Abstract
Oligonucleotide microarrays are a powerful tool for profiling the expression levels of thousands of genes. Different statistical methods for identifying differentially expressed genes can yield different results. To our knowledge, no experimental test has been performed to decide which method best identifies genes that are truly differentially expressed. We applied three statistical methods (dChip, t-test on log-transformed data, and Wilcoxon test) to identify differentially expressed genes in previously untreated patients with chronic lymphocytic leukemia (CLL). We used a training set of Affymetrix Hu133A microarray data from 11 patients with unmutated immunoglobulin (Ig) heavy chain variable region (VH) genes and 8 patients with mutated Ig VH genes. Differential expression was validated using semiquantitative real-time polymerase chain reaction assays and by validating models to predict the somatic mutation status of an independent test set of nine CLL samples. The methods identified 144 genes that were differentially expressed between cases of CLL with unmutated compared with mutated Ig VH genes. Eighty genes were identified by Wilcoxon test, 60 by t-test, and 65 by dChip, but only 11 were identified by all three methods. Greater agreement was found between the t-test and the Wilcoxon test. Differential expression was validated by semiquantitative real-time polymerase chain reaction assays for 83% of individual genes, regardless of the statistical method. However, the Wilcoxon test gave the most accurate predictions on new samples, and dChip, the least accurate. We found that all three methods were equally good for finding differentially expressed genes, but they found different genes. The genes selected by the nonparametric Wilcoxon test are the most robust for predicting the status of new cases. A comprehensive list of all differentially expressed genes can only be obtained by combining the results of multiple statistical tests.
Oligonucleotide microarrays are a powerful tool for profiling the expression levels of thousands of genes simultaneously. Numerous papers have been written describing the applications of this technology. In the most straightforward applications, one performs microarray experiments on multiple samples representing two different biologically interesting conditions and then produces a list of the genes that are differentially expressed.
There are a number of plausible statistical strategies for choosing this list of genes. For instance, the software program dChip, which is often used to analyze Affymetrix oligonucleotide array data, bases its identification of differentially expressed genes on the construction of a confidence interval for the fold change.1 The dChip statistical model works on the original scale of the data, assuming independent, identically distributed normal errors in the estimates of expression levels. In contrast, researchers using spotted cDNA microarrays have almost uniformly concluded that expression values must be log-transformed to achieve approximate normality of the error distributions. Thus, they commonly perform two-sample t-tests on the log-transformed data. It cannot be assumed that the dChip analysis, performed on the original scale data, yields the same or even a similar list of differentially expressed genes as the t-test performed on log-transformed data.
Both dChip and the t-test are examples of parametric statistical methods; that is, they assume that the distribution of the data can be completely described by a small number of parameters, in the same way that a normal distribution is determined by its mean and SD. Nonparametric methods do not make the same kinds of distributional assumptions. The Wilcoxon test, for example, works with the ranks instead of the measured intensities. Thus, it gives the same answer on the original data and on the log-transformed data. This test may not be as sensitive as a test based on a parametric model that correctly describes the true error structure, but it is also less likely to be led astray than a test that uses an incorrect error model.
To our knowledge, no experimental test has been performed to decide which of these three methods does the best job of identifying genes that are truly differentially expressed. In this paper, we apply all three methods to experiments performed using Affymetrix Hu133A oligonucleotide microarrays separated into a training set and a blinded, independent test set. The microarrays were hybridized with samples obtained from previously untreated patients with CLL. The training set included 8 cases with mutated VH genes and 11 cases with unmutated VH genes. The presence or absence of somatic mutations in the Ig VH genes separates patients into two prognostic groups; patients with unmutated VH genes (about 40% of patients) have a median survival of 8 years compared with 25 years for patients with mutated VH genes (about 60% of patients).2,3 We found that the lists of differentially expressed genes obtained using the different statistical methods showed substantial differences. We selected a subset of differentially expressed genes for validation experiments using semiquantitative real-time polymerase chain reaction (QRT-PCR) assays. We also used the gene lists to predict the mutation status in a test set of nine additional patient samples, and the predictions were validated by performing sequence analysis of the VH genes.
Materials and Methods
Sample Collection and RNA Preparation
CLL samples were collected from 28 untreated patients after obtaining informed consent. Total RNA was prepared from CD19-positive CLL cells as described previously.4
Evaluation of Ig VH Genes for Somatic Hypermutation
The somatic mutation status of the Ig VH genes was determined as described previously.4 Briefly, total RNA was reverse transcribed using an oligo-d(T) primer and a First-Strand cDNA Synthesis kit (Amersham Biosciences, Piscataway, NJ). The cDNA was amplified in a PCR reaction using a mixture of six 5′ VH leader primers that amplify all seven VH families, together with a 3′ constant region primer (Cμ) in the presence of reaction buffer, dNTPs (2.5 mmol/L), and HotStar TaqDNA polymerase (Qiagen, Inc., Valencia, CA). After incubation at 94°C for 15 minutes, the cDNA was amplified for 30 cycles of 94°C for 1 minute, 56°C for 1 minute, and 72°C for 1 minute. In cases that failed to amplify using this strategy, we used a mixture of VH Framework 1 primers (V BASE database; http://www.mrc-cpe.cam.ac.uk/PRIMERS.php?menu=901) and a 3′ JH consensus primer (5′-AACTGAGGAGACGGTGACC-3′). We performed two independent PCR amplification reactions for each sample. Amplified products were separated by agarose gel electrophoresis and purified using the GeneClean II kit (Qbiogene, Carlsbad, CA). The PCR products were sequenced directly using the 3′ PCR primer and an ABI Prism 3700 or 3730 DNA Analyzer (Applied Biosystems, Foster City, CA).
Target Preparation, Microarray Hybridization, Image Quantification, and Normalization
Target preparation, microarray hybridization, image quantification, and normalization were performed as described previously.5 Briefly, 5 μg of total RNA was reverse-transcribed in a 20-μl reaction with 200 U of SuperScript II (Invitrogen Corporation, Carlsbad, CA) and 100 pmol of T7-(dT)24 primer (5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGC GG-(dT)24–3′) in 1× first-strand buffer (Invitrogen) at 42°C for 1 hour. The second-strand synthesis was performed at 16°C for 2 hours, in the presence of Escherichia coli enzymes, DNA Polymerase I (40 U), DNA ligase (10 U), RNase H (2 U), and 1× second-strand buffer (Invitrogen). The double-stranded cDNA was blunt-ended using 20 U of T4 DNA polymerase, purified by phenol/chloroform extraction, and transcribed in the presence of biotin labeled-ribonucleotides, using the BioArray HighYield RNA transcript labeling kit (Enzo Laboratories) according to the manufacturer’s instructions. The biotin-labeled cRNA was purified using an RNeasy minicolumn (RNeasy kit; Qiagen) and fragmented at 94°C for 35 minutes in 1× fragmentation buffer (40 mmol/L Tris-acetate, pH 8.0, 100 mmol/L potassium acetate, and 30 mmol/L magnesium acetate).
The Affymetrix GeneChip system was used for hybridization, staining, and imaging of the arrays. Hybridization cocktails (300 μl) containing 15 μg of cRNA and exogenous hybridization controls were hybridized to Hu133A GeneChips (Affymetrix, Santa Clara, CA) overnight at 42°C. Hybridized fragments were detected using streptavidin linked to phycoerythrin (Molecular Probes, Eugene, OR). The GeneChips were scanned and imaged using Affymetrix Microarray Analysis Suite, version 5.0.
Data from all 28 microarrays were loaded into dChip version 1.3 for normalization and quantification.6 Normalization was performed using the default settings in the software. Expression values were quantified using the PerfectMatch-only model. The expression levels estimated by dChip were exported and loaded into S-Plus (Insightful Corp., Seattle, WA) for further analysis. Estimated values equal to 0 were replaced by the threshold value 0.01; this modification only affected 17 measurements in the entire data set.
QRT-PCR Assays
The QRT-PCR assays were performed using TaqMan technology and a PRISM 7000 Sequence Detector (Applied Biosystems). PCR was carried out in a 25-μl reaction volume that contained 50 ng of cDNA, 1× TaqMan Universal PCR Master Mix without AmpErase UNG, unlabeled gene-specific PCR primers, and a 6-carboxy fluorescein-labeled TaqMan MGB probe. The primer and probe sets were specific for the following genes: KIAA0892, AGPAT2, DDX27, NEU3, FSTL3, NOL5A, TRAF4, TNFRSF1B, HLA-DQB1, SPTAN1, BTN3A2, and APOD (Assays-on-Demand Gene Expression system; Applied Biosystems). Amplification of 18S ribosomal RNA (rRNA) was performed in all cases to normalize the gene expression values. The probe for 18S rRNA is labeled with VIC (Pre-Developed TaqMan Assay Reagents; Applied Biosystems). After an incubation at 95°C for 10 minutes, the cDNA was amplified for 40 cycles of denaturation at 95°C for 15 seconds and combined annealing/extension at 60°C for 1 minute. Each sample was analyzed in duplicate. Standard curves for each gene and 18S rRNA were constructed using serially diluted cDNA prepared from a Burkitt lymphoma cell line (GA-10). The standards were analyzed in triplicate. Sequence detection software (SDS version 1.7; Applied Biosystems) was used to analyze the fluorescence emission data after PCR. The threshold cycle (Ct) values of each sample and the standards were exported to Microsoft Excel for further analysis. The Ct represents the cycle number at which fluorescence passes a fixed threshold. Standard curves were generated by plotting the Ct versus the amount of target cDNA in each dilution. Gene expression levels in test samples were expressed as the ratio of the gene of interest to 18S rRNA expression.
Statistical Analysis of Microarray Data
Differential expression was assessed using three different methods: dChip, t-test, and Wilcoxon rank-sum test. First, dChip was used to compute 90% confidence intervals around estimates of the fold change. Genes were selected as differentially expressed if the lower bound of fold change was greater than 1.2-fold and if the difference in mean expression levels was greater than 100. Next, two-sample t-test statistics and their associated P values were computed for each probe set on transformed data after computing the base-two logarithm. To account for multiple testing, we modeled the P values as a β-uniform mixture.7 This model allowed us to estimate the false discovery rate (FDR);8 we selected genes as differentially expressed by choosing a P-value cutoff that ensured that FDR was <10%. Finally, we computed Wilcoxon rank-sum statistics for each probe set. To account for multiplicities, we used an empirical Bayes method to estimate the posterior probability of differential expression.9 We selected genes with a posterior probability of differential expression of at least 80%, based on the most conservative prior probability estimate that ensured that none of the posterior probability estimates became negative.
Models for predicting the mutation status of new cases were built from the training data of 19 patient samples by combining principal components analysis and linear discriminant analysis (LDA). For each method, we first selected the differentially expressed genes as described above. We then performed principal components analysis on the 19 samples using the selected genes. Next, we performed LDA using the first k principal components to construct predictors for different values of k. Test samples were projected into the principal component space, and their mutation status was predicted using LDA and a uniform prior.
Results
Identification of Differentially Expressed Genes Using Three Different Statistical Methods: dChip, t-Test, and Wilcoxon Test
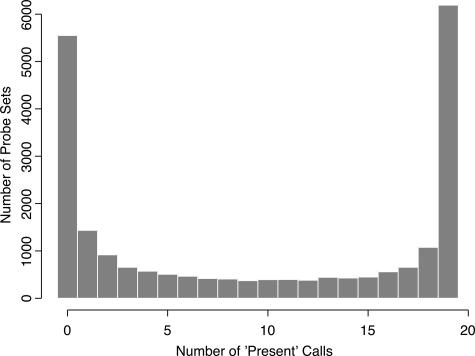
Probe sets were filtered to remove any probe set that was not called present at least once in the training set of 19 microarrays (Figure 1). This filtering step eliminated 5550 probe sets, leaving 16,733 probe sets for further analysis. First, we used dChip to compute 90% confidence intervals around estimates of fold change for each of the 16,733 probes set across the 19 training samples.1,10 We selected genes as differentially expressed if the lower bound of fold change was at least 1.2-fold and if the difference in mean expression was at least 100. (These parameters are the default settings for the software.) Using these parameters, we found 65 differentially expressed genes. Of these genes, 49 were overexpressed in unmutated samples, and 16 were overexpressed in mutated samples.
Figure 1.
Histogram of the number of times a probe set was called present in 19 microarray experiments. About one-fourth of the genes were never present, and about one-fourth were present in all samples.
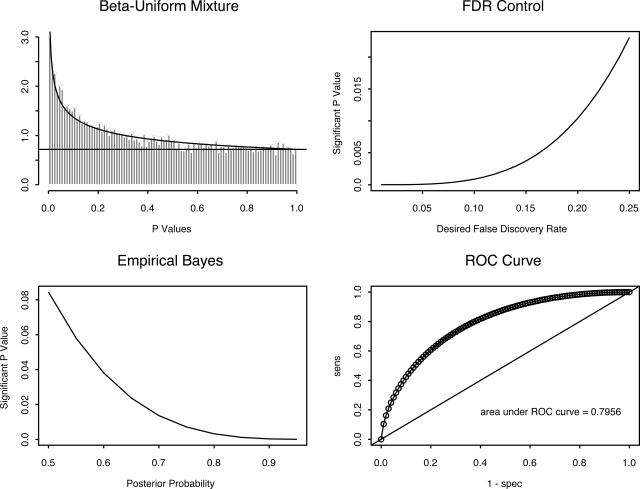
Next, we performed individual two-sample t-tests on the log-transformed expression values of 16,733 probe sets. We computed P values for each probe set, and we modeled the collection of P values as a β-uniform mixture.6 The β-uniform mixture model is based on the idea that P values for genes that are not differentially expressed should be uniformly distributed, whereas P values for genes that are differentially expressed should be strongly concentrated near zero (Figure 2). With this model, one can estimate the FDR associated with each possible cutoff on P values; we selected a cutoff by setting the FDR <10%. This bound on the FDR yielded a P-value cutoff of P < 0.00088, which corresponded to setting t > 4.01. Using this method, we found 60 differentially expressed genes, with 39 overexpressed in unmutated samples and 21 overexpressed in mutated samples.
Figure 2.
Analysis of the P values arising from 16,733 t-tests as a β-uniform mixture. Top left: Histogram of the observed P values, with overlaid curves representing the division into uniform and β contributions. Top right: Relationship between cutoff for P values and the false discovery rate. Bottom left: Relation between cutoff for P values and the posterior probability of differential expression. Bottom right: Receiver operating characteristics curve associated with selecting different P value cutoffs.
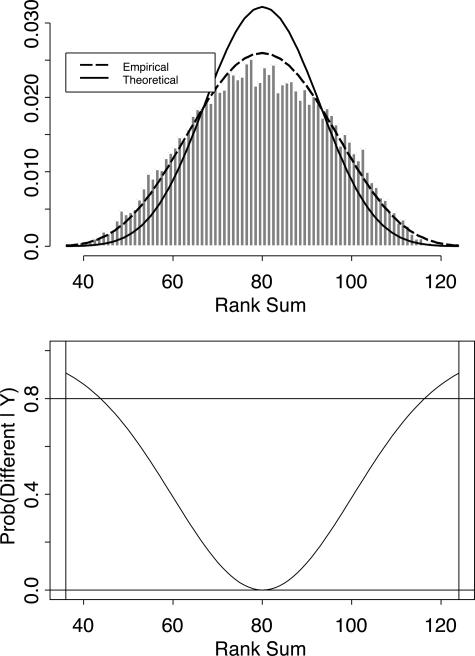
Finally, we computed Wilcoxon rank-sum statistics for all 16,733 probe sets. With 8 mutated and 11 unmutated samples, the possible rank-sum statistics range from 36 to 124, with a median of 80. We summarized the statistics by preparing a histogram of the number of times each rank sum was observed (Figure 3, top). The ratio of the theoretical Wilcoxon distribution to the observed distribution was fit using Poisson regression.9 Given a prior estimate, p0, of the number of genes that are not different between the two groups of samples, we computed the posterior probability that an observed rank-sum represented a differentially expressed gene (Figure 3, bottom). We chose the smallest prior proportion (p0 = 0.81) that ensured that all estimates of posterior probability remained non-negative. We selected genes as differentially expressed if their posterior probability was at least 80%. Using this criterion, we found 80 differentially expressed genes. Of these, 56 were overexpressed in unmutated samples, and 24 were overexpressed in mutated samples.
Figure 3.
Analysis of the Wilcoxon rank-sum statistics of 16,733 probe sets using an empirical Bayes method. Top: Histogram of the empirically observed distribution of rank-sum statistics, with an overlaid curve representing the theoretical distribution. Bottom: Posterior probability that an observed rank sum represents a differentially expressed gene.
Comparison between the Three Statistical Methods
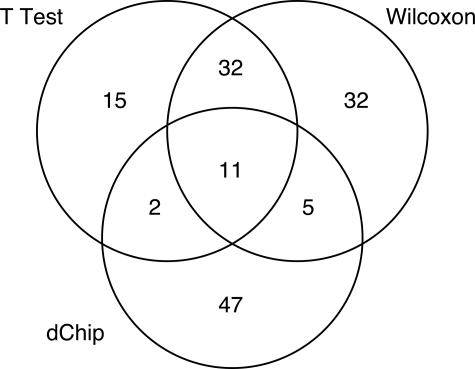
All three methods found more genes overexpressed in the unmutated samples than in the mutated samples. However, they identified different sets of differentially expressed genes. We have summarized the agreement between the three methods in a Venn diagram (Figure 4). Taken together, the methods identified 144 different probe sets as being differentially expressed, but only 11 of these probe sets were identified by all three methods. The 11 genes identified by all three statistical methods are listed in Table 1. None of these 11 genes have been reported previously to be differentially expressed in a microarray study of CLL. The mRNA for the gene ETV5 has been reported to be elevated in CLL patients,11 and the synthesis of GMP may be altered in CLL cells compared with normal peripheral blood lymphocytes.12,13 A complete list of all 144 genes is contained in Supplementary Table S1 at http://jmd.amjpathol.org/.
Figure 4.
Venn diagram showing the level of agreement between three different statistical methods for selecting differentially expressed genes from the same data set.
Table 1.
Genes Identified as Differentially Expressed between Mutated and Unmutated Cases of CLL by All Three Statistical Methods
| Probe set | t Statistic* | Gene | Description |
|---|---|---|---|
| 203348_s_at | −5.52 | ETV5 | ets variant gene 5 (ets-related molecule) |
| 205383_s_at | −4.75 | ZNF288 | Zinc finger protein 288 |
| 202150_s_at | −4.51 | NEDD9 | Neural precursor cell expressed, developmentally down-regulated 9 |
| 209155_s_at | 4.17 | NT5C2 | 5′-Nucleotidase, cytosolic II |
| 209186_at | 4.19 | ATP2A2 | ATPase, Ca2+ transporting, cardiac muscle, slow twitch 2 |
| 207668_x_at | 4.20 | TXNDC7 | Thioredoxin domain containing 7 |
| 212442_s_at | 4.78 | LASS6 | Longevity assurance homolog 6 |
| 203593_at | 4.95 | CD2AP | CD2-associated protein |
| 218029_at | 5.13 | FLJ13725 | Hypothetical protein FLJ13725 |
| 201088_at | 5.24 | KPNA2 | Karyopherin α2 (RAG cohort 1, importin α1) |
| 212652_s_at | 6.05 | SNX4 | Sorting nexin 4 |
Negative t statistics identify genes that are overexpressed in mutated samples; positive t statistics identify genes that are overexpressed in unmutated samples.
The best agreement between methods was between the t-test and the Wilcoxon test, which identified 43 differentially expressed genes in common. This overlap represents 72% of all 60 genes found by the t-test and 54% of all 80 genes found by the Wilcoxon test. In contrast, 47 (72%) of the 65 genes identified using dChip were not found by either of the other two methods.
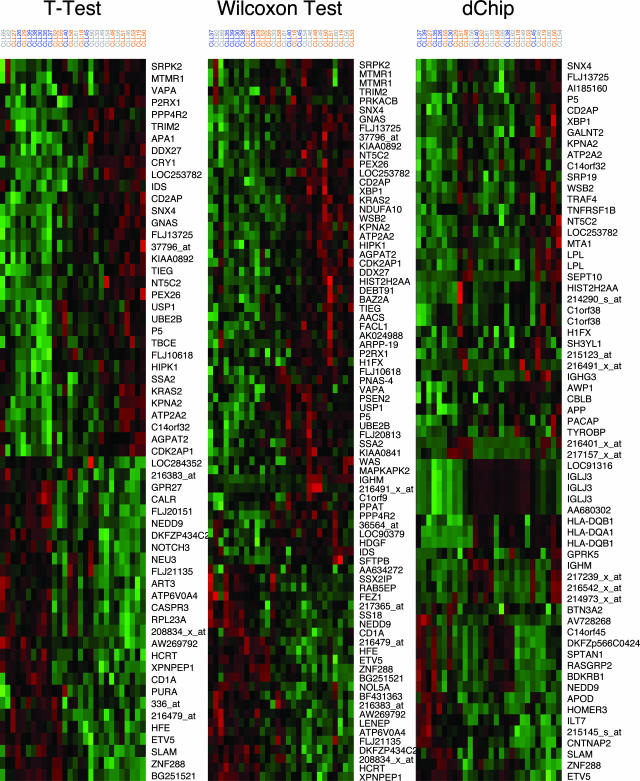
We performed two-way hierarchical clustering on the samples using each of the three sets of genes (Figure 5).14 For this analysis, both the CLL samples and the genes were clustered using average linkage and a distance metric based on the Pearson correlation coefficient between the log-transformed intensities. For display purposes only, we standardized the log-transformed intensities for each gene by subtracting the mean across the samples and dividing by the SD
Figure 5.
Results of two-way clustering of 28 samples using the genes found to be differentially expressed using three different statistical methods. The samples include 8 mutated samples (blue), 11 unmutated samples (orange), and 9 samples whose status was unknown (gray). Each row contains standardized log expression values for one gene.
Validation of Differentially Expressed Genes by QRT-PCR Assays
We selected 12 genes identified as differentially expressed by the different statistical methods in the training set of 19 cases for validation using QRT-PCR assays. To get the best test of the reliability of the methods, we only validated genes that were found by dChip but not by the t-test or Wilcoxon test, or vice versa. Because the agreement between the t-test and Wilcoxon test was so strong, we combined their results when selecting genes for validation. We investigated six genes selected by each test, three overexpressed and three underexpressed in mutated cases of CLL, in a randomly selected subset of the samples that contained four mutated and four unmutated CLL cases.
The results are summarized in Table 2. Ten of the 12 genes were successfully validated using QRT-PCR, that is, 1) the log-transformed measurements of gene expression levels determined using the microarrays were positively correlated with the QRT-PCR measurements, and 2) the sign of the t statistic computed using the QRT-PCR data agreed with the direction of expression change found in the microarray data. Of the two genes that failed to be validated by QRT-PCR, one (FSTL3) had been identified by the t-test or Wilcoxon test, and one (SPTAN1) had been identified by dChip.
Table 2.
Validation of Microarray Results by QRT-PCR
| Probe set | Gene | Array t stat.* | LBFC* | UBFC* | Subset t stat.* | QRT-PCR t stat.* | Correlation* |
|---|---|---|---|---|---|---|---|
| 212505_s_at | KIAA0892 | 4.07 | 1.06 | 1.24 | 2.12 | 0.87 | 0.17 |
| 210678_s_at | AGPAT2 | 4.30 | 1.18 | 1.61 | 3.00 | 1.96 | 0.51 |
| 219108_x_at | DDX27 | 4.76 | 1.07 | 1.28 | 3.64 | 1.44 | 0.42 |
| 206948_at | NEU3 | −4.67 | −1.07 | −1.26 | −8.18 | −2.88 | 0.79 |
| 203592_s_at | FSTL3 | −3.93 | −1.06 | −1.24 | −2.59 | 1.80 | −0.78 |
| 200874_s_at | NOL5A | −3.63 | −1.07 | −1.20 | −2.61 | −2.64 | 0.60 |
| 202871_at | TRAF4 | 2.37 | 1.21 | 1.90 | 2.71 | 1.51 | 0.12 |
| 203508_at | TNFRSF1B | 2.39 | 1.24 | 2.47 | 2.73 | 2.53 | 0.85 |
| 212999_x_at | HLA− DQB1 | 2.61 | 1.62 | 5.99 | 0.65 | 0.81 | 0.93 |
| 214925_s_at | SPTAN1 | −3.11 | −1.37 | −3.85 | −8.01 | 1.21 | −0.25 |
| 209846_s_at | BTN3A2 | −2.90 | −1.21 | −1.91 | −2.48 | −0.50 | 0.27 |
| 201525_at | APOD | −2.26 | −1.26 | −4.28 | −1.63 | −0.92 | 0.86 |
Array t stat., the t-statistic based on 19 training set microarray experiments; LBFC, the lower bound of the 90% confidence intervals of fold change as estimated by dChip; UBFC, the upper bound of the 90% confidence intervals of fold change as estimated by dChip; Subset t stat.: the t statistic based on the microarray data for the eight samples selected for QRT-PCR; PCR t stat.: the t statistic based on the QRT-PCR data; Correlation, the Pearson correlation coefficient between the microarray and the QRT-PCR data.
Negative t statistics identify genes that are overexpressed in mutated samples; positive t statistics identify genes that are overexpressed in unmutated samples.
Validation of Sets of Selected Genes by Predicting Mutation Status in an Independent Data Set
In addition to the 19 CLL samples in the training set, we performed microarray experiments on nine additional CLL samples whose VH mutation status was unknown at the time the clustering analysis was performed. These samples clustered in different ways depending on the set of genes selected as differentially expressed (Figure 5). We built models to predict the mutation status of the new samples separately for each method of gene selection. Then we performed sequence analysis to determine the mutation status of these nine additional samples and to validate the accuracy of the predictions. Sequence analysis demonstrated that seven were mutated and two were unmutated.
To predict the status of new samples, we performed LDA using the first few principal components derived from each set of selected genes. For each method, we chose the minimal number of principal components needed to explain 80% of the variation. This rule required four principal components for the t-test and Wilcoxon test, and five principal components for dChip. The analysis correctly predicted the mutation status of seven of nine samples (78%) using the Wilcoxon test, six of nine samples (67%) using the t-test, and five of nine samples (56%) using dChip. We also found that the number of principal components that we used for the analysis maximized the prediction accuracy of each method. In several cases, using fewer principal components caused the method to incorrectly predict the status of 1 or 2 of the training samples. In every case, using more principal components reduced the prediction accuracy on the validation set.
Validation of Differential Expression Using Additional Microarrays
Finally, we looked at all 144 genes that were found to be differentially expressed by at least one method using the original training set of 19 CLL samples. Our goal was to quantify the number of genes that still appeared to be differentially expressed after incorporating the data from the nine additional samples. We observed that many of the genes showed some evidence of differential expression by all three methods, regardless of the method used to select them. For example, for 136 genes, the unadjusted P value for the t-test was P < 0.05, and 141 genes had a lower bound of fold change >1. Based on this observation, we determined the number of genes that satisfied similar criteria for each method, both on the training set and on the full data set (Table 3). When we used stringent criteria for reproducibility, ie, a gene had to satisfy the same criterion on the full data set that was used to select it from the training set, the confirmation rates were modest (41.7% for the t-test, 36.3% for the Wilcoxon test, and 36.9% for dChip). When we used less stringent criteria, the confirmation rates were high (85.5% by the t-test, 86.8% by the Wilcoxon test, and 80.9% by dChip). The latter confirmation rates are compatible with the validation by QRT-PCR, which validated 10 of 12 selected genes (83.3%).
Table 3.
Number of Genes (out of 144) Satisfying Various Criteria in the Training Set of 19 Samples and Confirmed in the Full Data Set of All 28 Samples
| Criterion | Training samples | Confirmed (%) |
|---|---|---|
| t-test, P < 0.10 | 138 | 118 (85.5) |
| t-test, P < 0.05 | 136 | 111 (81.6) |
| t-test, P < 0.0009 | 60 | 25 (41.7) |
| Wilcoxon, P < 0.10 | 136 | 118 (86.8) |
| Wilcoxon, P < 0.05 | 129 | 101 (78.3) |
| Wilcoxon, P < 0.015 | 80 | 29 (36.3) |
| LBFC > 1.0 | 141 | 114 (80.9) |
| LBFC > 1.1 | 106 | 56 (52.8) |
| LBFC > 1.2 | 71 | 32 (45.1) |
| LBFC > 1.0 and D > 100 | 85 | 44 (51.8) |
| LBFC > 1.1 and D > 100 | 78 | 36 (46.2) |
| LBFC > 1.2 and D > 100 | 65 | 24 (36.9) |
LBFC, lower bound of fold change based on dChip 90% confidence interval; D, difference of mean expression between mutated and unmutated samples.
Discussion
We applied three different established statistical methods to microarray data to identify differentially expressed genes in a training set of 19 CLL samples, 11 with unmutated Ig VH genes and 8 with mutated Ig VH genes. Our analysis demonstrated that the three methods (the statistical model used by dChip, the t-test with correction for multiple testing using a β-uniform mixture model, and the Wilcoxon rank-sum test with correction for multiple testing using an empirical Bayes model) produced very different lists of differentially expressed genes. Of the 144 genes that were identified as differentially expressed by at least one method, only 11 were identified as differentially expressed by all three methods. Unsupervised clustering of the samples using genes selected by the different methods displayed different structures, and predictions of the mutation status on a blinded, independent set of nine CLL samples gave different results.
There are two broad classes of statistical methods: parametric and nonparametric. Parametric methods model the data using distributions specified by a small number of parameters. Nonparametric methods do not make the same kinds of distributional assumptions. The two parametric methods that we applied, dChip and t-test, use different error models. The model used by dChip to compute a confidence interval for estimates of fold change assumes that the errors are normally distributed on the original measurement scale. In contrast, the model that underlies the application of the t-test after log transformation assumes that the measurement errors are normal on the transformed scale. It is unlikely that both assumptions can be true simultaneously. The Wilcoxon test is a nonparametric method; it gives results that do not depend on the choice of a measurement scale. On our data, the most accurate predictions were achieved using genes selected by the nonparametric Wilcoxon test. The least accurate predictions were achieved using genes selected by dChip. Genes selected by the t-test gave intermediate accuracy, and this set of genes showed substantial agreement with the list produced by the Wilcoxon test. This finding suggests that neither parametric model perfectly describes the data, but that the t-test error model is closer to the truth than the dChip error model.
The best models of microarray data may ultimately include both additive errors (normal on the original scale) and multiplicative errors (normal on the log scale). There are, after all, numerous sources of variability in microarray studies. One source arises from the technology: when dChip quantifies gene expression using multiple probes in a probe set on a single array, the method estimates the technological variability. This technological variability could well be normally distributed on the original scale. However, a second and critically important source of variability across multiple microarrays is biological. The dChip model assumes that the biological variability is also normal on the original scale. Models used in the analysis of spotted cDNA arrays, like the t-test applied in this paper, have typically assumed that the biological variability is normal on the log scale. The first model of cDNA microarray data that explicitly incorporated both additive and multiplicative error terms was constructed by Rocke and Durbin.15 Their ideas were later extended to develop variance-stabilizing transformations for microarray data.16,17 Related ideas have been developed in a β-binomial model for microarray data with replications.18 To our knowledge, no one has yet applied similar ideas to the analysis of oligonucleotide array data.
Despite the differences in prediction accuracy, attempts to validate the differential expression of individual genes were equally successful regardless of the statistical method used to select the genes. Semiquantitative real-time PCR assays confirmed the differential expression of five of six genes found only by dChip and five of six genes found only by the t-test or Wilcoxon test. Validation by investigating the full set of 28 microarrays also had similar success rates regardless of the method applied. This finding suggests that all three methods can successfully identify individual genes that are differentially expressed and that a comprehensive list of all differentially expressed genes can only be obtained by combining the results of multiple statistical tests.
The 11 genes that were identified by all three statistical methods include several genes whose known functions suggest that they may play an important role in the biology of CLL. The genes overexpressed in unmutated cases of CLL include Karyopherin α2 (KPNA2) and CD2-associated protein (CD2AP). KPNA2 (also known as RCH1) is a nuclear transport protein that binds to the nuclear localization signal of several proteins and escorts them into the nucleus. KPN2A has been shown to bind RAG-1 and BSAP (Pax-5), proteins that are critical for B-cell development.19,20 CD2AP is an adapter protein that facilitates CD2 coupling to the actin cytoskeleton.21 CD2, a T-cell antigen that is aberrantly expressed by a subset of cases of CLL,22 is required for the molecular segregation that occurs at the contact site between the T-cell and the antigen-presenting cell and for full T-cell activation.21 The genes overexpressed in somatically mutated cases of CLL include NEDD9 and ZNF288. NEDD9 (also known as HEF1) is believed to be an important component in the cytoskeleton-linked signaling cascade. HEF1 is phosphorylated after ligation of β1 integrin or the B-cell receptor.23 ZNF288 (also known as DPZF) is a zinc finger protein that is highly homologous to BCL6 and is also located on the long arm of chromosome 3.24 It is expressed by dendritic cells, monocytes, B cells, T cells, and B-cell lymphoma cell lines.
The joint list of 144 differentially expressed genes is notable not only for what it includes, but for what it omits. Neither CD38 nor ZAP70 was found to be differentially expressed in the initial data set of 19 samples. Initial reports suggested that CD38 was strongly associated with mutation status in CLL patients,2 but later reports suggested instead that CD38 was an independent prognostic factor.25 Our findings support the latter interpretation. The t statistic for CD38 was t = −0.38 (P = 0.71) on the original 19 samples, and dChip estimated the fold change (FC) to be FC = −1.07 (90% CI = (−0.93, −1.22)). The results on the full set of 28 samples were comparable. ZAP70 has also been reported as strongly differentially expressed between mutated and unmutated CLL samples.26,27 ZAP70 is a T-cell/NK-cell signaling molecule that is expressed by the majority of cases of CLL with unmutated VH genes. In the original set of 19 samples, we found slight evidence of overexpression in the unmutated cases (t = 2.15, P = 0.046; FC = 1.71, 90% CI = (1.16, 2.61)) that was inadequate to satisfy any of our selection criteria. When we analyzed the full set of 28 microarrays, however, the evidence for differential expression of ZAP70 was much stronger (t = 3.33, P = 0.0027; FC = 1.70, 90% CI = (1.30, 2.32)). Although this finding supports the differential expression of ZAP70, it suggests that the magnitude of differential expression may be smaller or less consistent than previously reported.
Supplementary Material
Footnotes
Supported by a grant from the Commonwealth Foundation for Cancer Research and Mr. and Mrs. William H. Goodwin, Jr. The Microarray Core Facility is supported by Cancer Center Support grant 16672.
References
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application, Genome Biol. 2001;2:research0032.1–research0032.11. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, Buchbinder A, Budman D, Dittmar K, Kolitz J, Lichtman SM, Schulman P, Vinciguerra VP, Rai KR, Ferrarini M, Chiorazzi N. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;15:1848–1854. [PubMed] [Google Scholar]
- McCarthy H, Wierda WG, Barron LL, Cromwell CC, Wang J, Coombes KR, Rangel R, Elenitoba-Johnson KSJ, Keating MJ, Abruzzo LV. High expression of activation-induced cytidine deaminase (AID) and splice variants is a distinctive feature of poor prognosis chronic lymphocytic leukemia. Blood. 2003;101:4903–4908. doi: 10.1182/blood-2002-09-2906. [DOI] [PubMed] [Google Scholar]
- Gold D, Coombes K, Medhane D, Ramaswamy A, Ju Z, Strong L, Koo JS, Kapoor M. A comparative analysis of data generated using two different target preparation methods for hybridization to high-density oligonucleotide microarrays. BMC Genomics. 2004;5:2. doi: 10.1186/1471-2164-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pounds S, Morris SW. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics. 2003;19:1236–1242. doi: 10.1093/bioinformatics/btg148. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Efron B, Tibshirani R. Empirical bayes methods and false discovery rates for microarrays. Genet Epidemiol. 2002;23:70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. DNA-Chip Analyzer (dChip). Parmigiani G, Garrett ES, Irizarry R, Zeger SL, editors. New York: Springer-Verlag,; The analysis of gene expression datamethods and software. 2003:120–141. [Google Scholar]
- Korz C, Pscherer A, Benner A, Mertens D, Schaffner C, Leupolt E, Dohner H, Stilgenbauer S, Lichter P. Evidence for distinct pathomechanisms in B-cell chronic lymphocytic leukemia and mantle cell lymphoma by quantitative expression analysis of cell cycle and apoptosis-associated genes. Blood. 2002;99:4554–4561. doi: 10.1182/blood.v99.12.4554. [DOI] [PubMed] [Google Scholar]
- Carpentieri U, Monahan TM, Gustavson LP. Observations on the level of cyclic nucleotides in three population of human lymphocytes in culture. J Cyclic Nucleotide Res. 1980;6:253–259. [PubMed] [Google Scholar]
- Carlucci F, Tabucchi A, Pagani R, Marinello E. Synthesis of adenine and guanine nucleotides at the ‘inosinic branch point’ in lymphocytes of leukemia patients. Biochim Biophys Acta. 1999;1454:106–114. doi: 10.1016/s0925-4439(99)00032-0. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocke DM, Durbin B. A model for measurement error for gene expression arrays. J Comput Biol. 2001;8:557–569. doi: 10.1089/106652701753307485. [DOI] [PubMed] [Google Scholar]
- Durbin BP, Hardin JS, Hawkins DM, Rocke DM. A variance-stabilizing transformation for gene-expression microarray data. Bioinformatics. 2002;18 (Suppl 1):S105–S110. doi: 10.1093/bioinformatics/18.suppl_1.s105. [DOI] [PubMed] [Google Scholar]
- Geller SC, Gregg JP, Hagerman P, Rocke DM. Transformation and normalization of oligonucleotide microarray data. Bioinformatics. 2003;19:1817–1823. doi: 10.1093/bioinformatics/btg245. [DOI] [PubMed] [Google Scholar]
- Baggerly KA, Coombes KR, Hess KR, Stivers DN, Abruzzo LV, Zhang W. Identifying differentially expressed genes in cDNA microarray experiments. J Comput Biol. 2001;8:639–659. doi: 10.1089/106652701753307539. [DOI] [PubMed] [Google Scholar]
- Cuomo CA, Kirch SA, Gyuris J, Brent R, Oettinger MA. Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc Natl Acad Sci USA. 1994;91:6156–6160. doi: 10.1073/pnas.91.13.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovac CR, Emelyanov A, Singh M, Ashouian N, Birshtein BK. BSAP (Pax5)-importin alpha 1 (Rch1) interaction identifies a nuclear localization sequence. J Biol Chem. 2000;275:16752–16757. doi: 10.1074/jbc.M001551200. [DOI] [PubMed] [Google Scholar]
- Zaru R, Cameron TO, Stern LJ, Muller S, Valitutti S. TCR engagement and triggering in the absence of large-scale molecular segregation at the T cell-APC contact site. J Immunol. 2002;168:4287–4291. doi: 10.4049/jimmunol.168.9.4287. [DOI] [PubMed] [Google Scholar]
- Kampalath B, Barcos MP, Stewart C. Phenotypic heterogeneity of B cells in patients with chronic lymphocytic leukemia/small lymphocytic lymphoma. Am J Clin Pathol. 2003;119:824–832. doi: 10.1309/4AGU-T3LK-EURD-7T7K. [DOI] [PubMed] [Google Scholar]
- Manie SN, Beck AR, Astier A, Law SF, Canty T, Hirai H, Druker BJ, Avraham H, Haghayeghi N, Sattler M, Salgia R, Griffin JD, Golemis EA, Freedman AS. Involvement of p130(Cas) and p105(HEF1), a novel Cas-like docking protein, in a cytoskeleton-dependent signaling pathway initiated by ligation of integrin or antigen receptor on human B cells. J Biol Chem. 1997;272:4230–4236. doi: 10.1074/jbc.272.7.4230. [DOI] [PubMed] [Google Scholar]
- Zhang W, Mi J, Li N, Sui L, Wan T, Zhang J, Chen T, Cao X. Identification and characterization of DPZF, a novel human BTB/POZ zinc finger protein sharing homology to BCL-6. Biochem Biophys Res Commun. 2001;282:1067–1073. doi: 10.1006/bbrc.2001.4689. [DOI] [PubMed] [Google Scholar]
- Hamblin TJ, Orchard JA, Ibbotson RE, Davis Z, Thomas PW, Stevenson FK, Oscier DG. CD38 expression and immunoglobulin variable region mutations are independent prognostic variables in chronic lymphocytic leukemia, but CD38 expression may vary during the course of the disease. Blood. 2002;99:1023–1029. doi: 10.1182/blood.v99.3.1023. [DOI] [PubMed] [Google Scholar]
- Chen L, Widhopf G, Huynh L, Rassenti L, Rai KR, Weiss A, Kipps TJ. Expression of ZAP-70 is associated with increased B-cell receptor signaling in chronic lymphocytic leukemia. Blood. 2002;100:4609–4614. doi: 10.1182/blood-2002-06-1683. [DOI] [PubMed] [Google Scholar]
- Wiestner A, Rosenwald A, Barry TS, Wright G, Davis RE, Henrickson SE, Zhao H, Ibbotson RE, Orchard JA, Davis Z, Stetler-Stevenson M, Raffeld M, Arthur DC, Marti GE, Wilson WH, Hamblin TJ, Oscier DG, Staudt LM. ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile. Blood. 2003;101:4944–4951. doi: 10.1182/blood-2002-10-3306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.