Abstract
Mutations in the dystrophin gene result in both Duchenne and Becker muscular dystrophies (DMD and BMD). Approximately two-thirds of the affected patients have large deletions or duplications. Using the multiplex polymerase chain reaction and Southern blotting techniques, the detection of these larger mutations is relatively straightforward. Detection of the point mutations in the remaining one-third of the patients has been challenging, mainly due to the large gene size and lack of hotspots or prevalent mutations. However, with the addition of some of the newer molecular screening methods, it is becoming more feasible for clinical laboratories to test for point mutations in the larger genes like dystrophin. Here we review the clinical features, describe the mutation distributions, evaluate current molecular strategies, and illustrate how the genetic findings have impacted the current clinical diagnostics of Duchenne and Becker muscular dystrophies.
Duchenne and Becker muscular dystrophies (DMD and BMD) are X-linked, allelic, neuromuscular diseases characterized by progressive muscular weakness and degeneration of skeletal muscle. DMD is the most common X-linked recessive lethal disease with an incidence of ∼1 in 3500 newborns, and it has been estimated that approximately one third of the cases result from new mutations.1,2 Clinical symptoms of the disease are observed between 2 and 3 years of age. Most affected boys exhibit retarded motor development, with approximately half of them failing to walk until the age of 18 months. Other early onset characteristics include an unusual waddling gait, difficulties with running and jumping, lumbar lordosis, and calf enlargement.1,3 Weakness and wasting of muscle are progressive and symmetrical, affecting the lower limbs before the upper limbs and the proximal muscles before the distal muscles. Joint contractures are an important clinical manifestation, and by the age of 6 years most patients have contractures at the iliotibial bands, hip joints, and heel cords. Regenerating fibers becomes less frequent as the disease progresses and are eventually replaced by adipose and connective tissues, accounting for the pseudohypertrophic muscles. The affected children are usually wheelchair-bound by the age of 12 years. As the disease progresses, the contractures increasingly develop, leading to the asymmetrical spinal deformities.3 Most patients die at approximately the age of 20 of pneumonia related to chronic respiratory insufficiency.
Cardiac involvement is a consistent part of DMD. As many as 90% of DMD patients demonstrate electrocardiogram abnormalities.4 The heart exhibits fibrosis in the posterobasal portion of the left ventricular wall. Defects in the intra-atrial conduction system are more common than atrioventricular and infranodal disturbances. Despite known cardiac disease, most patients with DMD remain surprisingly free of cardiovascular symptoms.
Approximately 20% of affected patients will be mentally handicapped. The impairment of intellectual function appears to be nonprogressive and affects verbal ability more than performance.5 The cognitive impairment cannot be attributed solely to physical limitations, as similarly handicapped patients with spinal muscular atrophy do not have impaired intelligence. The neuropathological correlate for mental retardation in DMD has not been established; however, a specific isoform of the DMD protein has been shown to be expressed in the brain.
The allelic disorder BMD has a milder clinical course and a slower disease progression.6 BMD has been estimated to occur approximately one-tenth as frequently as DMD, with an incidence of ∼3 per 100,000 newborns. The majority of BMD patients initially experience difficulties between 5 and 15 years of age, although onset in the 3rd or 4th decade, or even later, can occur. By definition the affected patients remain ambulant until 16 years of age or later, thus allowing the clinical distinction from patients with DMD. Patients with BMD have a reduced life expectancy, but the majority of patients survive into at least the 4th or 5th decade. A well-recognized subgroup of patients with an intermediate course between those typical of Duchenne and Becker dystrophies are referred to as intermediate patients or outliers.7 These patients can usually be recognized by the age of 3 years by the relative preservation of strength in neck flexion (anti-gravity neck flexor muscles), whereas patients with DMD lack this ability throughout their entire life. The intermediate patients retain the ability to climb stairs and walk (after the age of 12 but not beyond 15 years) longer than patients with typical DMD.
Gene Studies
The DMD gene is one of the largest human genes identified, spanning more than 2000 kb of genomic DNA, and is composed of 79 exons that encode a 14-kb transcript that is translated into the protein dystrophin.8,9 Analysis of the amino acid sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Four distinct domains have been defined: 1) an amino terminus that associates with actin or an actin-like protein, 2) a rod domain consisting of long flexible rows of 24 α-helical repeats, 3) a cysteine-rich region, and 4) a unique carboxy terminus. Dystrophin tightly associates with a large oligomeric complex of sarcolemmal glycoproteins through its cysteine-rich domain and carboxy-terminus whereas the amino-terminal domain interacts with actin or an actin-like protein.10,11,12
By immunochemistry, dystrophin has been shown to be on the cytoplasmic face of the muscle cell membrane and at postsynaptic membrane specializations in neurons. Dystrophin comprises only 0.002% of total muscle protein but up to 5% of the membrane skeleton. Dystrophin is found in skeletal muscle, smooth muscle, cardiac muscle, and brain. There are slightly different forms of dystrophin mRNA in different tissues due to different transcription start sites and alternative splicing.13,14,15 Dystrophin’s exact function is not known, but it may be important in maintaining muscle membrane stability. Patients with DMD have very little or no detectable dystrophin whereas BMD patients have dystrophin of altered size and/or less quantity.16 However disease cause may be more complex than a simple loss of dystrophin. Studies have shown that several of the glyco-proteins that interact with dystrophin are also absent in these disorders.17,18,19 The dystrophin-associated proteins may be directly involved with the calcium flux in the dystrophic fibers. Thus, the loss of dystrophin may be the first of many steps that ultimately lead to muscular dystrophy.
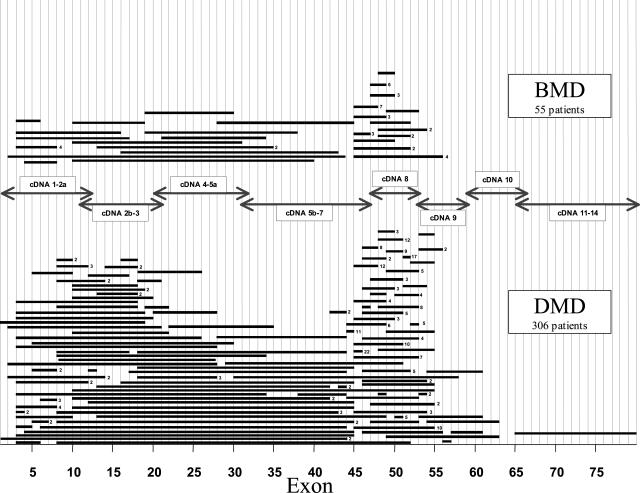
It has been observed that ∼60 to 65% of the mutations that cause DMD/BMD are large deletions in the dystrophin gene.20,21 The distribution of deletions within the DMD gene of DMD/BMD patients studied at The Ohio State University are shown in Figure 1. Blood specimens were obtained from affected patients who had been referred to the Ohio State Molecular Pathology Laboratory. The probands were diagnosed by standard clinical diagnostic criteria, including elevated creatine kinase levels and myopathic changes detectable on muscle biopsy. The deletions are nonrandomly distributed and occur primarily in the center (∼80%) and less frequently near the 5′ end (∼20%) of the gene. The 200-kb region covering intron 44, exon 45, and intron 45 is the major deletion breakpoint region of the gene. The majority of the larger deletions initiate at the 5′ end of the gene. The distribution of deletions (Figure 1) has been demonstrated in many populations and ethnic groups.
Figure 1.
Distribution of deletions in the dystrophin gene in DMD and BMD patients. Each bar represents a deletion observed in a patient. The number to the right of the deletion bar indicates the number of independent patients sharing deletions of the same exon. Analysis was performed by Southern blotting and multiplex PCR. Arrows indicate the cDNA probes used for Southern blotting.
There is no apparent correlation between the size or location of the deletion and the severity or progression of the disorder. One of the largest deletions (35 exons) we have identified is in a mild BMD patient. Furthermore, sequences deleted in DMD patients overlap with deletions in BMD patients. However, it was proposed that if a deletion disrupts the translational reading frame of the dystrophin mRNA triplet codons, then a C-terminally truncated nonfunctional protein will be synthesized, resulting in the more severe DMD.22 In the milder BMD, the deletion maintains the translational reading frame, and a semifunctional protein is produced. The reading frame hypothesis explains the phenotypic differences observed in ∼90% of the DMD/BMD cases. One major exception to the reading frame hypothesis has been the identification of BMD patients with the out-of-frame exon 3 to 7 deletion.23 It has been proposed that an alternate splicing mechanism or a new cryptic translational start site may account for the production of protein and the milder phenotype in these patients. A small number of DMD patients with in-frame deletions have also been identified. The more severe phenotype in these patients may be due to the overall effect of the deletion on the protein conformation or may be the result of message instability. We have found some phenotypic variability in several of our patients who share identical gene deletions. The out-of-frame deletion of exon 45, one of the most commonly observed DMD deletions, has also been associated with BMD phenotypes.24 Some genetic variability may be due to modifier genes that affect splicing or other molecules involved in destruction of damaged muscle fibers, muscular regeneration, or in the cellular response to different hormones.
The large gene size, particularly the introns that average 35 kb, may account for part of the high deletion rate. However, in addition to target size, other factors must be involved. The observed nonrandom deletion pattern may reflect domain-associated variation in chromosomal stability. For instance, complications related to the maintenance of replication, correct transcription, and proper splicing of such a large gene may play an extremely important role.
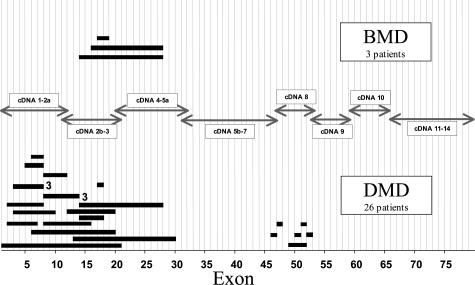
Partial gene duplications have been revealed in ∼5 to 10% of patients.25 Unlike the deletion distribution, we have found ∼80% of the duplications at the 5′ end of the gene and only 20% in the central region (Figure 2). The duplication distribution, like the deletion distribution, has also been demonstrated in different populations and ethnic groups. Out-of-frame duplications in DMD patients and in-frame duplications in BMD patients have been observed, suggesting that the reading frame hypothesis also holds true for duplications.25
Figure 2.
Distribution of duplications in the dystrophin gene in DMD and BMD patients. Each bar represents a duplication observed in a patient. The number right of the duplication bar indicates the number of independent patients sharing duplications of the same exon. Analysis was performed by Southern blotting. Arrows indicate the cDNA probes used for Southern blotting.
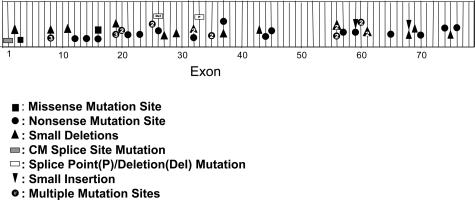
There are now reports of small mutations (point mutations and small deletions and duplications) detected in the dystrophin gene in the remaining DMD and BMD patients without large deletions or duplications.26,27 The distribution of small types of mutations within the dystrophin gene of DMD and BMD patients studied at The Ohio State University is shown in Figure 3 and Table 1. The majority of these mutations are unique to single or a few patients and result in truncated dystrophins lacking part or all of the C-terminus as a consequence of nonsense or frameshifting mutations. The truncated proteins are presumably unstable, and little or no dystrophin is produced. Although the point mutations are consistent with the reading frame hypothesis described for deletions and duplications, these types of mutations provide little information on structural/functional relationships in the dystrophin protein. Missense mutations are rare in the dystrophin gene, even in the milder BMD patients. Although several base changes causing significant amino acid substitutions have been reported in the dystrophin gene, the majority of these are polymorphic changes. The identification of mutations that do not cause protein truncation provides further insight into the function of dystrophin as well as defining the essential regions and conformations necessary for dystrophin stability. DMD missense mutations, which we previously described in exons 3 and 16, have supported the important role of an intact actin-binding domain and the proper conformation of the rod domain for dystrophin function.27
Table 1.
Summary of Small Mutations in the Dystrophin Gene
| Mutation | Phenotype | Exon | Description | Translational effect |
|---|---|---|---|---|
| 1 | CM | Prom + ex1 | IVS1 + 1 G>T splice point mutation | Splice site mutation |
| 2 | IMD | 2 | 58 del A | Frameshift |
| 3 | DMD | 3 | 161 T>G | Leu > Arg |
| 4 | DMD | 8 | 709 C>T | Gln > term |
| 5 | DMD | 8 | 721 C>T | Gln > term |
| 6 | DMD | 8 | 724 C>T | Gln > term |
| 7 | DMD | 8 | 775 del A | Frameshift |
| 8 | DMD | 11 | 1235 del T | Frameshift |
| 9 | IMD | 12 | 1438 G>T | Gly > term |
| 10 | DMD | 14 | 1702 C>T | Gln > term |
| 11 | DMD | 16 | 1934 A>G | Asp > Gly |
| 12 | DMD | 16 | 1974 del T | Frameshift |
| 13 | DMD | 19 | 2302 C>T | Arg > term |
| 14 | DMD | 19 | 2314 G>T | Glu > term |
| 15 | DMD | 19 | 2359 del C | Frameshift |
| 16 | DMD | 19 | 2368 C>T | Gln > term |
| 17 | DMD | 20 | 2475 G>A | Trp > term |
| 18 | DMD | 20 | 2512 C>T | Gln > term |
| 19 | DMD | 21 | 2788 A>T | Lys > term |
| 20 | DMD | 23 | 2954 T>A | Leu > term |
| 21 | DMD | 25 | 3304 C>T | Gln > term |
| 22 | BMD | 25 | 3413 G>A | Trp > term |
| 23 | DMD | 26 | IVS 25 to 8 to 3434 del TGTGGAAG/GT ins GTTT (10 base deletion, 4 base insertion, splice site is in the deletion) | Splice site mutation |
| 24 | DMD | 26 | 3523 C>T | Gln > term |
| 25 | DMD | 27 | 3630 del A | Frameshift |
| 26 | DMD | 29 | 4031 del 13 | Frameshift |
| 27 | DMD | 32 | 4375 C>T | Arg > term |
| 28 | DMD | 32 | 4414 del C | Frameshift |
| 29 | DMD | 32 | 4470 del AA | Frameshift |
| 30 | DMD | 33 | IVS 33 + 1 G>A | Splice site mutation |
| 31 | DMD | 35 | 4933 A>T | Lys > term |
| 32 | DMD | 35 | 5009 G>A | Trp > term |
| 33 | DMD | 37 | 5266 C>T | Gln > term |
| 34 | DMD | 37 | 5307 del C | Frameshift |
| 35 | DMD | 43 | 6202 del C | Frameshift |
| 36 | DMD | 44 | 6373 C>T | Gln > term |
| 37 | DMD | 45 | 6544 C>T | Gln > term |
| 38 | DMD | 56 | 8267 del A | Frameshift |
| 39 | DMD | 56 | 8269 G>T | Glu > term |
| 40 | DMD | 56 | 8311 del G | Frameshift |
| 41 | DMD | 56 | 8371 A>T | Lys > term |
| 42 | DMD | 57 | 8443 C>T | Gln > term |
| 43 | BMD | 59 | 8713 C>T | Arg > term |
| 44 | DMD | 59 | 8782 ins A | Frameshift |
| 45 | DMD | 60 | 8944 C>T | Arg > term |
| 46 | DMD | 60 | 9072 G>A | Trp > term |
| 47 | DMD | 61 | 9090 del C | Frameshift |
| 48 | DMD | 61 | 9154 del TTTC | Frameshift |
| 49 | DMD | 65 | 9380 C>G | Ser > term |
| 50 | DMD | 68 | 9823 ins A | Frameshift |
| 51 | DMD | 68 | 9852 del 10 | Frameshift |
| 52 | DMD | 69 | 9976 del T | Frameshift |
| 53 | DMD | 70 | 10141 C>T | Arg > term |
| 54 | BMD | 74 | 10504 G>T | Glu > term |
| 55 | DMD | 75 | 10562 del A | Frameshift |
| 56 | DMD | 76 | 10903 C>T | Gln > term |
CM, cardiomyopathy patient.
All mutations were screened initially by denaturing high performance liquid chromatography and heteroduplex analysis. A final sequencing step was then utilized to confirm the nature of all the positive screening tests.
Unlike the deletion hotspots occurring in the central and 5′ portion of the gene, the distribution of small mutations are more randomly distributed throughout the gene sequence (Figure 3). However, whereas less than 3% of the deletions are found 3′ of exon 55, we have found more than 30% of the small mutations in this same region of the gene. The extent of protein truncation caused by the 3′ mutations does not determine the phenotype because even the exon 76 point mutation results in the more severe DMD phenotype.
Figure 3.
Distribution of point mutations in the dystrophin gene in DMD and BMD patients. The numbers indicate the number of independent patients sharing a mutation in the same exon. Table 1 specifies the actual nucleotide changes. CM, cardiomyopathy splice point mutation.
There have now been several cases of dystrophin gene mutations resulting in X-linked dilated cardiomyopathy (XLDC). XLDC is a familial heart disease that presents in young males as a rapidly progressive heart failure, without clinical signs of skeletal muscle disease. The XLDC mutations occur in the dystrophin muscle promoter region or exon 1, which results in phenotypic rescue by an alternative promoter in all tissues except the heart, thus causing a cardiac condition rather than a skeletal myopathy.28 We recently identified a mutation at the 5′ splice site of dystrophin intron 1 in an XLDC patient (Figure 3 and Table 1). This mutation was first reported by Milasin and colleagues,29 who showed that compensatory transcription in the muscle of other dystrophin isoforms were able to prevent the myopathy but that absence of these isoforms in the heart resulted in the dilated cardiomyopathy.
Molecular Diagnostics
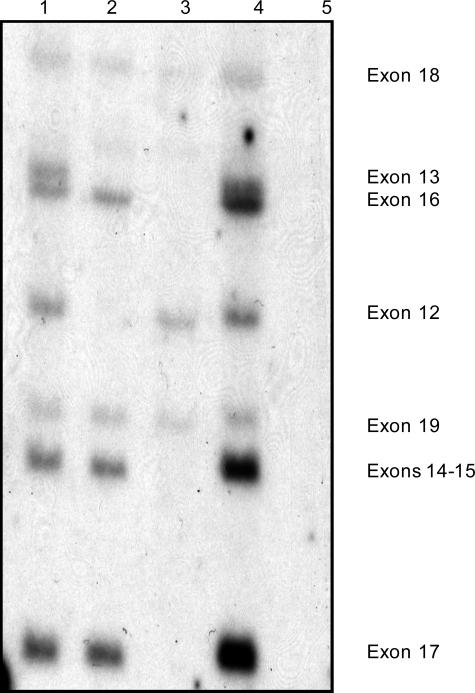
The analysis of gene mutations has greatly improved diagnosis, carrier detection, and genetic counseling. With the ability to identify deletions and duplications in ∼70% of affected patients, accurate direct DNA testing can be used for these cases. By using full-length dystrophin cDNA clones to probe Southern blots, it is possible to directly detect deletions and duplications. The cDNA probes detect the site of the mutation itself, so meiotic recombination events are irrelevant. Therefore, the chance of diagnostic error is greatly reduced. The digested and blotted DNA is sequentially hybridized with seven to nine cDNA probes, which cover the complete 14-kb transcript. Approximately 10 exons are scored for each cDNA hybridization. However, as shown in Figure 1, the deletions are primarily located in two hotspots; therefore, the majority of deletions can be identified by four cDNA probe hybridizations (1-2a, 2b-3, 5b-7, and 8). The deletions are simply detected by examination of Southern blots for the presence or absence of each exon containing genomic restriction fragments that hybridize to the cDNA probe (Figure 4). A male control is included on all Southern blots to show the proper location and intensity of the restriction fragments. A duplication is revealed by an increased hybridization intensity of one or more DNA fragments when compared to the male control (Figure 4). Duplications should always be confirmed using a second different restriction enzyme digestion, and the autoradiogram should be scanned by densitometry.
Figure 4.
Southern hybridization using a dystrophin cDMD probe that hybridizes to exons 12 to 19 (DNA digested with HindIII). Lane 1 is an unaffected male control. The DMD patient in lane 2 has a gene deletion of exons 12 to 13. The DMD patient in lane 3 has a gene deletion of exons 13 to 17. The DMD patient in lane 4 has a gene duplication of exons 14 to 17. The DMD patient in lane 5 has a gene deletion of exons 12 to 19.
The most commonly used restriction enzyme for DMD analysis is HindIII because the restriction pattern for all 79 exons is known and the majority of exons are on single fragments. BglII and EcoRI are also commonly used enzymes. If a duplication or deletion starts or ends within the restriction enzyme exon fragment, an altered sized fragment will be detected (Figure 5). The altered fragments are known as junction fragments, or J-bands, and are found in ∼5% of the deletions. The J-bands can be helpful in determining the origin of the mutation and in carrier determinations; however, normal restriction enzyme polymorphisms can also generate new altered fragments. We have found several dystrophin gene HindIII polymorphisms in the African-American population so care should be taken not to confuse these with deletions.30 The use of a second restriction enzyme often allows the distinction between junction fragments generated from deletions from polymorphic variants. We have also found a polymorphism, with a frequency of ∼20% in African-Americans, which alters both the exon 8 and 9 HindIII and BglII restriction fragments.30 To avoid misinterpretation of this of this common African-American polymorphism, polymerase chain reaction (PCR) of the exons 8 and 9 is an excellent confirmation.
Figure 5.
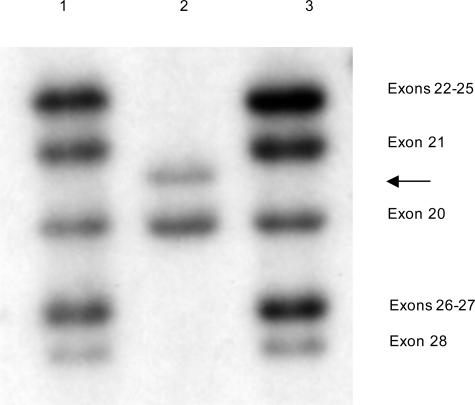
Southern hybridization using a dystrophin cDMD probe that hybridizes to exons 20 to 28 (DNA digested with HindIII). The DMD patient in lane 2 is deleted for exons 22 to 28 and an exon 21 junction fragment has resulted (arrow).
The Southern blotting technique requires isotope, requires high molecular weight DNA, and is tedious and time consuming. Alternatively, rapid and efficient deletion screening can be performed by the multiplex PCR.31 The technique allows one to amplify specific deletion-prone exons within the DMD gene up to a million-fold from nanogram amounts of genomic DNA. The exon products are discriminated from one another by size after gel electrophoresis. When any one of the coding sequences is absent from a patient sample, no ethidium bromide-stained amplification product, corresponding to the specific exon, is present on the gel (Figure 6). Multiplex PCR, using primer sets for ∼20 different exons, can now detect ∼98% of the deletions in the dystrophin gene.31,32
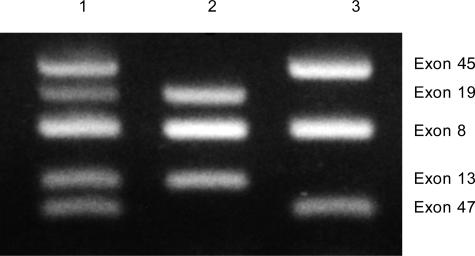
Figure 6.
Multiplex DNA amplification of DNA from DMD patients. Lane 1, Unaffected male control. Lane 2, DMD patient deleted for exons 45 and 47. Lane 3, DMD patient deleted for exons 13 and 19.
In contrast to Southern blotting, which may require several cDNA hybridizations and take several weeks to obtain results, the multiplex PCR assay can be completed in 1 day. This makes the technique ideal for prenatal diagnosis, when time is critical. Prenatal diagnosis is performed on DNA extracted either by chorionic villus sampling or by amniocentesis. When the deletion has already been identified in an affected male, the multiplex PCR deletion test is straightforward and highly accurate for female relatives. However, an important issue in prenatal testing is the confirmation that the fetal DNA sample is not contaminated with maternal DNA. Contamination from the mother could potentially mask a deletion present in the male fetus. The absence of maternal DNA should be demonstrated by typing the fetal DNA sample with a polymorphic marker for which the mother is heterozygous. The presence of a single maternal allele in the fetus thus confirms the lack of maternal contamination.
We have found that multiplex PCR and Southern blotting complement each other and we therefore test all patients using both methods. There are several reasons for our strategy. First, the identification of duplications by standard multiplex conditions and ethidium bromide staining is technically difficult because it is during the exponential phase that the amount of amplified products is proportional to the abundance of starting DNA. This occurs when the primers, nucleotides, and Taq polymerase are in large excess over that of the template concentration. In our experience, after the completion of an adequate number of cycles25,26,27,28,29,30 to visualize the PCR products on an ethidium bromide-stained gel, the PCR reaction is no longer in the exponential quantitative range and the duplicated exons appear little or no brighter than the normal single copy exons. By using densitometry and multiple restriction digests, we have found the detection duplications by Southern blotting to be relatively straightforward. However, the recent utilization of automated DNA fragment analysis using multiplex PCR with fluorescently labeled primers has allowed more accurate detection of duplications. Second, Southern blotting allows determination of all deletion and duplication endpoints, which is important in determining the effect of the mutation on the reading frame. Because the majority of labs tend to assess ∼20 to 25 deletion prone exons by multiplex PCR, it is not possible to obtain all endpoints by PCR alone. Third, the Southern blot technique allows for the detection of junction bands. Last, we have found it to be a good quality control practice to confirm all mutations by two separate analyses. A new technique was recently described to detect both deletions and duplications, combining both multiplex PCR and probe hybridization. The multiplex amplifiable probe hybridization is based on the quantitative recovery of probes after their hybridization to immobilized DNA.33 The probes are recovered by simultaneous PCR amplification, which produces different sized products, and are analyzed on a 96-capillary sequencer. Therefore, changes in peak heights reflect either gene deletions or duplications. The technique has been shown to be accurate and labor efficient.
Carrier Studies
The identification of a deletion in a DMD patient not only confirms the diagnosis but also allows one to perform accurate carrier detection in the affected family. Carrier status is determined by gene dosage, which is determined by whether a female at risk exhibits no reduction or 50% reduction in hybridization intensity in those bands that are deleted for the affected male.34,35 A 50% reduction (single-copy intensity) for the deleted band or bands on the autoradiograph indicates a deletion on one of her X chromosomes and she would therefore be a carrier. Dosage determinations can be made from Southern blot or quantitative polymerase chain reaction. Because cosmid clones are available for several of the dystrophin exons, fluorescence in situ hybridization can also be used for carrier detection in families with a known dystrophin deletion.36
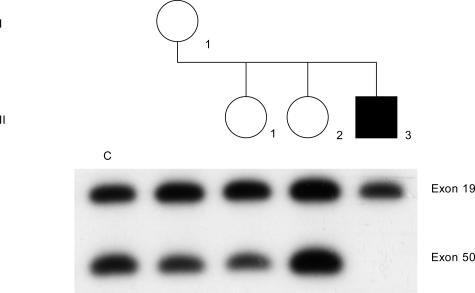
Results from a case study using quantitative PCR and dosage testing are shown in Figure 7. A DMD patient was found to have a molecular deletion for exon 50. This was an isolated case of the disease, so the mother and two daughters were tested for the deletion. To obtain quantitative results, PCR products must be measured during the exponential phase of the amplification process. Therefore, exon 50 in the mother, daughters, proband, and a normal female control were amplified for 12 cycles and hybridized with the corresponding cDNA probe, with the autoradiogram shown in Figure 7. Exon 19 serves as an internal control because this is an exon that is not deleted in the patient. The female control reveals the normal two-copy intensity of each exon and is included on all dosage determinations. Rather than directly comparing single bands, band ratios are calculated as a means of decreasing the error caused by differences in the amount of amplified product in each lane. The exon 50:19 ratio in the mother (I-1) is approximately half the normal control ratio. These ratios were confirmed by densitometer. Therefore, the mother is a carrier of the exon 50 deletion and the proband (II-3) is not the result of a new spontaneous mutation. Daughter II-1 also had an exon 50:19 ratio consistent with positive carrier status, whereas daughter II-2 had a normal noncarrier exon 50:19 ratio.
Figure 7.
Carrier determination by gene dosage. The affected son (II-3) is deleted for exon 50. Exon 19 is the internal standard because the affected son is not deleted for this exon. The mother (I-1) and the daughter (II-1) show a 50% reduction of hybridization of exon 50:19 dosage ratio compared to the noncarrier female control (c), and are therefore carriers. Daughter (II-2) is a noncarrier because her exon 50:19 dosage ratio is equivalent to the female control (c). The samples were subjected to 14 PCR cycles and the PCR products were Southern blotted and hybridized with the corresponding cDNA probes 2b-3 and 8.
Dosage determinations permit direct carrier analysis and eliminates the inherent problems of the restriction fragment length polymorphism technique (recombinations, noninformative meioses, unavailability of family members, and spontaneous mutations). This is important because, unlike the affected males, the heterozygous females are generally asymptomatic, and creatine kinase is only elevated in ∼50 to 60% of known carriers.
When the dosage analysis indicates that the mother does not have a deletion, she still has an uncertain risk of carrier status, owing to the possibility of germline mosaicism.37 Cases of germline mosaicism in DMD have been reported in which a deletion is transmitted to more than one offspring by a mother who shows no evidence of the mutation in her somatic cells. Cases of germline mosaicism have important implications for counseling. First and most obvious is the need to perform carrier studies on all daughters of deletion cases. The sisters of DMD patients may be carriers and should be investigated independently of the outcome of the mother. Furthermore, a negative deletion result in a mother does not rule out a recurrence risk for future pregnancies, and prenatal screening should still be offered. Because it depends on the size of the mutant clone in the mosaic mother, the exact recurrence risk in germline carriers is unknown. However, in these cases the risk is significantly increased relative to what had been initially perceived as a new mutation with a low-recurrence risk. It has been estimated that mothers of apparently sporadic DMD cases, when the mutation is not present in her somatic DNA, have a 20% risk of being a germline carrier.38 Therefore, the mother has a 5% risk of having an affected son.
With the high rate of mutation, possibly as a result of the large intron serving as a source of genetic recombination leading to deletions and duplications, we recently investigated the origin of the deletion in isolated cases. The proportion of mothers of isolated cases found to be carriers of the deletion, by dosage or junction fragment analysis, was ∼44% (42 of 96). The remaining mothers did not carry the mutation identified in their affected son, consistent with a de novo mutation in one or a portion of their germ cells. It is possible that a number of mothers may actually be somatic mosaics, and the deletion may have been revealed by testing other tissues. On further investigation we found that the origin of the mutation differed depending on the location of the deletion. For deletions initiating at the 5′ end of the gene (exon 1 to 20), 18 of 32 of the mothers were carriers of the deletion whereas for deletions more distal in location (exons 43 to 55), 24 of 64 mothers were carriers. Our results are consistent with those of Passos-Bueno and colleagues39 who showed that deletions in the proximal part of the gene have a higher probability of becoming a familial inherited mutation, whereas distal deletions are more often sporadic.
In the 35% of families with undefined mutations, carrier detection and prenatal diagnosis depend on linkage analysis. The method relies on the co-inheritance of the disease gene with those DNA polymorphic variations known to be located very close to or within the disease gene. Thus, even when the responsible gene mutation remains unknown, the linkage technique allows one to trace the mutation through an affected family and make predictions about the inheritance of the disorder. Microsatellite sequences, which correspond to short tandem repeats (di-, tri-, or tetranucleotides) and tend to be highly polymorphic in repeat number, have been found in several locations in the DMD gene and have significantly improved linkage analysis.40,41,42 The microsatellites vary in allele length and can easily be tested by PCR. Although the indirect approach can provide valuable information, it is limited by the possibility of recombination between the microsatellite sequence and the unknown mutation, the presence of sporadic mutations, and the availability of family members. The intragenic recombination rate over the entire length of the DMD gene was estimated to be as high as 12%.43 The high recombinational error rate can be overcome by using markers at both ends of the gene. By using at least four microsatellite markers evenly distributed across the gene, the ability to identify a recombination event is increased. However, the results are still often extremely limited for extended family members of isolated cases of the disease, due to the possibility of the occurrence of a new mutation. Linkage indicates only whether the female at risk inherited the same X chromosome as the affected male, not whether she is a carrier the defective gene. Furthermore, because the gene mutation remains unidentified, a correct diagnosis is essential. This is extremely important with patients presenting with the milder BMD because this phenotype can overlap with other neuromuscular disorders. The diagnosis can usually be made clinically on the basis of symptoms and signs at presentation, increased creatine kinase levels, and myopathic findings. A family history in conjunction with the clinical findings would strongly suggest the diagnosis of DMD or BMD. However, if there is any question of the diagnosis, the Western blot assay of the dystrophin protein on a muscle biopsy specimen should be considered to confirm the diagnosis.
Point Mutation Detection in the Dystrophin Gene
As previously described, using multiplex PCR and Southern blotting, large genomic deletions and duplications have been identified in approximately two-thirds of the DMD/BMD population. The other mutations are due to smaller types of mutations within the dystrophin gene and require some type of sequencing-based strategy. In most routine diagnostic services, these mutations remain undetected because sequencing the entire gene is both expensive and labor intensive. However the identification of these mutations is not only important for the confirmation of the diagnosis but also for the determination of accurate carrier and prenatal studies. Due to the high mutation rate in the dystrophin gene, carrier testing based on indirect linkage results is often limited for extended family members of isolated cases of the disease. Knowledge of the exact causative mutation allows determination of the origin of the mutation in families with simplex cases of the disease.
Using a variety of screening methods (single strand conformational polymorphism, denaturing high performance liquid chromatography, heteroduplex analysis, denaturing gradient gel electrophoresis, detection of virtually all mutations, protein truncation test) performed primarily in research settings, several studies have now identified smaller types of mutations in the dystrophin gene. Although some common mutations have been found, most mutations have been unique (private mutations) to single or few patients and are distributed throughout the gene with no mutational hotspots (Figure 3). The majority of the mutations have been shown to affect only one or a few nucleotides and result in protein truncation, lacking part or all of the C-terminus. It is clear from numerous studies that the testing of the nondeletion/duplication patients, due to the large gene size and the lack of a point mutation hotspot, is laborious and expensive. Because the majority of mutations result in protein truncation, the protein truncation test has been successfully used by some investigators to detect point mutations in the DMD gene.44,45 Using de novo protein synthesis from RNA extracted from the patient, the coding region is screened for truncating types of mutations. The RNA is reverse transcribed and the cDNA is then PCR amplified with a primer that facilitates in vitro transcription by T7-RNA polymerase. A translation step then generates peptide fragments that are analyzed on gels for the identification of shorter fragments indicative of a truncation. The major limitation of the protein truncation test is that it requires dystrophin RNA, which is most abundant in the muscle, and therefore muscle biopsies are the specimen of choice. Muscle biopsies are not always available from affected patients, and RNA extracted from lymphocytes is more difficult to use because its presence is very low.
Although a number of the current strategies have been shown to be very sensitive for detecting small alterations in the very large dystrophin gene, the majority of these methods cannot distinguish mutations from polymorphic variations. A final sequencing step is required to confirm the nature of all positive screening tests. In our point mutation studies on nondeletion cases (Figure 3 and Table 1), we initially screened each DMD exon by denaturing high performance liquid chromatography and only exons that demonstrated aberrant peaks were sequenced. We found the denaturing high performance liquid chromatography screen to be both sensitive and labor efficient. However, for the testing of the nondeletion/duplication patients to be performed in the routine molecular diagnostic laboratory, more high-throughput sequencing techniques are necessary. Recently a single condition amplification/internal primer sequencing technique was described for point mutation detection in the dystrophin gene.46 The method relied on amplification of dystrophin gene exons at a single set of PCR conditions followed by sequencing using a second set of internal primers. The analysis was both automated and high throughput, with all of the dystrophin exons being sequenced within 3 working days at a reasonable cost. The key features of this system, being sequence-based and automated, increase its desirability and potential for application in a routine molecular diagnostic laboratory.
By using one of the current methods of point mutation analysis, detection rates can now be increased from ∼70% (by deletion and duplication studies alone) to greater than 90%. As a result of the improved testing sensitivity, a diagnostic muscle biopsy for the measurement of dystrophin levels is not necessary in the majority of cases. The molecular testing not only replaces the invasive muscle biopsy test, and its general discomfort, but is also cost effective.47 It is apparent, however, from the majority of point mutation studies, that not all mutations are identified. Some of the undetected mutations may reside in the large dystrophin introns or in regulatory regions. In these cases a muscle biopsy may be helpful in establishing an accurate diagnosis.
Conclusions
As a result of the discovery of the dystrophin gene and elucidation of the mutational spectrum, clinical diagnostic testing for DMD and BMD has significantly improved. Until an effective treatment is found to cure or arrest the progression of these diseases, prevention of new cases through accurate diagnosis and carrier and prenatal testing is of the utmost importance. In the future molecular therapies (such as anti-sense oligonucleotides, antibiotics, or chimeric RNA/DNA) will be applied according to the specific dystrophin mutation. This will require a complete mutation analysis and identification of all types of dystrophin mutations.
Footnotes
Supported by a grant from the Muscular Dystrophy Association.
References
- Moser H. Duchenne muscular dystrophy: pathogenetic aspects and genetic prevention. Hum Genet. 1984;66:17–40. doi: 10.1007/BF00275183. [DOI] [PubMed] [Google Scholar]
- Emery AEH. Muscle histology and creatine kinase levels in the fetus in Duchenne muscular dystrophy. Nature. 1977;266:472–473. doi: 10.1038/266472a0. [DOI] [PubMed] [Google Scholar]
- Emery AEH. New York: Oxford University Press; Duchenne Muscular Dystrophy. (ed 2.) 1993 [Google Scholar]
- Farah MG, Evans EB, Vignos PJ. Echocardiographic evaluation of left function in Duchenne’s muscular dystrophy. Am J Med. 1980;69:248–252. doi: 10.1016/0002-9343(80)90385-x. [DOI] [PubMed] [Google Scholar]
- Leibowitz D, Dubowitz V. Intellect and behavior in Duchenne muscular dystrophy. Dev Med Neurol. 1981;23:577–590. doi: 10.1111/j.1469-8749.1981.tb02039.x. [DOI] [PubMed] [Google Scholar]
- Blythe H, Pugh RJ. Muscular dystrophy in childhood: the genetic aspect: a field study in the Leeds region of clinical types and their inheritance. Ann Hum Genet. 1958;23:127–163. doi: 10.1111/j.1469-1809.1958.tb01457.x. [DOI] [PubMed] [Google Scholar]
- Brooke MH, Fenichel G, Griggs RC, Mendell JR, Moxley RT. Clinical investigation in Duchenne dystrophy. 2. Determination of the “power” of therapeutic trials based on the natural history. Muscle Nerve. 1983;6:91–97. doi: 10.1002/mus.880060204. [DOI] [PubMed] [Google Scholar]
- Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C, Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Brown RH, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Campbell KP, Kahl SD. Association of dystrophin and an integral membrane glycoprotein. Nature. 1989;338:259–262. doi: 10.1038/338259a0. [DOI] [PubMed] [Google Scholar]
- Ervasti JM, Campbell KP. Membrane organization of the dystrophin-glycoprotein complex. Cell. 1992;66:1121–1131. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Ozawa E. Glycoprotein complex anchoring dystrophin to sarcolemma. J Biochem. 1990;108:748–752. doi: 10.1093/oxfordjournals.jbchem.a123276. [DOI] [PubMed] [Google Scholar]
- Chelly J, Hamard G, Koulakoff A, Kaplan JC, Kahn A, Berwald-Netter Y. Dystrophin gene transcribed from different promoters in neuronal and glial cells. Nature. 1990;344:64–65. doi: 10.1038/344064a0. [DOI] [PubMed] [Google Scholar]
- Feener CA, Koenig M, Kunkel LM. Alternative splicing of dystrophin mRNA generates isoforms at the carboxy terminus. Nature. 1989;338:509–511. doi: 10.1038/338509a0. [DOI] [PubMed] [Google Scholar]
- Byers TJ, Lidov GW, Kunkel LM. An alternative dystrophin transcript specific to peripheral nerve. Nat Genet. 1993;4:77–81. doi: 10.1038/ng0593-77. [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Fischbeck K, Brown RH, Johnson M, Medori R, Loike JD, Harris JB, Waterston R, Brooke M, Specht L, Kupsky W, Chamberlain J, Caskey T, Shapiro F, Kunkel LM. Dystrophin characterization in muscle biopsies from Duchenne and Becker muscular dystrophy patients. N Engl J Med. 1988;318:1363–1368. doi: 10.1056/NEJM198805263182104. [DOI] [PubMed] [Google Scholar]
- Ohlendieck K, Matsumura K, Ionasescu VV, Towbin JA, Bosch EP, Weinstein SL, Sernett SW, Campbell KP. Duchenne muscular dystrophy: deficiency of dystrophin associated proteins in the sarcolemma. Neurology. 1993;43:795–800. doi: 10.1212/wnl.43.4.795. [DOI] [PubMed] [Google Scholar]
- Matsumura K, Campbell KP. Dystrophin-glycoprotein complex: its role in the molecular pathogenesis of muscular dystrophies. Muscle Nerve. 1994;17:2–15. doi: 10.1002/mus.880170103. [DOI] [PubMed] [Google Scholar]
- Campbell KP. Three muscular dystrophies: loss of cytoskeleton-extracellular matrix linkage. Cell. 1995;80:675–579. doi: 10.1016/0092-8674(95)90344-5. [DOI] [PubMed] [Google Scholar]
- Forest S, Cross GS, Speer A, Gardner-Medwin D, Burner J, Davies K. Preferential deletion of exons in Duchenne and Becker muscular dystrophies. Nature. 1987;329:638–640. doi: 10.1038/329638a0. [DOI] [PubMed] [Google Scholar]
- Darras BT, Blattner P, Harper JF, Spiro AJ, Alter S, Franke U. Intragenic deletions in 21 Duchenne muscular dystrophy (DMD)/ Becker muscular dystrophy (BMD) families studied with the dystrophin cDNA: location of breakpoints on HindIII and BglII exon-containing fragment maps, meiotic and mitotic origin of mutations. Am J Hum Genet. 1988;43:620–629. [PMC free article] [PubMed] [Google Scholar]
- Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- Malhotra SB, Hart KA, Klamut HJ, Thomas NST, Bodrug SE, Burghes AHM, Bobrow M, Harper PS, Thompson MW, Ray PN, Worton RG. Frame-shift deletions in patients with Duchenne and Becker muscular dystrophy. Science. 1988;242:755–759. doi: 10.1126/science.3055295. [DOI] [PubMed] [Google Scholar]
- Prior TW, Bartolo C, Papp AC, Snyder PJ, Sedra MS, Burghes AHM, Kissel JT, Luquette MH, Tsao C-Y, Mendell JR. Dystrophin expression in a Duchenne muscular dystrophy patient with a frameshift deletion. Neurology. 1997;48:486–488. doi: 10.1212/wnl.48.2.486. [DOI] [PubMed] [Google Scholar]
- Hu X, Ray PN, Murphy E, Thompson MW, Worton RG. Duplicational mutation at the Duchenne muscular dystrophy locus: its frequency, distribution, origin and phenotype/genotype correlation. Am J Hum Genet. 1990;46:682–695. [PMC free article] [PubMed] [Google Scholar]
- Roberts RG, Gardner RJ, Bobrow M. Searching for the 1 in 2,400,000: a review of dystrophin gene point mutations. Hum Mutat. 1994;4:1–11. doi: 10.1002/humu.1380040102. [DOI] [PubMed] [Google Scholar]
- Prior TW, Bartolo C, Pearl DK, Papp AC, Snyder PJ, Sedra MS, Burghes AHM, Mendell JR. Spectrum of small mutations in the dystrophin coding region. Am J Hum Genet. 1995;57:22–33. [PMC free article] [PubMed] [Google Scholar]
- Muntoni F, Melis MA, Ganau A, Dubowitz V. Transcription of the dystrophin gene in normal tissues and skeletal muscle of a family with X-linked dilated cardiomyopathy. Am J Hum Genet. 1995;56:151–157. [PMC free article] [PubMed] [Google Scholar]
- Milasin J, Muntoni F, Sererini GM, Bartoloni L, Vatta M, Krajinovic M, Mateddu A, Angelini C, Camerini F, Falaschi A, Mestroni L, Giacca M, the Heart Muscle Disease Group A point mutation in the 5′ splice of the dystrophin gene first intron responsible for X-linked dilated cardiomyopathy. Hum Mol Genet. 1996;5:73–79. doi: 10.1093/hmg/5.1.73. [DOI] [PubMed] [Google Scholar]
- Prior TW, Papp AC, Snyder PJ, Burghes AHM, Wallace BH. A HindIII/BglII dystrophin gene polymorphism in the Black population. Hum Genet. 1992;89:687–688. doi: 10.1007/BF00221965. [DOI] [PubMed] [Google Scholar]
- Chamberlain JS, Gibbs RA, Ranier JE, Nga Nguyen PN, Caskey CT. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucl Acids Res. 1988;16:11141–11156. doi: 10.1093/nar/16.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs AH, Koenig M, Boyce FM, Kunkel LM. Detection of 98% of DMD/BMD gene deletions by PCR. Hum Genet. 1990;86:45–48. doi: 10.1007/BF00205170. [DOI] [PubMed] [Google Scholar]
- White S, Kalf M, Liu Q, Villerius M, Engelsma D, Kriek M, Vollebreg E, Bakker B, van Ommen GJB, Breuning MH, den Dunnen JT. Comprehensive detection of genomic duplications and deletions in the DMD gene, by use of multiplex amplifiable probe hybridization. Am J Hum Genet. 2002;71:365–374. doi: 10.1086/341942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darras BT, Koenig M, Kunkel LM, Francke U. Direct method for prenatal diagnosis and carrier detection in Duchenne/Becker muscular dystrophy using the entire dystrophin cDNA. Am J Med Genet. 1988;29:713–726. doi: 10.1002/ajmg.1320290341. [DOI] [PubMed] [Google Scholar]
- Prior TW, Friedman KJ, Highsmith WE, Perry TR, Silverman LM. Molecular probe protocol for determining carrier status in Duchenne and Becker muscular dystrophies. Clin Chem. 1990;36:441–445. [PubMed] [Google Scholar]
- Calvano S, Memeo E, Piemontese MR, Melchionda S, Bisceglia L, Gasparini P, Zelante L. Detection of dystrophin deletion carriers using FISH analysis. Clin Genet. 1997;52:17–22. doi: 10.1111/j.1399-0004.1997.tb02509.x. [DOI] [PubMed] [Google Scholar]
- Bakker E, Veenema H, den Dunnen JT, van Broeckhoven CH, Grootscholten P, Bonten EJ, van Ommen GJB, Pearson PL. Germinal mosaicism increases the recurrence risk for ‘new’ Duchenne muscular dystrophy mutations. J Med Genet. 1989;26:553–559. doi: 10.1136/jmg.26.9.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Essen AJ, Abbs S, Baiget M, Bakker E, Boileau C, van Broeckhoven CH, Bushby K, Clarke A, Claustres M, Covone AE. Parental origin and germline mosaicism of deletions and duplications of the dystrophin gene: a European Study. Hum Genet. 1992;88:249–257. doi: 10.1007/BF00197255. [DOI] [PubMed] [Google Scholar]
- Passos-Bueno MR, Bakker E, Knepers ALJ, Takata RI, Rapaport D, den Dunnen JT, Zatz M, van Ommen GJB. Different mosaicism frequencies for proximal and distal Duchenne muscular dystrophy (DMD) mutations indicate difference in etiology and recurrence risk. Am J Hum Genet. 1992;51:1150–1155. [PMC free article] [PubMed] [Google Scholar]
- Clemens PR, Fenwick RG, Chamberlain JS, Gibbs RA, de Andrade M, Chakraboty R, Caskey CT. Carrier detection and prenatal diagnosis in Duchenne and Becker muscular dystrophy families, using dinucleotide repeat polymorphisms. Am J Hum Genet. 1991;49:951–960. [PMC free article] [PubMed] [Google Scholar]
- Oudet C, Helig R, Hanauer A, Mandel JL. Nonradioactive assay for new microsatellite polymorphisms at the 5′ end of the dystrophin gene, and estimation of intragenic recombination. Am J Hum Genet. 1991;49:311–319. [PMC free article] [PubMed] [Google Scholar]
- King SC, Roche AL, Passos-Bueno MR, Takata R, Zatz M, Cocburn D, Seller A, Stapleton PM, Love DR. Molecular characterization of further dystrophin gene microsatellites. Mol Cell Probes. 1995;9:361–370. doi: 10.1016/s0890-8508(95)91700-4. [DOI] [PubMed] [Google Scholar]
- Abbs S, Roberts RG, Mathew CG, Bentley DR, Bobrow M. Accurate assessment of intragenic recombination frequency within the Duchenne muscular dystrophy gene. Genomics. 1990;7:602–606. doi: 10.1016/0888-7543(90)90205-9. [DOI] [PubMed] [Google Scholar]
- Roest PAM, Roberts RG, Sugino S, van Ommen GJB, den Dunnen JT. Protein truncation test (PTT) for rapid detection of translation-terminating mutations. Hum Mol Genet. 1993;2:1719–1721. doi: 10.1093/hmg/2.10.1719. [DOI] [PubMed] [Google Scholar]
- Gardner RJ, Bobrow M, Roberts RG. The identification of point mutations in Duchenne muscular dystrophy patients using reverse transcript PCR and the protein truncation test. Am J Hum Genet. 1995;57:311–320. [PMC free article] [PubMed] [Google Scholar]
- Flanigan KM, Niederhausern AV, Dunn DM, Alder J, Mendell JR, Weiss RB. Rapid direct sequence analysis of the dystrophin gene. Am J Hum Genet. 2003;72:931–939. doi: 10.1086/374176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JR, Buzin CH, Feng J, Yan J, Serrano C, Sangani DS, Wall C, Prior TW, Sommer SS. Diagnosis of Duchenne dystrophy by enhanced detection of small mutations. Neurology. 2001;57:645–650. doi: 10.1212/wnl.57.4.645. [DOI] [PubMed] [Google Scholar]