomas
| Reference | 1 | 2 | 3 | 4 | 5 | 8 | Total | % |
|---|---|---|---|---|---|---|---|---|
| Total number of EGFR mutated cases | n = 10 | n = 22 | n = 23 | n = 46* | n = 39 | n = 111 | n = 251 | |
| Exon 19 deletion | 6 | 15 | 9 | 16 | 18 | 52 | 116 | 46.2 |
| Exon 21 L858R | 2 | 5 | 13 | 22 | 17 | 49 | 108 | 43.0 |
| Exon 18 G719S or G719C | 1 | 2 | 0 | 2† | 3 | 4§ | 12 | 4.8 |
| Exon 20 | 0 | 0 | 0 | 2 | n.t. | 5¶ | 7 | 2.8 |
| Exon 21 non-L858R | 1 | 0 | 1 | 4‡ | 1 | 0 | 7 | 2.8 |
| Exon 19 insertion | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0.4 |
Rare cases with a combination of a hotspot mutation (exon 19 deletion or exon 21 L858R) with a nonhotspot mutation are listed only once, under the hotspot mutation. Very rare cases with a combination of two nonhotspot mutations are listed only under one of the two mutations and further details provided in the footnotes. There are no cases reported with two different hotspot mutations.
One case with an EGFR nonsense mutation not included.
Both cases also had exon 20 mutations.
One case also had another exon 21 non-L858R mutation.
One case also had an exon 18 non-G719 mutation.
One case had two exon 20 point mutations.
So far, screening for these mutations has been based on direct sequencing or single-strand conformation polymorphism analysis.5 We report here the development and validation of polymerase chain reaction (PCR)-based assays for the two predominant EGFR mutations. These assays offer ease of scoring and higher analytical sensitivity and eliminate the need for sequencing. Thus, they provide a robust and accessible approach to the rapid identification of most lung cancer patients who are likely to respond to specific EGFR inhibitors.
Materials and Methods
Samples and Cell Lines
Tumor specimens were obtained through protocols approved by the Institutional Review Board of Memorial Sloan-Kettering Cancer Center. Thirty-nine lung cancer samples were studied with both assays, including all cases reported to have exon 19 or 21 mutations in our initial sequencing study that had material available for further analysis.3 Dilutions for sensitivity studies were performed by mixing DNA extracted from positive control lung cancer cell lines NCI-H1650 or NCI-H1975 (American Type Culture Collection, Rockville, MD) into EGFR-germline lymphoma DNA. Each mixture contained 100 ng of total DNA with the proportion of cell line DNA ranging from 100 to 0.8%.
EGFR Exon 19 Deletion Assay
Because 99% of exon 19 mutations reported to date have been short in-frame deletions (Table 1), we designed an assay based on length analysis of fluorescently labeled PCR products. A 207-bp genomic fragment including all of exon 19 was amplified using primers EGFR-Ex19-FWD1 and EGFR-Ex19-REV1 (Table 2). The reverse primer was labeled with the 6-FAM fluorophore (6-FAM emits fluorescence with a peak wavelength of 522 nm). The PCR reaction mix was made up as follows: HotStarTaq DNA polymerase and 10 × buffer (Qiagen, Valencia, CA), EGFR-Ex19-FWD1 and EGFR-Ex19-REV1 primers (15 pmol each), genomic DNA template (100 ng), PCR Carry-Over prevention kit reagents (N-glycosylase + dUTP) (Applied Biosystems, Foster City, CA), remaining dNTPs, MgCl2 (0.5 mmol/L and sterile distilled water (to 50 μl). The PCR was performed as follows: 50°C × 2 minutes (to complete the PCR Carry-Over prevention procedure), 95°C × 15 minutes (to inactivate N-glycosylase and activate TaqDNA polymerase), followed by 40 cycles of 95°C × 0.5 minutes, 60°C × 1 minute, 72°C × 1 minute, and a final extension of 72°C × 10 minutes. For DNA extracted from frozen tissue, 35 cycles was sufficient. PCR product intensity was checked on a 2% agarose gel. If PCR product intensity was strong (equal or stronger than size marker), a 1 in 50 dilution was made of which 1 μl was added into 20 μl of formamide plus 1 μl of Genescan 400HD size standard (Applied Biosystems). If the PCR product intensity was weak (band fainter than size marker), up to 1 μl of undiluted PCR product was added to 20 μl or formamide and 1 μl of Genescan 400HD size standard. The samples were denatured at 95°C for 5 minutes and cooled on ice for 5 minutes. They were then subjected to capillary electrophoresis using POP4 polymer with an excitation wavelength of 494 nm and a detection wavelength of 522 nm on an ABI 3100 Avant genetic analyzer (Applied Biosystems).
Table 2.
Primers Used for Mutation Assays and Direct Sequencing
| Name | Sequence |
|---|---|
| EGFR-Ex19-FWD1 | GCACCATCTCACAATTGCCAGTTA |
| EGFR-Ex19-REV1 | Fam-AAAAGGTGGGCCTGAGGTTCA |
| EGFR-Ex21-FWD1 | CCTCACAGCAGGGTCTTCTCTGT |
| EGFR-Ex21-REV1 | Fam-TCAGGAAAATGCTGGCTGACCTA |
| EGFR-Ex19-FWD-seq | CCCAGCAATATCAGCCTTAGGTG |
| EGFR-Ex19-REV-seq | CCACTAGAGCTGGAAAGGGAAAGA |
| EGFR-Ex21-FWD-seq | TCCATTCTTTGGATCAGTAGTCACTAAC |
| EGFR-Ex21-REV-seq | GCTCACACTACCAGGAGACCCT |
All sequences are 5′ to 3′. All the primers were synthesized by Integrated DNA Technologies (Coralville, IA).
EGFR Exon 21 L858R Mutation Assay
The 2573T>G mutation creates a new Sau96I restriction site, GGNCC (Figure 1) that can be used as the basis for a PCR restriction fragment length polymorphism (PCR-RFLP) assay design. Another Sau96I restriction site upstream in exon 21 provides an internal restriction enzyme digestion control. The digested fluorescently labeled PCR products are analyzed by capillary electrophoresis. The precise product sizing possible by this approach allows unambiguous and sensitive identification of the 173-bp digested wild-type product and 87-bp digested mutant PCR product. Specifically, a 222-bp genomic fragment including all of exon 21 was amplified using primers EGFR-Ex21-FWD1 and EGFR-Ex21-REV1 (Table 2). Again, the reverse primer was labeled with the 6-FAM fluorophore. The PCR reaction mix was made up as above, but with EGFR-Ex21-FWD1 and EGFR-Ex21-REV1 primers (15 pmol each). The PCR was performed as follows: 50°C × 2 minutes (to complete PCR Carry-Over prevention procedure), 95°C × 15 minutes (to inactivate N-glycosylase and activate TaqDNA polymerase), followed by 95°C × 0.5 minutes, 60°C × 1 minute, 72°C × 1 minute, and a final extension of 72°C × 10 minutes for 40 cycles if the DNA was from paraffin blocks, 35 cycles if the DNA was from frozen tissue. PCR product intensity was checked on a 2% agarose gel. The PCR products were then purified using PCR Kleen Spin column (Bio-Rad, Hercules, CA) at 735 × g for 2 minutes. This step was not included in the procedure for the exon 19 deletion analysis, but was found to be necessary in the exon 21 assay to reduce baseline noise and increase sensitivity after restriction enzyme digestion. The Sau96I digestion reaction was then performed at 37°C for 2 hours and consisted of the following: 5 μl of PCR product, 2 μl of 10× NEBuffer 4 (New England Biolabs, Beverly, MA), 2 μl (10 U) of Sau96I restriction enzyme (New England Biolabs), and 11 μl of sterile distilled water. After digestion, 1 μl of undiluted Sau96I-digested PCR product was added to 20 μl of formamide and 1 μl of Genescan 400HD size standard. The samples were denatured at 95°C for 5 minutes, cooled on ice for 5 minutes, and subjected to capillary electrophoresis as described above.
Figure 1.
EGFR exon 21 L858R mutation assay design showing primer locations and native and mutant Sau96I restriction enzyme sites.
Direct Sequencing
Exons 19 and 21 were amplified using HotStarTaq DNA polymerase and primers EGFR-Ex19-FWD-seq and EGFR-Ex19-REV-seq, and EGFR-Ex21-FWD-seq and EGFR-Ex21-REV-seq, respectively. The PCR products were purified using PCR Kleen Spin columns and sequenced using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) according to the manufacturer’s protocol on an ABI 3100 Avant genetic analyzer running ABI Prism DNA sequence analysis software (Applied Biosystems).
Results
EGFR Exon 19 Deletion Assay
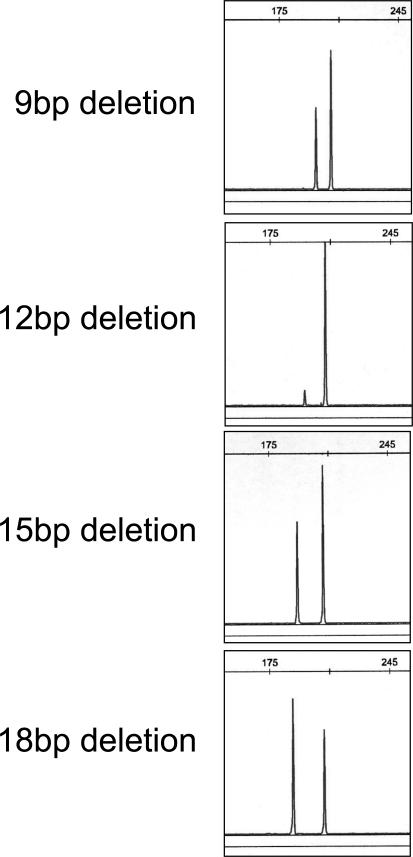
The assay was performed as described in Materials and Methods. We scored the test as positive when the expected product of 207 bp was accompanied by a distinct peak at 9, 12, 15, 18, or 24 bp below the normal product in the electrophoretogram. Conservatively, we only accepted mutant peaks that were greater than a minimum cutoff, designated arbitrarily at least fivefold above local background noise. Examples of 9-, 12-, 15-, and 18-bp deletions are shown in Figure 2. All cases with deletions also showed a germline product of 207 bp. Exon 19 deletion with loss of the remaining normal allele appears rare but preferential PCR amplification of the shorter deleted product could result in rare cases with a very small germline product of 207 bp. Furthermore, the nonneoplastic cells present in lung cancer samples should always provide a template for PCR amplification of the germline allele. Negative cases showed the expected germline product of 207 bp and the absence of any peak above background noise at 9, 12, 15, 18, 21, or 24 bp below the normal product in the electrophoretogram. Negative controls (including placenta and 21 lymphoma samples) consistently showed the germline product only.
Figure 2.
EGFR exon 19 deletion assay: examples of detection of 9-, 12-, 15-, and 18-bp deletions in EGFR exon 19. The variable relative heights of the deleted and germline peaks may reflect different proportions of tumor cells in these clinical tumor DNAs, or different levels of copy number alterations in the mutated EGFR allele.
The H1650 lung adenocarcinoma cell line contains an exon 19 deletion.6 Serial dilutions of H1650 cell line DNA into normal DNA were analyzed using the exon 19 deletion assay. The exon 19 deletion was readily detected in the presence of 6.25% H1650 DNA (Figure 3). In comparison, detection of the exon 19 deletion by direct sequencing was only readily possible down to a dilution of 12.5% H1650 DNA (Supplemental Figure 1 at http://jmd.amjpathol.org/). We should note that because of ploidy differences between cancer cell lines and nonneoplastic cells, sensitivities based on dilutions of DNA should be viewed only as approximations of the absolute sensitivity based on dilutions of cells, however this should not affect relative comparisons of the analytical sensitivities of different techniques.
Figure 3.
Sensitivity analysis of EGFR exon 19 deletion assay using serial dilutions of H1650 cell line DNA into normal DNA. Note that for dilutions of 25% and less the scale has been adjusted to highlight the mutant peak.
The observation that the deleted peak is approximately twice the height of the germline peak in pure H1650 DNA suggests that the deleted allele may be duplicated in this cell line. Thus the analytical sensitivities estimated using this cell line might be somewhat higher than for lung cancer cells with a nonduplicated mutant allele. Low-level amplification of EGFR in concert with EGFR mutation has been described in some tumors and another lung adenocarcinoma cell line shows high-level EGFR amplification and exon 19 deletion.7 We and others8 have also observed that many clinical samples show evidence of increased copies of the mutant allele, which in practical terms can further raise the technical sensitivity of the assay.
EGFR Exon 21 L858R Mutation Assay
The same cases were also screened for the exon 21 L858R mutation by a PCR-RFLP assay based on a new Sau96I restriction site created by the L858R mutation (2573T>G). The Sau96I-digested fluorescently labeled PCR products were analyzed by capillary electrophoresis. Examples of undigested, digested germline, and digested mutant PCR products are shown in Figure 4. All cases showed a germline product of 173 bp. Digestion of both Sau96I sites was complete in all runs as shown by the absence of a 222-bp undigested peak. Any case in which the peak corresponding to the mutated allele was less than five times the local background noise in the electrophoretogram (for example, see peak at 1.6% tumor cells in Figure 5) was considered at risk for a false-positive result and was repeated at least once using two to five times more template. Negative controls showed the expected germline digested product of 173 bp and the absence of any peak above background noise at 87 bp. Negative controls (including placenta and 21 lymphoma samples) consistently showed the germline product only.
Figure 4.
EGFR exon 21 L858R mutation assay: examples of undigested, digested germline, and digested mutant PCR products.
Figure 5.
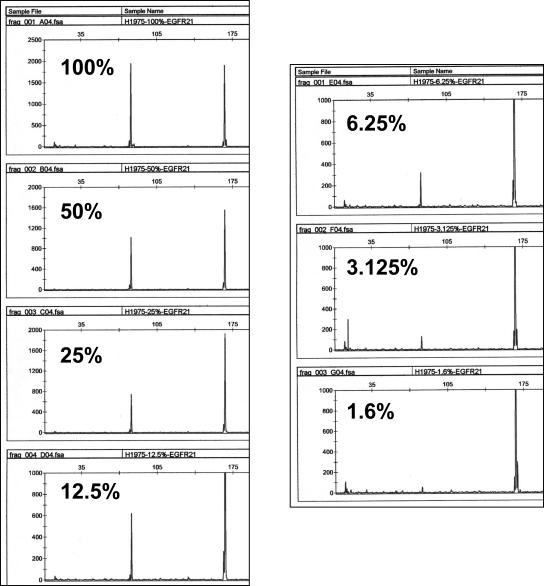
Sensitivity analysis of EGFR exon 21 L858R mutation assay using serial dilutions of H1975 cell line DNA into normal DNA. Note that for dilutions of 12.5% and less the scale has been adjusted to highlight the mutant peak.
The H1975 lung adenocarcinoma cell line contains the exon 21 L858R mutation.6 Serial dilutions of H1975 cell line DNA into normal DNA were analyzed using the exon 21 L858R mutation assay. The observation that the 87-bp and 173-bp peaks are approximately equal in the presence of 100% H1975 DNA suggests that mutant and germline alleles are present in equal proportions in this cell line. The exon 21 L858R mutation was readily detected by this assay in the presence of 3.125% H1975 DNA (Figure 5). In comparison, detection of the exon 21 L858R mutation by direct sequencing was only possible down to a dilution of 6.25% H1975 DNA (Supplemental Figure 2 at http://jmd.amjpathol.org/).
Comparison to Direct Sequencing
We then tested 39 lung cancer samples with both assays and compared the results to direct sequencing. To provide an adequate test of the sensitivity of our assays, this series of cases was enriched for mutated cases based on previous sequencing data (and is therefore not representative of mutation prevalence). The exon 19 deletion assay was positive in 15 of 39 cases and the exon 21 L858R assay was positive in 14 of 39 cases. No cases were positive for both assays. In four cases (two exon 19, two exon 21), the PCR-based assays detected mutations not apparent by direct sequencing. In an additional two cases positive by our exon 21 assay, direct sequencing was only equivocally positive (results not shown). The results are summarized in Table 3.
Table 3.
Comparison of EGFR Exon 19 and 21 Mutation Status as Determined by Present PCR-Based Assays and Direct Sequencing in 39 Lung Adenocarcinomas
| Case |
EGFR exon 19
|
EGFR exon 21 L858R
|
||
|---|---|---|---|---|
| Deletion assay | Direct sequencing | Mutation assay | Direct sequencing | |
| 1 | 15-bp deletion | nt 2235–2249 15-bp deletion | Negative | Negative |
| 5T | Negative | Negative | Positive | Positive |
| 12 | 15-bp deletion | nt 2235–2249 15-bp deletion | Negative | Negative |
| 14 | 15-bp deletion | nt 2239–2256 18-bp del. + 3-bp ins. | Negative | Negative |
| 15 | Negative | Negative | Positive | Positive |
| 18 | 15-bp deletion | nt 2235–2249 15-bp deletion | Negative | Negative |
| 21 | Negative | Negative | Positive | Positive |
| 34 | 15-bp deletion | nt 2236–2250 15-bp deletion | Negative | Negative |
| 65T | Negative | Negative | Positive | Positive |
| 67 | Negative | Negative | Positive | Weak Positive |
| 76 | Negative | Negative | Positive | Negative |
| 77 | 12-bp deletion | Negative | Negative | Negative |
| 79 | Negative | Negative | Negative | Negative |
| 80 | Negative | Negative | Negative | Negative |
| 83 | Negative | Negative | Negative | Negative |
| 84 | Negative | Negative | Negative | Negative |
| 85 | Negative | Negative | Negative | Negative |
| 86 | Negative | Negative | Negative | Negative |
| 87 | Negative | Negative | Positive | Positive |
| 88 | Negative | Negative | Positive | Positive |
| 89 | 15-bp deletion | nt 2235–2249 15-bp deletion | Negative | Negative |
| 90 | Negative | Negative | Positive | Negative |
| 91 | Negative | Negative | Negative | Negative |
| 92 | 15-bp deletion | nt 2235–2249 15-bp deletion | Negative | Negative |
| 93 | Negative | Negative | Negative | Negative |
| 94 | 15-bp deletion | nt 2235–2249 15-bp deletion | Negative | Negative |
| 96 | Negative | Negative | Negative | Negative |
| 98T | Negative | Negative | Positive | Positive |
| 99 | Negative | Negative | Negative | Negative |
| 134T | Negative | Negative | Positive | Positive |
| 230T | 15-bp deletion | nt 2236–2250 15-bp deletion | Negative | Negative |
| 71a | Negative | Negative | Positive | Weak Positive |
| 72a | Negative | Negative | Positive | Positive |
| A | 9-bp deletion | nt 2238–2248 11-bp del. + 2-bp ins. | Negative | Negative |
| B | Negative | Negative | Positive | Positive |
| C | 9-bp deletion | nt 2238–2248 11-bp del. + 2-bp ins. | Negative | Negative |
| D | 9-bp deletion | Negative | Negative | Negative |
| E | 9-bp deletion | nt 2239–2248 10-bp del. +1-bp ins. | Negative | Negative |
| F | 18-bp deletion | nt 2239–2258 20-bp del. + 2-bp ins. | Negative | Negative |
nt, nucleotide; bp, base pairs; del., deletion; ins., insertion.
Discussion
Because all but 1 of the 117 exon 19 mutations reported to date have been short in-frame deletions, we designed an assay based on length analysis of fluorescently labeled PCR products on a capillary electrophoresis device. The precise PCR product sizing possible by this approach allows confirmation that the number of nucleotides deleted is a multiple of 3 as expected for in-frame deletions (Figure 2). It has been previously shown that this assay design is as accurate as and more sensitive than direct sequencing for the detection of in-frame deletion (or insertion) mutations9 (in the context of exon 11 KIT mutations in gastrointestinal stromal tumors). Although there is a theoretical risk that an in-frame deletion could create a novel stop codon (with opposite biological consequences), this has not been reported in more than 100 exon 19 deletions sequenced to date. Interestingly, the only other type of mutation so far reported in exon 19, an 18-bp insertion found in a single tumor,8 should also be detected by the present assay. Indeed, we have recently observed another such case in our clinical testing (unpublished data).
Using this assay on ∼200 clinical samples throughout a 6-month period, we have observed 9-, 12-, 15-, 18-, and 24-bp deletions in exon 19 of EGFR. By far the most common deletion size is 15 bp. A peculiar observation is that 21-bp deletions have so far not been reported and 12-bp deletions are rare. Novel deletions (eg, 6 bp, 21 bp, or 27 bp) not previously characterized in terms of function or association with therapy response should be considered of uncertain significance although their functional impact may be similar if they affect the LREA motif that forms the core of all presently described deletions. It should also be noted that, although a minority of tumors have been reported to carry two mutations (Table 1), the two hotspot mutations in exons 19 and 21 have never been found together.
Our assays found ∼10% (4 of 39) more mutations than were detected by direct sequencing. Another two cases were only equivocally positive by direct sequencing (Table 3). Thus, the overall potential false-negative rate for direct sequencing compared to our assays is 15% (6 of 39), and this may be an underestimate because our study group was enriched for cases with mutations based on previous sequencing data. We attribute this to the greater analytical sensitivity of our assays. That this should be significant in the setting of lung cancer samples is not surprising given the often considerable admixture of nonneoplastic elements in these tumors.
Based on the aggregate data from four studies,1,2,3,5 ∼10% of lung adenocarcinomas from American or European patients who are unselected for response to EGFR inhibitors contain EGFR mutations. This percentage also matches the response rate of ∼10% seen in single agent trials of gefitinib or erlotinib performed before the discovery of EGFR mutations.10,11 However, there appear to be striking ethnic differences in the prevalence of EGFR mutations. It had been previously observed that responses to gefitinib were significantly more frequent in Japanese non-small cell lung cancer patients than in non-Japanese patients.11 That unusual clinical observation is now explained by the finding of a fourfold higher prevalence of EGFR mutations in Asian non-small cell lung cancer patients, ie, ∼40%.4,8
It is important to note that the use of the clinical term non-small cell lung cancer may result in unnecessary testing. This is because the vast majority of EGFR mutations are found in moderately to well differentiated adenocarcinomas, in particular those with partial or complete bronchioloalveolar features. The same histological features had been shown to correlate with response to EGFR inhibitors before the discovery of these mutations.12 In contrast, mutations appear exceedingly rare in large cell carcinomas2,5,7 and adenosquamous carcinomas.4,8 Lastly, numerous pure squamous carcinomas (>500), including one with apparent response to gefitinib,3 have been tested but were uniformly negative.4,5,8
Other strong correlates of EGFR mutations (and response to EGFR inhibitors) include female sex and never smoker status.3,5,8,12 It is presently estimated that females are approximately three times as likely as males to have mutation-positive tumors and never smokers are at least five times as likely to have mutation-positive tumors as past or present smokers.13 However, the interrelationships between these different factors have not yet been fully explored by multivariate analyses.
The two assays described here can be used as first line assays in all cases submitted for EGFR mutation analysis. The advantages of this overall approach to EGFR mutation screening is that the two types of mutation accounting for 90% of all mutations (exon 19 deletion and exon 21 L858R mutation) are detected by techniques that are faster (ie, 1 day versus 2 days) and more sensitive than direct sequencing, allowing for prompt initiation of treatment in most of the patients likely to respond to EGFR inhibitors. If both assays are negative, these first line assays can be followed by assays based on direct sequencing of EGFR exons 18, 20, and 21 to detect the remaining 10% of EGFR mutations. The predictive value of these assays could be further enhanced by combining them with testing for KRAS exon 2 mutations. Recent data from our center and elsewhere have shown that KRAS exon 2 mutations, present in ∼15 to 30% of lung adenocarcinomas, rarely, if ever, co-exist with EGFR mutations.8,14 This is biologically consistent because KRAS is downstream in the EGFR signaling pathway. It is therefore not unexpected that KRAS-mutated lung cancers fail to respond to EGFR inhibitors, a clinically important observation.14
Although there is currently still no consensus on the role of EGFR mutation screening in patient management, it is quite possible that testing of tumor tissue for EGFR mutations may soon be indicated in most or all patients with moderately to well differentiated lung adenocarcinoma,13 to aid in selecting therapy in neoadjuvant, adjuvant, and advanced/metastatic settings. In American or European centers, at least 10% of lung adenocarcinomas will harbor EGFR mutations and 90% of these will be detectable using the two EGFR mutation assays described here. Mutational analysis of EGFR is likely to become the mainstay of laboratory testing in this setting because other parameters such as EGFR immunoreactivity or EGFR gene amplification show at best only weak correlation with response to EGFR inhibitors.7,15,16 Immunohistochemistry with antibodies to phosphorylated EGFR remains to be systematically studied in this setting but the potential of this approach is uncertain given the observation that mutant EGFR proteins generally do not show simple constitutive autophosphorylation in vitro.1,3 The search for EGFR mutations in isolated cases of other cancers that have responded to EGFR inhibitors is in progress in many research laboratories. A wide variety of other cancers have already been screened with negative results.13 However, it remains possible that such studies could define new indications for EGFR testing in clinical molecular pathology laboratories in the coming months or years.
Supplementary Material
Acknowledgments
We thank Dr. Harold Varmus for advice and support; Jennifer Doherty for technical assistance; Josh Somar for technical assistance; and Drs. Maureen F. Zakowski, Rohit Bhargava, and Violetta Barbashina for related work.
Footnotes
Supported by the Chest and Lungevity Foundations of the American College of Chest Physicians and by an anonymous donor (to W.P.).
References
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski MF, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGF receptor gene mutations are common in lung cancers from “never smokers” and correlate with sensitivity of tumors to gefitinib (Iressa) and erlotinib (Tarceva). Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SF, Liu HP, Li LH, Ku YC, Fu YN, Tsai HY, Chen YT, Lin YF, Chang WC, Kuo HP, Wu YC, Chen YR, Tsai SF. High frequency of epidermal growth factor receptor mutations with complex patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin Cancer Res. 2004;10:8195–8203. doi: 10.1158/1078-0432.CCR-04-1245. [DOI] [PubMed] [Google Scholar]
- Marchetti A, Martella C, Felicioni L, Barassi F, Salvatore S, Chella A, Camplese PP, Iarussi T, Mucilli F, Mezzetti A, Cuccurullo F, Sacco R, Buttitta F. EGFR mutations in non-small-cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol. 2005;23:857–865. doi: 10.1200/JCO.2005.08.043. [DOI] [PubMed] [Google Scholar]
- Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- Amann J, Kalyankrishna S, Massion PP, Ohm JE, Girard L, Shigematsu H, Peyton M, Juroske D, Huang Y, Stuart SJ, Kim YH, Pollack JR, Yanagisawa K, Gazdar A, Minna JD, Kurie JM, Carbone DP. Aberrant epidermal growth factor receptor signaling and enhanced sensitivity to EGFR inhibitors in lung cancer. Cancer Res. 2005;65:226–235. [PubMed] [Google Scholar]
- Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- Emile JF, Lemoine A, Bienfait N, Terrier P, Azoulay D, Debuire B. Length analysis of polymerase chain reaction products: a sensitive and reliable technique for the detection of mutations in KIT exon 11 in gastrointestinal stromal tumors. Diagn Mol Pathol. 2002;11:107–112. doi: 10.1097/00019606-200206000-00007. [DOI] [PubMed] [Google Scholar]
- Kris MG, Natale RB, Herbst RS, Lynch TJ, Jr, Prager D, Belani CP, Schiller JH, Kelly K, Spiridonidis H, Sandler A, Albain KS, Cella D, Wolf MK, Averbuch SD, Ochs JJ, Kay AC. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- Fukuoka M, Yano S, Giaccone G, Tamura T, Nakagawa K, Douillard JY, Nishiwaki Y, Vansteenkiste J, Kudoh S, Rischin D, Eek R, Horai T, Noda K, Takata I, Smit E, Averbuch S, Macleod A, Feyereislova A, Dong RP, Baselga J. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer. J Clin Oncol. 2003;21:2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- Miller VA, Kris MG, Shah N, Patel J, Azzoli C, Gomez J, Krug LM, Pao W, Rizvi N, Pizzo B, Tyson L, Venkatraman E, Ben Porat L, Memoli N, Zakowski M, Rusch V, Heelan RT. Bronchioloalveolar pathologic subtype and smoking history predict sensitivity to gefitinib in advanced non-small-cell lung cancer. J Clin Oncol. 2004;22:1103–1109. doi: 10.1200/JCO.2004.08.158. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller VA. EGFR mutations, small molecule kinase inhibitors, and non-small cell lung cancer: current knowledge and future directions. J Clin Oncol. 2005;23:2556–2568. doi: 10.1200/JCO.2005.07.799. [DOI] [PubMed] [Google Scholar]
- Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, Zakowski MF, Heelan RT, Kris MG, Varmus HE. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLos Medicine. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra HS, Cavina R, Latteri F, Zucali PA, Campagnoli E, Morenghi E, Grimaldi GC, Roncalli M, Santoro A. Analysis of epidermal growth factor receptor expression as a predictive factor for response to gefitinib (‘Iressa’, ZD1839) in non-small-cell lung cancer. Br J Cancer. 2004;91:208–212. doi: 10.1038/sj.bjc.6601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SW, Hwang PG, Chung DH, Kim DW, Im SA, Kim YT, Kim TY, Heo DS, Bang YJ, Kim NK. Epidermal growth factor receptor (EGFR) downstream molecules as response predictive markers for gefitinib (Iressa, ZD1839) in chemotherapy-resistant non-small cell lung cancer. Int J Cancer. 2005;113:109–115. doi: 10.1002/ijc.20550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.