Abstract
We present the case of a 6-year-old male who received an allogeneic bone marrow transplant as part of treatment for acute lymphoblastic leukemia. The patient relapsed 5 months after transplantation and received additional chemotherapy. He acquired an angioinvasive fungal infection that required transfusion of granulocytes. Approximately 5 weeks after relapsing (181 days after transplant), a bone marrow specimen was taken for molecular engraftment analysis and flow cytometry to assess graft loss as well as residual disease. The engraftment results generated by the multiple short tandem repeat loci tested were inconsistent, and alleles were present at several loci that were of neither patient nor donor origin. An error in specimen identification was initially considered. Further investigation into the circumstances surrounding procurement of the patient’s bone marrow aspirate revealed that the patient had received a granulocyte transfusion approximately 10 hours before the bone marrow specimen was taken. In addition, morphological and flow cytometric analyses of the same bone marrow aspirate demonstrated a significant degree of peripheral blood contamination. We determined that the unknown alleles in the bone marrow engraftment specimen were derived from the donor of the transfused granulocytes. This case illustrates that white cell transfusion can lead to erroneous bone marrow engraftment results, particularly if only one microsatellite locus is used to monitor engraftment.
It is well-established that polymerase chain reaction (PCR) amplification of polymorphous short tandem repeat (STR) loci using standard commercially available kits is an accurate and efficient method to determine bone marrow engraftment (BME) in patients with hematopoietic malignancies status after allogeneic bone marrow transplantation1,2,3,4 (for reviews, see Refs. 5 and 6). This methodology was originally designed for forensic purposes; however, it is increasingly being used by clinical laboratories for patient samples secondary to its sensitivity and relatively rapid turnaround time.
Although this technology has streamlined the BME evaluation of patients and has even enabled reliable estimations of minimal residual disease,7 the results are potentially complicated by the chromosomal abnormalities inherent to many hematopoietic malignancies. For example, gain or loss of chromosomal material can affect STR loci used to calculate percent engraftment and lead to potential errors.8,9
As an additional source of difficulty in the interpretation of BME analysis, we present a case of a 6-year-old boy with relapsed acute lymphoblastic leukemia (ALL) who required transfusions of granulocytes for an angioinvasive fungal infection. The case illustrates that foreign cells introduced by transfusion represent another potential pitfall in the interpretation of engraftment results.
Materials and Methods
Approval from the institutional review board of the Johns Hopkins Hospital was granted to present this study (exemption 03-12-22-01e). In addition, approval from the institutional review board of the Johns Hopkins Hospital and of the American Red Cross was granted to perform anonymous testing on the granulocyte donor (exemption 04-12-27-01e).
DNA Isolation
DNA was isolated from all peripheral blood and bone marrow specimens using the Qiagen EZ1 automated nucleic acid extraction instrument and reagents according to the manufacturer’s instructions (Qiagen, Valencia, CA). After nucleic acid quantification by spectrophotometric analysis at 260 and 280 nm, appropriate dilutions of DNA from each specimen were made for microsatellite analysis.
Microsatellite Analysis
PCR amplification was performed using the AmpFlSTR Profiler Plus kit (Applied Biosystems, Foster City, CA) that detects nine microsatellite loci as well as the Amelogenin locus (Table 1).10 Thermal cycling conditions and capillary electrophoresis were performed according to the manufacturer’s instructions. In brief, the PCR conditions included an initial denaturation step at 95°C for 11 minutes followed by 28 PCR cycles. Each cycle included a denaturation step at 94°C for 1 minute, an annealing step at 59°C for 1 minute, and a polymerization step at 72°C for 1 minute. The final extension step consisted of incubation at 60°C for 45 minutes. After amplification, 1 μl of multiplex PCR product was mixed with 9 μl of deionized formamide/GeneScan-500 [ROX] size standard solution (Applied Biosystems) per the manufacturer’s protocol. Samples were denatured at 95°C for 2 minutes and placed on ice for at least 1 minute before analysis on the ABI 3100 capillary electrophoresis instrument (Applied Biosystems).
Table 1.
STR Loci Amplified in the AmpFlSTR Profiler Plus Kit10
| STR locus | Chromosomal location | Size (bp) | Dye label (peak color in electropherogram) |
|---|---|---|---|
| D3S1358 | 3p | 114–142 | 5-FAM (Blue) |
| vWA | 12p12-pter | 157–197 | 5-FAM (Blue) |
| FGA | 4q28 | 219–267 | 5-FAM (Blue) |
| Amelogenin | X:p22.1–22.3 | 107,113 | JOE (Green) |
| D8S1179 | 8 | 128–168 | JOE (Green) |
| D21S11 | 21 | 189–243 | JOE (Green) |
| D18S51 | 18q21.3 | 273–341 | JOE (Green) |
| D5S818 | 5q21–31 | 135–171 | NED (Black) |
| D13S317 | 13q22–31 | 206–234 | NED (Black) |
| D7S820 | 7q11.21–22 | 258–294 | NED (Black) |
Calculation of Engraftment
Before bone marrow transplantation, donor and patient specimens were analyzed using the AmpFlSTR Profiler Plus kit loci. In this case, the patient received his transplant from a matched, unrelated donor. In total, six loci were informative for follow-up analyses. At our institution, two informative loci are routinely used for detection of recipient and donor alleles. D3S1358 and D8S1179 were chosen as the informative loci for detection of recipient alleles. For the identification of donor alleles, D3S1358 and D18S51 were chosen as the informative loci.
Results
Case Summary
The patient is a 6-year-old Caucasian male diagnosed with precursor B acute lymphoblastic leukemia. He relapsed during treatment on standard maintenance chemotherapy 13 months after diagnosis. A second remission was achieved with induction chemotherapy and a Capizzi II intensification regimen. After cyclophosphamide conditioning and total body irradiation, the patient received a matched unrelated donor bone marrow transplant 5 months after his initial relapse. His posttransplant course was complicated by mild skin and gastrointestinal graft-versus-host disease and Enterococcal bacteremia.
The patient subsequently developed pancytopenia and was diagnosed with a second relapse of precursor B acute lymphoblastic leukemia, 145 days after transplant. He was then enrolled in a trial of a clofarabine chemotherapy regimen for refractory and relapsed ALL. This postchemotherapy course was complicated by significant tumor lysis syndrome that required admission to the intensive care unit for supportive care and treatment of a severe angioinvasive fungal infection of his right hand by a Rhizopus species. Because the patient was neutropenic, he received multiple transfusions of granulocytes in an attempt to contain the fungal infection; however, he ultimately required a below-the-elbow amputation for infection control.
Engraftment Analysis
Before bone marrow transplant, STR analysis of donor and patient samples identified six loci that were determined to be informative for follow-up analysis (Figure 1, A and B). The D3S1358 and D8S1179 loci were chosen for follow-up analyses in the setting of predominantly donor DNA; D3S1358 and D18S51 were chosen for follow-up analyses in the setting of predominantly patient DNA. Because of the patient’s history of relapse before bone marrow transplant, his engraftment status was aggressively monitored. Initial results after transplant demonstrated the presence of predominantly donor DNA (Table 2). The patient subsequently developed pancytopenia and was diagnosed with a second relapse 145 days after transplant. Morphological and flow cytometric analysis demonstrated a precursor B lymphoblast population (CD19+, CD10+) accounting for approximately 70 to 75% of the total cellularity. Engraftment analysis revealed that nearly 80% of the cells were derived from the patient (Table 2).
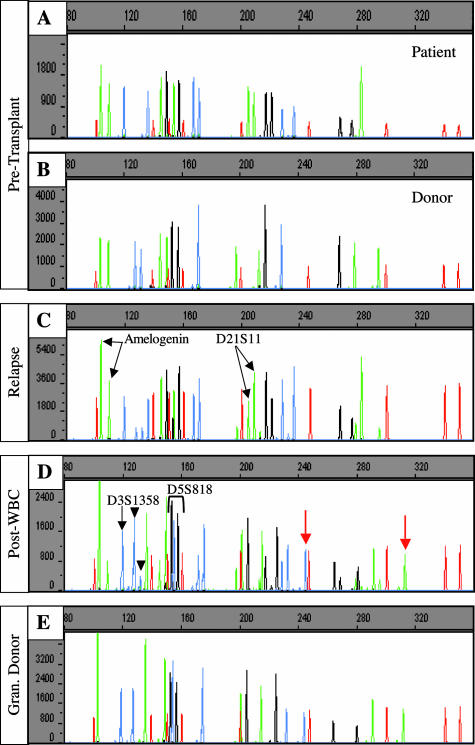
Figure 1.
Capillary electropherograms of STR analyses. The x axis represents fragment size in bases and the y axis represents fluorescence intensity. The red peaks represent the internal size standards. The blue, green, and black peaks designate two alleles from the Amelogenin locus and nine microsatellite markers (blue: D3S1358, vWA, and FGA; green: D8S1179 and D21S11; black: D18S51, D5S818, D13S317, and D7S820). A and B: Pretransplant analyses of STR loci for the patient (A) and donor (B). C: Bone marrow engraftment analysis during relapse. Note the differences in peak intensity at the Amelogenin locus, green peaks at 103 and 109 bp, and at the D21S11 locus, green peaks at 205 and 209 bp (arrows). D: Bone marrow engraftment analysis of the specimen procured status after granulocyte transfusion. At locus D3S1358, the two peaks presumed to be from donor alleles (127 and 131 bases; black arrowheads) are of unequal peak intensities; one of the presumed patient alleles (119 bases) is present (black arrow) at a significant peak intensity; however, the second patient allele (135 bases) is not present. In addition, alleles are present that are of neither patient nor donor origin (red arrows). However, the alleles at locus D5S818 (black peaks at 153 and 157 bases) correspond to the donor alleles. E: Analysis of a peripheral blood specimen from the granulocyte donor. The allele pattern is identical to the dominant alleles identified in the patient’s specimen after granulocyte transfusion (D). Note that at locus D5S818, the granulocyte donor and the bone marrow donor (B) have identical alleles (black peaks at 153 and 157 bases).
Table 2.
Patient Engraftment Analyses
| Specimen date (days after transplant) | Sample type | Engraftment results
|
|
|---|---|---|---|
| Patient | Donor | ||
| 32 | Bone marrow | <5% | >95% |
| 58 | Bone marrow | <5% | >95% |
| 96 | Bone marrow | 100% | |
| 145 | Bone marrow | 74–78% | 22–26% |
| 181 | Bone marrow | Alleles not consistent with donor or patient | |
| 198 | Bone marrow | 1–7% | 93–99% |
Engraftment analysis of the patient’s relapse specimen demonstrated some changes in peak intensity at the Amelogenin and D21S11 loci when compared with the patient’s specimen drawn before transplant (Figure 1, A and C). This phenomenon suggested the presence of genomic instability in the face of relapse. In fact, karyotypic analysis demonstrated numerous cytogenetic abnormalities in the patient’s bone marrow (data not shown), including additional copies of chromosomes 4, 14, 17, 21, and X. The additional copies of chromosomes X and 21 are likely to be responsible for the observed discrepancies in peak heights at the Amelogenin locus and at the D21S11 locus.
Because of his relapse status, the patient was treated aggressively with a clofarabine chemotherapeutic regimen. He became progressively neutropenic and developed an angioinvasive fungal infection with a Rhizopus species. Approximately 5 weeks after relapsing (181 days after transplant), a bone marrow specimen was taken for molecular engraftment analysis and flow cytometry to assess for graft loss as well as residual disease. The peak heights of the engraftment results generated by the multiple STR loci tested were inconsistent (Figure 1D). At locus D3S1358, the two peaks presumed to be from donor alleles (127 and 131 bases; Figure 1D, black arrowheads) were of unequal intensities, and only one of the two patient alleles was present (Figure 1D, black arrow). As described above, allelic imbalance of patient alleles can result from chromosomal abnormalities in tumor cells; however, this would not explain allelic imbalance in donor alleles. In addition, several loci were evaluated that demonstrated alleles that were of neither patient nor donor origin (Figure 1D, red arrows). However, at locus D5S818, only two alleles were present, which were consistent with the donor’s alleles.
Initially, the possibility of an error in specimen identity was considered. A new aliquot of DNA was extracted from the bone marrow sample, and the analysis was repeated, yielding similar results (data not shown). To further exclude the possibility of mistaken identity of the specimen, DNA samples from all of the patients seen in the bone marrow clinic on the same day were reviewed. None of these patients demonstrated allele patterns consistent with those identified in the specimen.
Further investigation into the circumstances surrounding procurement of the patient’s bone marrow aspirate revealed that the patient had received a transfusion of granulocytes (9.1 × 108/kg) to treat his fungal infection approximately 10 hours before the bone marrow specimen was taken. In addition, morphological examination of the same bone marrow aspirate demonstrated a preponderance of mature lymphocytes and mature myeloid elements (data not shown). Flow cytometric analysis of the marrow aspirate demonstrated an essential absence of B-cell precursors and CD71+ events corresponding to nucleated red cell precursors (data not shown). The myeloid elements were also essentially all mature. These morphological and flow cytometric findings were consistent with a significant degree of peripheral blood contamination. Taken together, the clinical history and the morphological and flow cytometric data suggested that the unknown alleles identified in the engraftment analysis were derived from transfused granulocytes introduced via peripheral blood contamination of the marrow aspirate specimen. To test this hypothesis, a peripheral blood specimen from the granulocyte donor was procured and analyzed. The alleles identified in the granulocyte donor’s specimen were consistent with the dominant peaks identified in the patient’s specimen after granulocyte transfusion, confirming that the unknown alleles in the patient’s specimen were derived from the transfused granulocytes (Figure 1, D and E). In light of these data, we reanalyzed the patient’s specimen after granulocyte transfusion and determined that the granulocyte donor accounted for approximately 80% of the DNA in the sample. The minor allele components, accounting for roughly 20% of the DNA in the specimen, appeared to originate from the bone marrow donor. Interestingly, at locus D5S818, the granulocyte donor and bone marrow donor had identical alleles (P ∼ 0.1).
The patient had an additional follow-up bone marrow aspirate drawn 198 days after transplant that was analyzed for engraftment. The results demonstrated that the marrow was composed of approximately 93 to 99% donor cells without evidence of aberrant alleles (Table 2). In addition, the patient received a peripheral blood stem cell transplant from the same unrelated donor without incident 229 days after his initial bone marrow transplant.
Discussion
Molecular analysis of bone marrow in patients after an allogeneic transplant is an important tool for confirming engraftment and detecting chimerism. Frequent monitoring is an important means to identify patients at high risk for relapse and to allow early clinical intervention.7 Several laboratory methods have been used for BME analysis; however, PCR amplification of polymorphous STR loci is the choice of many clinical laboratories because it is sensitive, quantitative, and relatively rapid to perform1,2,3,4 (for reviews, see Refs. 5 and 6).
Although amplification of microsatellites is an informative method of BME analysis, this technique is also subject to errors incurred as a result of the genomic instability not uncommonly encountered in patients with hematopoietic malignancies. There have been two reports of loss of an informative STR allele secondary to chromosomal instability in patients with chronic myelogenous leukemia.8,9 These clinical scenarios could have resulted in the erroneous conclusion that the patient’s marrow was composed of essentially all donor cells if only one marker had been used for engraftment analysis,
In the current case of a 6-year-old boy with relapsed ALL status after matched, unrelated donor bone marrow transplantation, one of the markers (D21S11) that had been determined to be informative in pretransplant analysis demonstrated inconsistent peak heights in the relapsed bone marrow specimen (Figure 1B). Karyotypic analysis demonstrated an additional copy of chromosome 21, the location of the D21S11 microsatellite marker. This case thus asserts that gain of chromosomal material, as well as loss, can lead to errors in the calculation of percent engraftment, especially when only one locus is used.
The present case also illustrates an additional potential source of complexity in the interpretation of BME analyses—granulocyte transfusion. In this case, the patient had received a dose of 9.1 × 108 granulocytes/kg, which was infused approximately 10 hours before aspiration of bone marrow taken for morphological, phenotypic, and engraftment studies. Because there was peripheral blood contamination of the marrow specimen, a significant number of donor granulocytes were inadvertently introduced into the marrow specimen. Fortunately, most patients undergoing bone marrow engraftment analysis have not received granulocyte transfusions. However, platelet and red blood cell products are commonly given in this setting. These products are often leukoreduced, which specifies that the leukocyte number be less than 5 × 106 for the entire product. If a patient received multiple units of these products (20), the total number of granulocytes transfused would be ≤1% of the amount transfused in this case. Thus, the likelihood that transfusions of platelets and/or red blood cell products would introduce enough leukocytes to complicate engraftment analysis is quite low.
Although standard practice suggests that BME analysis can be determined based on results of at least one locus,6 the present case and previous studies8,9 serve as cautionary notes to this practice. In this case, the bone marrow donor and the granulocyte donor had identical alleles at STR locus D5D818. Thus, engraftment analysis would have yielded erroneous results if only this locus had been used. This problem is, in part, due to the fact that the probability of identity of the STR loci is relatively high (approximately 0.03 to 0.1, a relatively low level of informativity). Given that the probability that two random people will have identical alleles at a given locus is approximately 1 in 20, the use of a single locus often may not identify such mixes and may even fail to identify analysis of an entirely incorrect specimen. To avoid these problems, we recommend that at least two informative microsatellite loci be routinely analyzed to determine the percentage of bone marrow engraftment. In addition, this case also reiterates the importance of assembling all available patient data including morphological examination and clinical information as well as ancillary studies such as cytogenetics and flow cytometry when evaluating specimens for BME.8
Note Added in Proof
Subsequent to submitting this manuscript, our laboratory identified an additional case in which bone marrow engraftment analysis was complicated by granulocyte transfusion(s). This case was an adult who failed to recover bone marrow cell counts after bone marrow transplantation, and who received multiple granulocyte transfusions. Approximately 30 days after bone marrow transplantation, both a peripheral blood and bone marrow specimen were submitted for BME analysis. The dominant alleles in the peripheral blood specimen corresponded to neither the donor nor the patient and are presumed to be derived from the transfused granulocytes. Interestingly, the bone marrow specimen was predominantly patient in orgin but contained a minor allele component (approximately 5%) that matched the dominant allele pattern identified in the patient’s peripheral blood specimen. These results are consistent with the peripheral blood containing a significant amount of transfused granulocytes and the bone marrow specimen containing a small amount of peripheral blood contamination.
References
- Nuckols JD, Rashedd BKA, McGlennen RC, Bigner SH, Stenzel TT. Evaluation of an automated technique for assessment of marrow engraftment after allogeneic bone marrow transplantation using a commercially available kit. Am J Clin Pathol. 2000;113:135–140. doi: 10.1309/QP7P-J49V-8Q15-36MT. [DOI] [PubMed] [Google Scholar]
- Schichman SA, Suess P, Vertino AM, Gray PS. Comparison of short tandem repeat and variable number tandem repeat genetic markers for quantitative determination of allogeneic bone marrow transplant engraftment. Bone Marrow Transplant. 2002;29:243–248. doi: 10.1038/sj.bmt.1703360. [DOI] [PubMed] [Google Scholar]
- Thiede C, Bornhäuser M, Ehninger G. Evaluation of STR informativity for chimerism testing: comparative analysis of 27 STR systems in 203 matched related donor recipient pairs. Leukemia. 2004;18:248–254. doi: 10.1038/sj.leu.2403212. [DOI] [PubMed] [Google Scholar]
- Thiede C, Florek M, Bornhäuser M, Ritter M, Brendel C, Ehninger G, Neubauer A. Rapid quantification of mixed chimerism using multiplex amplification of short tandem repeat markers and fluorescence detection. Bone Marrow Transplant. 1999;23:1055–1060. doi: 10.1038/sj.bmt.1701779. [DOI] [PubMed] [Google Scholar]
- Thiede C, Bornhäuser M, Ehninger G. Strategies and clinical implications of chimerism diagnostics after allogeneic hematopoietic stem cell transplantation. Acta Haematol. 2004;112:16–23. doi: 10.1159/000077555. [DOI] [PubMed] [Google Scholar]
- Van Deerlin VMD, Leonard DGB. Bone marrow engraftment analysis after allogeneic bone marrow transplantation. Clin Lab Med. 2000;20:197–225. [PubMed] [Google Scholar]
- Thiede C, Bornhäuser M, Oelschlägel U, Brendel C, Leo R, Daxberger H, Mohr B, Florek M, Kroschinsky F, Geissler G, Naumann R, Ritter M, Prange-Krex G, Lion T, Neubauer A, Ehninger G. Sequential monitoring of chimerism and detection of minimal residual disease after allogeneic blood stem cell transplantation (BSCT) using multiplex amplification of short tandem repeats. Leukemia. 2001;15:293–302. doi: 10.1038/sj.leu.2401953. [DOI] [PubMed] [Google Scholar]
- Schichman SA, Lin P, Gilbrech LJ, Gray PS, Wilson CS, Sawyer JR. Bone marrow transplant engraftment analysis with loss of an informative allele. J Mol Diagn. 2002;4:230–232. doi: 10.1016/S1525-1578(10)60708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Sheldon S, Akel N, Killeen AA. Chromosomal aneuploidy in leukemic blast crisis: a potential source of error in interpretation of bone marrow engraftment analysis by VNTR amplification. Mol Diagn. 1999;4:153–157. doi: 10.1016/s1084-8592(99)80039-3. [DOI] [PubMed] [Google Scholar]
- Applied Biosystems Foster City, CA: Applied Biosystems,; AmpFlSTR Profiler Plus PCR Amplification Kit User’s Manual. 2000:1–6. [Google Scholar]