Abstract
Sentinel lymph node (SLN) status is highly predictive of overall axillary lymph node involvement in breast cancer. Historically, SLN-positive patients have undergone axillary lymph node dissection in a second surgery. Intraoperative SLN analysis could reduce the cost and complications of a second surgery; however, existing histopathological methods lack standardization and exhibit poor sensitivity. Rapid molecular methods may lead to improved intraoperative diagnosis of SLN metastasis. In this study, we used a genome-wide gene expression analysis of breast and other tissues to identify seven putative markers for detecting breast cancer metastasis. We assessed the utility of these markers for identifying clinically actionable metastases in lymph nodes through reverse transcriptase-polymerase chain reaction analysis of SLNs from 254 breast cancer patients. Polymerase chain reaction signals were compared to pathology on a per-patient basis. The optimal two-gene combination, mammaglobin and cytokeratin 19, detected clinically actionable metastasis in breast SLNs with 90% sensitivity and 94% specificity. Application of stringent criteria for identifying presumptive hematoxylin- and eosin-positive samples increased sensitivity and specificity to 91 and 97%, respectively. This study represents the first comprehensive demonstration of the utility of gene expression markers for detecting clinically actionable breast metastases. An intraoperative molecular assay using these markers has the potential to significantly reduce second surgeries for patients undergoing SLN dissection.
Breast cancer is second only to lung cancer in mortality among women worldwide.1 In the care of this significant disease, the evaluation of blood, bone marrow, and lymph nodes for the presence of metastatic cells is an important component of disease characterization and management.2,3,4,5,6 The method for assessment of these peripheral tissues is largely dependent on histological and cytological methods. Detection of metastasis in lymph nodes is typically accomplished by hematoxylin and eosin (H&E) and antibody staining of lymph node sections.7,8 Analysis of bone marrow samples, although still in development, currently involves antibody staining of cytological smears. For some time, there has been discussion about the potential for the use of molecular biological tools to supplement or to improve existing methods.9 Molecular methods such as reverse transcriptase-polymerase chain reaction (RT-PCR) offer increased analytical sensitivity compared with standard histological methods.10,11,12 In addition, nucleic acid-based detection methods such as real-time PCR offer the potential of rapid and sensitive point-of-care testing and the application of objective quantitative assay cut-offs.
A clear intersection between these features is the intraoperative assessment of sentinel lymph nodes (SLNs) for the presence of clinically actionable metastasis, a level of metastasis that would be expected to consistently lead to a subsequent axillary lymph node (ALN) dissection. In the SLN dissection procedure,13,14,15 radioactive and colored tracers are injected around the tumor to identify the SLN(s). A lack of metastasis in SLN is highly predictive of a lack of overall ALN metastasis.16,17 If SLN metastasis is detected by histological analysis, the remaining ALNs are typically removed in a secondary surgical procedure as a therapeutic measure and to facilitate patient staging. Because of the high frequency of postsurgical morbidity18,19 associated with axillary lymph node dissection (ANLD), the SLN dissection procedure has emerged as the standard of care. To streamline the SLN process, many institutions have adopted intraoperative SLN analysis methods such as imprint cytology and frozen section analysis; however, these histological methods suffer from poor and variable sensitivity and a lack of standardization.20,21,22 The development of a rapid RT-PCR assay based on gene expression markers of metastasis would represent a major step forward for the field of intraoperative SLN analysis and for the application of molecular diagnostic technology at the point of surgery.
To fulfill this need, a gene marker is required that satisfies the analytical requirements for this assay. An ideal marker would have close to 100% specificity and sensitivity substantially greater than current intraoperative methods. Based on existing literature, it is likely that multiple markers may be required to achieve optimal correlation with histology. Here, we describe a study commencing with a genome-wide survey for breast tissue-specific and breast cancer status markers, concluding with in-depth characterization of candidate markers in sentinel lymph nodes, to identify the ideal markers for the detection of clinically actionable metastases in SLNs.
Materials and Methods
Selection of Markers from an Expressed Sequence Tag (EST) Database
Putative breast-specific and breast cancer status markers were identified using the Incyte EST database. Initial screening of markers was based on analysis of 325,539 ESTs from 35 normal breast tissue libraries, 179,640 ESTs from 25 breast tumor libraries, and 332,558 ESTs from 40 peripheral blood libraries. For each gene analyzed, the frequency of its ESTs in normal breast tissue, breast tumor, and peripheral blood was determined. If more than one EST was observed in the peripheral blood libraries, the marker was eliminated from further consideration. For each EST from genes that passed this initial screen, a Fisher’s exact test was used 1) to compare its frequencies in normal breast and peripheral blood; 2) to compare its frequencies in breast tumor and peripheral blood; and 3) to compare its frequencies in normal breast and breast tumor. If the P values of both comparisons 1 and 2 were less than 0.01 and the level of expression in breast tissue and breast tumor was higher than in peripheral blood, the gene was selected for analysis as a potential breast tissue-specific marker. If the P values of all comparisons (1, 2, and 3) were less than 0.01 and the expression level in breast tumor was higher than in breast tissue, the EST was selected as a potential breast cancer status marker. For each of the potential breast tissue-specific markers, the number of ESTs observed in colon, lung, and ovarian libraries was determined. If more than one EST was found in any one of the tissues, the marker was eliminated. Selected markers were used to search literature databases. Markers previously implicated in breast cancers were given highest priority, although a subset of the markers tested were previously unpublished.
DNA Microarray Samples and Methods
Five hundred and fifty-two primary cancer, 29 benign proliferative epithelial lesions, and 143 normal snap-frozen human tissue specimens were obtained from Genomics Collaborative, Inc. (Cambridge, MA), Clinomics Biosciences, Inc. (Pittsfield, MA), Washington University (St. Louis, MO), and Erasmus Medical Center (Rotterdam, The Netherlands). Tissue was collected and studied under a protocol approved by the respective Institutional Review Board. Each sample was examined for its histopathological features, preservation, and tumor content.
From cancer samples with >70% tumor cells, benign and normal samples were dissected and homogenized with mechanical homogenizer (IKA T8 Ultra-Turrax, Staufen, Germany) in Trizol reagent (Invitrogen, Carlsbad, CA). Total RNA was isolated from Trizol reagent and precipitated at −20°C with isopropyl alcohol. RNA pellets were washed with 75% ethanol, resuspended in water, and stored at −80°C until use. RNA integrity was examined with Agilent 2100 Bioanalyzer RNA 6000 Nano Assay (Agilent Technologies, Inc., Palo Alto, CA). This study used the Human U133a high-density array (Affymetrix, Santa Clara, CA). RNA amplification, labeling, hybridization, and scanning were all performed by the standard protocols from Affymetrix. The arrays were stained with streptavidin-phycoerythrin (Molecular Probes, Eugene, OR) and scanned using the GeneArray scanner from Affymetrix. Image analysis was performed by using GeneChip MAS 5.0 software from Affymetrix. Expression levels of the probe sets were measured in the RNA samples covering normal, benign, and cancerous tissues from breast, colon, lung, ovarian, prostate, and peripheral blood leukocytes from healthy donors. A quality control procedure was then performed to eliminate the poor signal chips. The first criterion was that the percentage of the genes with a “present” call on a chip needed to be >33%. The second was that the scaling factor of a chip needed to be smaller than 15 when scaled to an average target intensity of 600. The distribution of samples that met these criteria is summarized in Table 1. This set of samples was used to identify putative genes through a process of negative selection filters. To exclude genes expressed in leukocytes from consideration, any probe set with a detection P value <0.05 in at least one leukocyte sample by the default setting of the Affymetrix MAS 5.0 software was removed from further analysis. In total, 7567 probe sets were selected. Next, any probe set that did not at least once fall below the 0.05 detection P value limit in any of the samples other than leukocyte was excluded. The resulting gene set contained 4660 probe sets.
Table 1.
Sample Summary for Microarray Analysis
| Number of Samples
|
||||
|---|---|---|---|---|
| Tumor | Normal | Benign | Total | |
| Breast | 318 | 14 | 6 | 338 |
| Colon | 128 | 39 | 0 | 167 |
| Lung | 45 | 38 | 1 | 84 |
| Ovary | 22 | 32 | 0 | 54 |
| Prostate | 39 | 8 | 22 | 69 |
| Blood | 0 | 12 | 0 | 12 |
To select individual genes specific for the breast samples, the following procedure was performed: 1) genes were selected that had “absent” calls in the blood samples; 2) the 30th, 50th, and 70th percentiles of gene expression signal in breast cancer samples were determined for each gene, as well as the maximum observed expression in other tissues; 3) genes were selected as candidate breast tissue-specific markers if the 30th, 50th, and 70th percentiles of gene expression signal in breast cancer samples was higher than the maximum level of gene expression in other tissues; and 4) genes were selected as candidate breast cancer status markers if the 70th percentile of breast cancer expression exhibited a minimum twofold higher expression in the breast cancer samples versus the maximum expression level observed in normal breast tissue.
Clinical Tissue Sample for PCR Screening Analysis
A total of 129 breast tissue and RNA samples were procured from a variety of commercial sources. For tissue samples, RNA was prepared by a standard Trizol methodology. The breast RNA samples tested included: 19 normal breast, 12 benign breast disease, 12 ductal carcinoma in situ, 2 lobular carcinoma in situ, and all common types of invasive breast cancer (55 ductal, 19 lobular, 5 mucinous, 3 tubular, and 2 medullary). Twenty-four tissue RNA samples from lung, colon, rectum, and ovary (6 normal and 18 cancerous; procured from Biochain, Inc.) were used as tissue specificity samples, along with a white blood cell (WBC) total RNA pool derived from 24 female blood donors with no history of cancer. Reverse transcription was carried out with the Superscript II First Strand cDNA Synthesis kit (Invitrogen), essentially as per manufacturer’s instructions. SYBR Green-based quantitative PCR (at least two amplicons per gene) was carried out using universal amplification/detection conditions developed at Veridex. For all tissue RNAs, the assumption was made that each cell contains 10 pg of total RNA. For WBCs, the level of RNA was empirically determined to be ∼0.5 pg/cell.
Sentinel Lymph Node Patient Samples
Sentinel lymph node RNA samples were made available from the East Carolina University/Anne Arundel Medical Center multicenter trial. This trial is designed to determine the utility of qualitative PCR detection of molecular markers in lymph nodes to predict breast cancer relapse in patients undergoing SLN dissection procedures. All patients underwent a complete axillary dissection after the SLN dissection. As part of the trial protocol, all SLN false-negative patients (H&E-positive patients with no H&E-positive SLNs) were excluded from the trial. The protocol was approved by the Institutional Review Board of each participating site. For these evaluations, alternating serial sections of the SLNs were allotted for permanent section H&E histological analysis or quick-frozen for gene expression studies. RNA was extracted from tissue representing approximately one-half of each node using a Trizol method as previously described.11 The SLN RNA samples were received by Veridex and tested for expression of RNA markers in a blinded fashion (in the absence of any patient identifiers or associated clinical and pathological information).
Pathological Analysis of Sentinel Lymph Nodes
SLN tissue sections allotted for standard pathology were fixed in formalin, processed, and embedded in paraffin. Permanent section H&E histological analysis was carried at four levels on all blocks, with three unstained sections set aside at each level. Most, but not all, sites carried out AE1:3 cytokeratin immunohistochemical analysis on all nodes negative for metastases by H&E, following the lab’s standard protocols.
Lymph Node RNA Quality Control
RNA quality was assessed by Agilent, spectroscopy, and housekeeping gene PCR analyses. The RNA 6000 Nano Assay (Agilent Technologies) was carried out according to manufacturer’s protocol. Spectroscopic analysis of RNA was carried out using the Gene Spec III UV-VIS spectrophotometer (MiraiBio, Inc., Alameda, CA). Housekeeping gene PCR was performed as described in the PCR section. We empirically determined that housekeeping gene analysis was most predictive of sample adequacy. RNA samples were considered to be of poor quality if either of the housekeeping genes gave signals that were at least 3.5 cycles above the mean of lymph node samples tested. If at least one node from a patient was deemed to be of poor quality, the patient was removed from the study unless considered PCR positive for at least one of the breast markers. There were 254 patients and 503 lymph nodes included in the final data set from this study.
Reverse Transcription and Quantitative Polymerase Chain Reaction for Lymph Node RNA Samples
The RNA was reverse transcribed before PCR. Five micrograms of RNA and 25 ng/μl Oligo dT(12–18) (Invitrogen) were incubated at 65°C for 7 minutes and then placed on ice. The following was then included in the above mix: 1× Superscript first strand buffer, 10 mmol/L dithiothreitol, 0.5 mmol/L dNTP, 10 U/μl M-MLV (Moloney murine leukemia virus) reverse transcriptase (RT) (Invitrogen), 0.25 U/μl RNasin (Promega, Madison, WI), and in a final reaction volume of 20 μl. The mix was then incubated at 37°C for 60 minutes and then at 95°C for 5 minutes. The cDNA was then diluted 1:5 in water and stored at −20°C.
TaqMan assays were developed and/or validated for the following seven gene expression markers: mammaglobin (MG),23,24,25 cytokeratin 19 (CK19),26 prolactin-induced protein (PIP),10 B305D (C-form and A-form),27,28 γ-aminobutyric acid A receptor, pi (GABA-pi),27,28 B726,27,28 and prostate-derived ets transcription factor (PDEF).29,30 Porphobilinogen deaminase31 and β-actin were used as housekeeping genes. Primers and hydrolysis probes for each assay are listed in Table 2. All assays were designed to be specific for RNA targets, although the CK19 design was demonstrated to have low-level cross-reactivity with CK19 pseudogene DNA. β-Actin primers and probe sequences were as recommended by Applied Biosystems (Foster City, CA) (TaqMan β-actin control reagents, part 401846). All probes were used at a final concentration of 0.04 μmol/L, and all primers were at 0.3 μmol/L. Quantitation of gene-specific RNA was carried out on the ABI Prism 79000HT sequence detection systems (Applied Biosystems). A standard curve was carried out with each thermocycler run that consisted of cDNA generated from target gene in vitro transcript that was serially diluted into carrier cDNA from pig lymph node. No target controls were also included in each assay run to ensure a lack of environmental contamination. All samples and controls were run in triplicate.
Table 2.
TaqMan Assay Primer and Probe Sequences
| Gene | Type | Sequence (5′ to 3′) |
|---|---|---|
| MG | F | CAAACGGATGAAACTCTGAGCAATGTTGA |
| R | TCTGTGAGCCAAAGGTCTTGCAGA | |
| P | TGTTTATGCAATTAATATATGACAGCAGTCTTTGTG | |
| CK19 | F | AGATGAGCAGGTCCGAGGTTA |
| R | CCTGATTCTGCCGCTCACTATCA | |
| P | ACCCTTCAGGGTCTTGAGATTGAGCTGCA | |
| PDEF | F | GCCGCTTCATTAGGTGGCTCAA |
| R | AGCGGCTCAGCTTGTCGTAGTT | |
| P | AAGGAGAAGGGCATCTTCAAAATTGAGGACTCAGC | |
| B305D C-form | F* | TCTGATAAAGGCCGTACAATG |
| R* | TCACGACTTGCTGTTTTTGCTC | |
| P* | ATCAAAAAACAAGCATGGCCTCACACCACT | |
| B305D A-form | F | GTATCTTCTCAAGATCTGGAAAG |
| R | AAGTCTTGTTCTGGATTGCTGT | |
| P | AGTCATCATCATGTAATTTGCCAGTTACT | |
| PIP | F | GCTTGGTGGTTAAAACTTACC |
| R | TGAACAGTTCTGTTGGTGTA | |
| P | CTGCCTGCCTATGTGACGACAATCCGG | |
| GABA-pi | F* | CAATTTTGGTGGAGAACCCG |
| R* | GCTGTCGGAGGTATATGGTG | |
| P* | CATTTCAGAGAGTAACATGGACTACACA | |
| B726 | F* | GCAAGTGCCAATGATCAGAGG |
| R* | ATATAGACTCAGGTATACACACT | |
| P* | TCCCATCAGAATCCAAACAAGAGGAAGATG | |
| Porphobilinogen deaminase | F | CTGCTTCGCTGCATCGCTGAAA |
| R | CAGACTCCTCCAGTCAGGTACA | |
| P | CCTGAGGCACCTGGAAGGAGGCTGCAGTGT |
F, forward primer; R, reverse primer; P, probe.
Two microliters of cDNA template was added into each 50-μl reaction. Fifty cycles of PCR were performed with TaqMan PCR Core Reagents (Applied Biosystems) using 0.0375 U/μl AmpliTaq Gold; 1× buffer A; 5 mmol/L MgCl2; 0.2 mmol/L each of dCTP, dATP, and dGTP; 0.4 mmol/L dUTP; 0.01 U/μl AmpErase UNG; 8% glycerol (Sigma, St. Louis, MO); and 0.01% Tween 20. The following cycling parameters were followed: 1 cycle at 50°C for 2 minutes; 1 cycle at 95°C for 10 minutes; and 50 cycles of 95°C for 15 seconds, 60°C for 1 minute (58°C for B726 only), and 68°C for 1 minute.
Data Analysis
Data were reported in threshold cycles (Ct). The Ct is defined as the cycle at which a statistically significant increase in normalized reporter emission is seen. Each 1 cycle increase in Ct represents a relative two-fold increase in the expression level. Samples were unblinded at the conclusion of the PCR testing. H&E and immunohistochemistry results, recurrence, and additional pathological data were made available at such time. Ct cut-offs were established for determination of positive lymph node status using nonweighted multivariate receiver operating characteristic (ROC) analysis. The performances of individual gene markers and marker combinations were determined on a per patient basis, using H&E-positive histopathological detection of metastases (in the absence of supporting immunohistochemistry) as the gold standard. Results from B305D A-form and C-form testing were pooled for analysis purposes, because common primers can be developed that detect both gene family members.
Determination of Confidence Intervals for Sensitivity and Specificity
For the marker combination that was considered to give optimal performance (CK19 + mammaglobin), bootstrap analysis was carried out to determine 95% confidence ranges for the sensitivity and specificity using the marker Ct cut-offs determined in this analysis. The resulting confidence intervals were based on 5000 bootstrapping samples.
Results
Identification of Gene Set for Clinical Testing
The goal of this study was to identify gene expression markers that had the potential to act as breast-specific markers and/or as markers of cancerous breast tissue. Such markers would have utility for the detection of breast cancer metastasis in several bodily compartments, including lymph nodes, bone marrow, and blood. Several methods were used to identify markers specifically expressed in breast tissues and/or substantially up-regulated in cancerous breast tissue versus normal and benign breast tissues. Our marker identification strategy included microarray analysis, analysis of EST libraries in the Incyte database, and searches of primary literature to identify candidate markers for further analysis. Markers that passed initial inclusion criteria were then passed through a series of RT-PCR-based filters to identify optimal breast tissue markers and breast cancer status markers. A subset of these genes then underwent further large-scale validation to demonstrate clinical utility for the detection of clinically actionable metastasis in breast SLNs (for process flow chart, see Figure 1). Table 2 provides the names of the putative markers selected for RT-PCR analysis and also summarizes the results of the testing that was done with these markers.
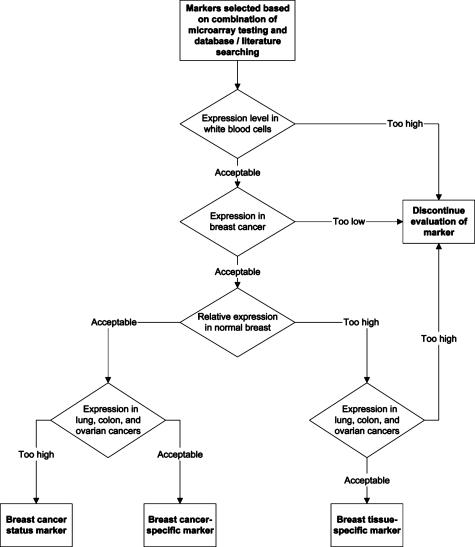
Figure 1.
Marker selection process. Identification of breast-specific markers and/or breast cancer-status markers was carried out using a series of four filters: 1) expression in WBCs, 2) expression level in breast cancer tissues, 3) expression level in normal/benign breast tissues, and 4) expression in other epithelial cancers. Markers had to pass the first two criteria and either of the last two criteria to be selected.
All selected markers were initially screened by RT-PCR for absolute expression level in a WBC RNA pool. Markers with significant expression (>0.1 copy/cell) in the WBC pool were eliminated from further screening. Markers that passed this criterion were then screened with 69 breast tissue RNA samples (16 normal/benign and 53 cancer) and 24 total RNA samples from normal and cancerous colon, rectal, lung, and ovarian tissues. Markers were selected for continued testing as breast tissue-specific markers if the following two criteria were met: 1) expression level in breast tissue >5 copies/cell in >40% of the breast samples tested (based on the assumption of 10 pg RNA per cell); and 2) expression levels ≤5 copies/cell in >90% of ovarian, lung, and colorectal cancer tissue RNAs tested. Genes demonstrating promise as breast tissue-specific markers were subsequently tested with additional samples to increase the overall breast tissue RNA data set to 129 samples (19 normal breast, 12 benign breast disease, and 98 breast cancer RNA samples). Markers were initially screened as potential breast cancer status markers and were selected for additional testing if the following two criteria were met: 1) expression level in breast tissue >5 copies/cell in >40% of the breast samples tested (based on the assumption of 10 pg RNA per cell); and 2) detection of >20% of cancerous samples using cut-offs that detected <15% of the benign and normal breast samples tested (specificity >85%). Markers selected for additional testing were subsequently tested with the 129 breast tissue RNA samples described above and the sensitivity/specificity criteria were further raised to >30 and >90%, respectively.
Based on the testing described above, the following five breast tissue-specific genes were selected for further testing as potential markers for the detection of metastasis of breast cancer to sentinel lymph nodes: MG, PIP, B305D, GABA-pi, and B726. These markers have all been previously published as having utility in the de-tection of metastasis of breast cancer to lymph nodes.10,23,28 Even though B305D did not pass all of our screening criteria, it was selected for additional testing because it had previously been demonstrated to complement three of the other markers selected for testing.27,28 In addition, two of the breast cancer status markers identified, CK19 and PDEF, were selected for further testing in lymph nodes. These genes were selected because 1) our microarray and RT-PCR data demonstrated that they are expressed at significant levels in nearly all breast cancers; 2) our data demonstrated that they are up-regulated in approximately 30 to 40% of breast cancers; and 3) both genes have previously been documented to have utility for the detection of lymph node metastases in breast cancer patients.26,32 PDEF also demonstrated mean expression in breast tissue that was higher than in the other tissues tested (even though the marker did not meet our criteria for breast tissue specificity). Based on the testing described above, the seven markers selected for additional testing offered the best opportunity to identify an optimal gene for the detection of metastasis of breast cancer to sentinel lymph nodes.
Performance of Individual Gene Markers
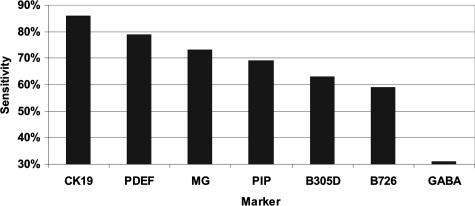
The seven markers selected above were tested on all available sentinel lymph nodes from approximately 300 sentinel lymph node dissection patients. The purpose of this testing was to identify genes whose expression closely correlated with the histological detection of clinically actionable metastasis. Initial analysis of the marker expression data clearly identified a small number of pathology-negative samples that demonstrated high levels of signal for several of the markers tested. This observation, combined with the recognition that the histological methods used in this study (total of three sections analyzed by H&E, representing a 100-μm section of the lymph node) would be expected to miss a subset of samples that contained metastases >0.2 mm, led us to test a variety of specificity cut-offs in the analysis of marker performance. Because of the presence of significant expression of at least three genes in 6% (11 of the 183) pathology-negative patients, a specificity of 94% was chosen for assessing the sensitivity of the individual markers (Figure 2). Based on this criterion, the highest single gene sensitivity was observed with the epithelial cell marker CK19 (86% sensitivity), followed by PDEF (79% sensitivity) and the breast-specific marker mammaglobin (73% sensitivity).
Figure 2.
Sensitivity of markers in lymph nodes. Individual marker sensitivity was assessed in lymph node tissues by RT-PCR. A specificity of 94% was used to identify individual markers that offered optimal performance for detection of clinically actionable metastases.
Performance of Marker Combinations
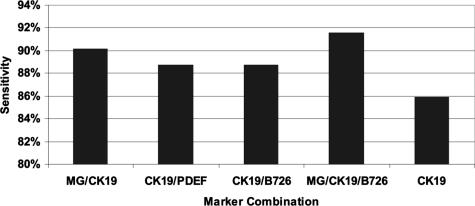
To increase sensitivity, we sought to determine the optimal set of two or more genes for correlation with histology. This optimal gene set would have the potential to form the basis of an intraoperative assay for the detection of metastasis in breast sentinel lymph nodes. For the same reasons described above for the single marker testing, optimal sensitivity of marker combinations was also determined at a specificity of 94%, using nonweighted ROC analysis (Figure 3). Based on these criteria, the optimal marker combination was mammaglobin plus CK19, which demonstrated 90% sensitivity at 94% specificity (Table 4). Only one three-gene combination, MG/CK19/B726, demonstrated an improvement in performance, with B726 complementing one of the seven MG/CK19-negative samples. Additional markers did not complement MG/CK19/B726 in the remaining six PCR false-negative samples. These results suggest that the majority of observed PCR false negatives likely resulted from uneven sampling during the splitting of sample between PCR and pathology, rather than from lack of expression of the individual markers being tested. This conclusion is supported by the fact that the testing of the primary tumors from four of the molecular false-negative patients demonstrated substantial expression of mammaglobin and/or CK19 in all four tumors tested (data not shown). Additional support for this hypothesis is provided by the fact that five of the seven false-negative samples were characterized as micrometastases by histological analysis.
Figure 3.
Sensitivity of marker combinations in lymph nodes. Two-marker and three-marker combinations were assessed in lymph node tissues by RT-PCR. A specificity of 94% was used to identify marker combinations that offered optimal performance for detection of clinically actionable metastases.
Table 4.
Performance of Two-Gene Combination Relative to Standard Histology
| Formalin-fixed paraffin-embedded H&E (+) without immunohistochemistry
|
|||
|---|---|---|---|
| + | − | ||
| MG/CK19 | + | 64 | 11 |
| − | 7 | 172 | |
| Total | 71 | 183 | |
Sensitivity, 90%; Specificity, 94%; positive predictive value, 85%; negative predictive value, 96%.
We determined the two-gene combination of mammaglobin and CK19 to be the optimal marker set for a variety of reasons. First, the impact of the third gene B726 was minimal and not convincing—the one sample detected by B726 was weakly positive with B726 and weakly negative with CK19. Second, addition of another gene to a multiplexed assay would likely have a negative impact on assay performance. Finally, the low expression of B726 made it an unattractive target for an intraoperative assay, due to the need to extend the number of thermocycles run to determine B726 positivity (the Ct threshold used for B726 was >5 cycles higher than the thresholds used for mammaglobin and CK19).
Further analysis of the 11 pathology-negative samples that were MG/CK19 positive is summarized in Tables 4and 5. Several pieces of evidence strongly suggest that the majority of these samples referred to as molecular false positives are in reality true positives missed by pathology. The frequency of RT-PCR-positive results was similar between the pathology-positive patients and the pathology-negative, mammaglobin/CK19-positive patients for all markers tested; in contrast, the frequency of RT-PCR-positive results was 9- to 42-fold lower in pathology-negative, mammaglobin/CK19-negative samples and consistently <5% across all markers (Table 5). The mean number of markers testing positive was identical between the pathology-positive and the pathology-negative, mammaglobin/CK19-positive samples; in both cases, this level was 33-fold higher than in pathology-negative, mammaglobin/CK19-negative samples. Several of the samples referred to as molecular false positive are positive with all or nearly all of the seven markers tested (Table 6). In many cases, several markers were highly expressed (Ct values at least five cycles below the Ct cut-off for positivity). Several of these genes are up-regulated in breast cancer tissues and should not be detected unless a large number of cancerous cells were present in the sample. One of the samples categorized as molecular false positive was actually determined to be H&E positive on reexamination after initial identification by IHC (following an initial diagnosis of H&E negative). This sample, identified with an asterisk in Table 6, was positive for five of the markers tested. The fact that the criteria for presumptive positivity failed to call this sample positive suggests that the criteria for identifying presumptive positives may be excessively stringent.
Table 5.
Performance of RT-PCR Markers
| Marker | Histology positive (%) | MG/CK19 positive, histology negative (%) | MG/CK19 negative, histology negative (%) |
|---|---|---|---|
| MG | 73 | 82 | NA |
| CK19 | 86 | 82 | NA |
| PDEF | 79 | 73 | 1.7 |
| PIP | 69 | 73 | 1.7 |
| B305D | 70 | 73 | 2.3 |
| GABA | 31 | 36 | 4.1 |
| B726 | 59 | 45 | 3.5 |
| Markers (+) | 5.2 | 5.2 | 0.16 |
Table 6.
Identification of Presumptive Positive Patients
| Patient | MG | VBM1 | Total no. of markers positive | Markers strongly positive | Presumptive positive |
|---|---|---|---|---|---|
| 1 | ++ | ++ | 7 | 6 | + |
| 2 | ++ | 5 | 4 | + | |
| 3 | ++ | ++ | 7 | 3 | + |
| 4 | ++ | + | 7 | 3 | + |
| 5 | ++ | ++ | 4 | 3 | + |
| 6 | ++ | + | 4 | 1 | + |
| 7 | ++ | 2 | 1 | ||
| 8 | ++ | 2 | 1 | ||
| 9 | + | + | 6 | 0 | |
| 10* | + | + | 5 | 0 | |
| 11 | + | 1 | 0 |
+, PCR positive (Ct ≤ cut-off); ++, strongly PCR positive (>5 cycles below Ct cut-off); Presumptive positive, PCR positive with ≥4 markers, strongly PCR positive with ≥1 marker.
Sample detected by H&E only after immunohistochemistry.
We stratified molecular marker-positive, H&E-negative patients on the likelihood that the patients’ nodes truly contained significant metastases that were missed by standard pathology. Patients were deemed to be presumptively H&E positive if at least four of the seven markers were positive, with at least one marker strongly positive (Ct value at least five cycles below the Ct cut-off for positivity). Applying these criteria, 6 of the 11 molecular positives were presumptive H&E positives (Table 6). When compared with both H&E positivity and presumptive H&E positivity, the adjusted performance of mammaglobin and CK19 was 91% sensitivity and 97% specificity (Table 7).
Table 7.
Performance of Two-Gene Combination Relative to Presumptive Positivity
| Formalin-fixed paraffin-embedded H&E (+) or presumptive (+)
|
|||
|---|---|---|---|
| + | − | ||
| MG/CK19
|
+
|
70
|
5 |
| − | 7 | 172 | |
| Total | 77 | 177 | |
Sensitivity, 91%; Specificity, 97%; PPV, 93%; NPV, 96%.
Determination of Confidence Intervals for Performance Parameters of Optimal Two-Gene Combination
Bootstrap analysis was used to determine 95% confidence intervals for sensitivity and specificity for the marker Ct cut-offs that gave the optimal observed sensitivity at 94% specificity (Ct = 31.7 for mammaglobin and Ct = 30.9 for CK19, observed sensitivity of 90%). This analysis demonstrated 95% confidence intervals of 83 to 97% for sensitivity and 90 to 97% for specificity. Applying the same type of analysis to the data set that included presumptive H&E-positive results (observed sensitivity and specificity of 91 and 97%, respectively) led to 95% confidence intervals of 84 to 97% for sensitivity and 95 to 99.4% for specificity. The individual marker cut-offs that gave optimal performance were unchanged.
Discussion
We have used a global strategy for identifying optimal markers for the metastasis of breast cancer. To achieve this, we have used a series of filters to sequentially reduce the number of candidate breast tissue and/or breast cancer status gene expression markers tested to 14. Seven of these markers underwent further validation for the detection of breast metastasis in SLNs. The ability to test a significant number of markers on a high-density chip has the potential to dramatically improve the probability of selecting markers that meet specific diagnostic requirements. Our results strongly suggest that microarray analysis is an ideal method of predicting success for markers previously published as having desirable expression behavior. Eight of the 10 literature markers also identified by microarray analysis passed our screening criteria for utility as breast tissue-specific or breast cancer status markers. In contrast, only 3 of the 13 markers identified by literature but not confirmed by microarray analysis were deemed to provide acceptable performance.
In addition to demonstrating the utility of combining literature and microarray methods in the marker selection process, we also report here the identification of an optimal two-gene expression marker set for detection of clinically actionable metastasis in breast sentinel lymph nodes. The two specific markers identified, mammaglobin and CK19, have been previously published as potentially useful components of multimarker panels for the identification of occult metastasis in breast lymph nodes.12,32,33 However, previous studies have also concluded that both markers are expressed in the lymph nodes of 20 to 40% of patients categorized as lymph node negative by standard histological analysis.10,11 It has also been reported that lymph nodes from a high percentage of cancer-free patients express CK19.34,35 This report represents the first statistically significant data set demonstrating the feasibility of using a combination of molecular markers for the detection of clinically actionable metastases in breast lymph nodes. We have subsequently developed a rapid RT-PCR assay using mammaglobin and CK19 that generates a result in ≤30 minutes from lymph node tissue (J. Backus, G. Green, M. Xu, J. Painter, S. Varde, unpublished data).
One of the challenges in accurately determining the specificity of this molecular assay is the relatively high false-negative rate observed with standard H&E pathology interpretation. Estimates of the true false-negative rates for the detection of metastases range from 5 to 15%.36,37 Analysis of the seven molecular markers tested demonstrated that the majority of mammaglobin/CK19 molecular false positives were also positive with several other breast tumor markers not expressed in the vast majority of H&E-negative samples. Samples demonstrating significant expression of mammaglobin and/or CK19 have a high probability of also expressing the other markers tested, including B305D, GABA, B726, and PDEF, markers previously demonstrated to have increased expression in cancerous breast tissues.27,28 A significant conclusion from this testing is that any clinical study designed to determine correlation between a molecular assay and standard H&E pathology needs to employ comprehensive H&E analysis to minimize the risk of not detecting clinically significant metastases, leading to false-negative H&E results that will be subsequently interpreted as molecular false-positive results.
A major criticism of RT-PCR detection of occult metastasis is that it can be too sensitive, detecting as little as one copy of RNA for genes that can be expressed at levels of >1000 copies/cell. By using quantitative PCR with thresholds based on clinical samples, a clinically significant level of RNA expression can be determined, minimizing the risk of false-positive results. The threshold used for mammaglobin in this testing, for instance, correlates to the presence of approximately 10,000 copies of RNA in the amplification reaction, which uses a median of approximately 0.03% of the total RNA in a typical tissue sample. Assuming expression of 1000 copies of mammaglobin mRNA per cell, a positive sample would represents a minimum of approximately 70,000 cells expressing mammaglobin in the original sample. This is more than 10-fold more cells than would be expected to be present in a 0.2-mm micrometastasis, suggesting that the thresholds developed are consistent with the detection of clinically significant metastasis, although biased toward specificity at the cost of sensitivity. We believe this performance bias to be appropriate for application of the gene set to an intraoperative assay designed to direct additional axillary lymph node dissection. This performance bias would also be expected to minimize the risk of detecting benign lesions that are infrequently observed in breast lymph nodes. These lesions are typically small and only visualized with the assistance of immunohistochemistry.
In summary, we have used a whole-genome approach, using a series of filters, to identify gene expression markers that correlate with standard histological detection of breast lymph node metastasis with high sensitivity and specificity. A variety of data and a large body of literature evidence support the conclusion that most of the discrepancies that do exist between the RT-PCR assay and standard permanent section H&E results represent sampling errors between the two methods used. We further demonstrate that analysis of two marker genes, mammaglobin and CK19, can form the basis of a rapid RT-PCR assay for the detection of clinically actionable metastases. We have subsequently developed a rapid assay based on this marker set and are in the process of validating the clinical performance of this assay. Such an assay has the potential to lead to a significant reduction in the number of unnecessary second surgeries to remove axillary lymph nodes in sentinel lymph node-positive patients.
Table 3.
Markers Tested and Testing Summary
| Breast marker type
|
Identification method
|
Mode of marker failure
|
Marker adequate? | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tissue | Cancer | Incyte | Micro-array | Literature | Low breast expression | High WBC expression | Tissue cross-reactivity | Inadequate up-regulation | ||
| Putative breast-specific markers | ||||||||||
| Mammaglobin | X | X | X | X | NA | Y | ||||
| PIP | X | X | X | X | NA | Y | ||||
| Small breast epithelial mucin | X | X | X | NA | Y | |||||
| Gamma-aminobutyric acid A receptor, pi | X | X | X | NA | Y | |||||
| Breast tumour-specific gene B726P | X | X | NA | Y | ||||||
| ML-1 protein | X | X | X | X | NA | N | ||||
| FLJ22418 fis, clone HRC08590 | X | X | X | X | NA | N | ||||
| Iroquois-class homeodomain protein (IRX-2A) | X | X | X | NA | N | |||||
| Breast tumour-specific gene B311D | X | X | X | NA | N | |||||
| Breast tumour-specific gene B305D | X | X | X | NA | N | |||||
| Breast tumour-specific gene B533S | X | X | X | NA | N | |||||
| Putative breast cancer-specific markers | ||||||||||
| PDEF | X | X | X | X | X | X | N/Y | |||
| FLJ22945 fis, clone KAT09065 | X | X | X | X | N | |||||
| Putative breast cancer status markers | ||||||||||
| Nonspecific cross-reacting antigen (CEACAM-6) | X | X | X | X | NA | Y | ||||
| Grb7V protein | X | X | X | X | NA | Y | ||||
| Polymorphic epithelial mucin | X | X | X | X | NA | Y | ||||
| CK19 | X | X | X | NA | Y | |||||
| Troponin T1, skeletal, slow | X | X | NA | Y | ||||||
| S100-type calcium binding protein A14 | X | X | NA | Y | ||||||
| S100 calcium-binding protein A7 (psoriasin 1) | X | X | NA | Y | ||||||
| Erb-b-2 (Her2/neu) | X | X | NA | Y | ||||||
| Secretory Protein (P1.B) | X | X | X | NA | X | N | ||||
| Integrin β-5 Subunit | X | X | X | X | NA | X | N | |||
| Trefoil factor 3, intestinal (TFF3) | X | X | X | X | NA | X | N | |||
| Glycoprotein hormones, α polypeptide (CGA) | X | X | X | NA | X | N | ||||
| Carboxypeptidase B1 | X | X | NA | X | N | |||||
| Fatty acid binding protein, brain-type (hB-FABP) | X | X | NA | X | N | |||||
| UROC28 | X | X | X | NA | X | N | ||||
| Survivin | X | X | NA | X | N | |||||
| Epidermal growth factor receptor (EGFR) | X | X | NA | X | N | |||||
| Plasminogen activator, urokinase receptor (uPAR) | X | X | NA | X | N | |||||
| Synuclein, γ (breast cancer-specific gene BCSG1) | X | X | NA | X | N | |||||
Acknowledgments
We acknowledge Michelle Homick and Peggy Kriger for their efforts that supported the initial marker selection and assay feasibility, Yi Zhang for the boot-strapping analysis, Tim Jatkoe for the ROC analysis, George Green for assistance with the marker flow diagram, and Ron Mazumder and Karen Gray for critical reading and feedback on this manuscript. We also thank Barbara Zehentner and Ray Houghton for consultation during this testing and for design of the PIP TaqMan probe used in this testing.
References
- Anonymous Treatment of early-stage breast cancer: NIH consensus conference. J Am Med Assoc. 1991;265:391–395. [PubMed] [Google Scholar]
- Valgussa P, Bonadonna G, Veronesi U. Patterns of relapse and survival following radical mastectomy. Cancer. 1978;41:1170–1178. doi: 10.1002/1097-0142(197803)41:3<1170::aid-cncr2820410355>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Fisher B, Bauer M, Wickeehan W, Redmond CK, Fisher E. Relationship of number of positive axillary nodes to the prognosis of patients with primary breast cancer: an NSABP update. Cancer. 1983;52:1551–1557. doi: 10.1002/1097-0142(19831101)52:9<1551::aid-cncr2820520902>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Braun S, Pantel K, Muller P, Janni W, Hepp F, Kentenich CR, Gastroph S, Wischnik A, Dimpfl T, Kindermann G, Riethmuller G, Schlimok G. Cytokeratin-positive cells in the bone marrow and survival of patients with stage I, II, or III breast cancer. N Engl J Med. 2000;342:525–533. doi: 10.1056/NEJM200002243420801. [DOI] [PubMed] [Google Scholar]
- Braun S, Pantel K. Clinical significance of occult metastatic cells in bone marrow of breast cancer patients. The Oncologist. 2001;6:125–132. doi: 10.1634/theoncologist.6-2-125. [DOI] [PubMed] [Google Scholar]
- Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- Siegel RJ. Surgical pathology of lymph nodes in cancer staging: routine and specialized techniques. Surg Oncol Clin N Am. 1996;5:25–31. [PubMed] [Google Scholar]
- Cote RJ, Peterson HF, Chaiwun B, Gelber RD, Goldhirsch A, Castiglione-Gertsch M, Gusterson B, Neville AM. Role of immunohistochemical detection of lymph-node metastases in management of breast cancer. Lancet. 1999;354:896–900. doi: 10.1016/s0140-6736(98)11104-2. [DOI] [PubMed] [Google Scholar]
- Slade MJ, Smith BM, Sinnett HD, Cross NC, Coombes RC. Quantitative polymerase chain reaction for the detection of micrometastases in patients with breast cancer. J Clin Oncol. 1999;17:870–879. doi: 10.1200/JCO.1999.17.3.870. [DOI] [PubMed] [Google Scholar]
- Lockett MA, Baron PL, O’Brien PH, Elliott BM, Robison JG, Maitre N, Metcalf JS, Cole DJ. Detection of occult breast cancer micrometastases in axillary lymph nodes using a multimarker reverse transcriptase-polymerase chain reaction panel. J Am Coll Surg. 1998;187:9–16. doi: 10.1016/s1072-7515(98)00130-6. [DOI] [PubMed] [Google Scholar]
- Min CJ, Tafra L, Verbanac KM. Identification of superior markers for polymerase chain reaction detection of breast cancer metastases in sentinel lymph nodes. Cancer Res. 1998;58:4581–4584. [PubMed] [Google Scholar]
- Gillanders WE, Mikhitarian K, Hebert R, Mauldin PD, Palesch Y, Walters C, Urist MM, Mann GB, Doherty G, Herrmann VM, Hill AD, Eremin O, El-Sheemy M, Orr RK, Valle AA, Henderson MA, Dewitty RL, Sugg SL, Frykberg E, Yeh K, Bell RM, Metcalf JS, Elliott BM, Brothers T, Robison J, Mitas M, Cole DJ. Molecular detection of micrometastatic breast cancer in histopathology-negative axillary lymph nodes correlates with traditional predictors of prognosis: an interim analysis of a prospective multi-institutional cohort study. Ann Surg. 2004;6:828–837. doi: 10.1097/01.sla.0000128687.59439.d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton DL, Wen D-R, Wong JH, Economou JS, Cagle LA, Storm FK, Foshag LJ, Cochran AJ. Technical details of intraoperative lymphatic mapping for early stage melanoma. Arch Surg. 1992;127:392–399. doi: 10.1001/archsurg.1992.01420040034005. [DOI] [PubMed] [Google Scholar]
- Krag DN, Weaver DL, Alex JC, Fairbank JT. Surgical resection and radiolocalization of the sentinel lymph node in breast cancer using a gamma probe. Surg Oncol. 1993;2:335–339. doi: 10.1016/0960-7404(93)90064-6. [DOI] [PubMed] [Google Scholar]
- Giuliano AE, Kirgan DM, Guenther JM, Morton DL. Lymphatic mapping and sentinel lymphadectomy for breast cancer. Ann Surg. 1994;220:391–398. doi: 10.1097/00000658-199409000-00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner RR, Ollila DW, Krasne DL, Giuliano AE. Histopathological validation of the sentinel lymph node hypothesis for breast carcinoma. Ann Surg. 1997;226:271–278. doi: 10.1097/00000658-199709000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMasters KM, Giuliano AE, Ross MI, Reintgen DS, Hunt KK, Byrd DR, Klimberg VS, Whitworth PW, Tafra LC, Edwards MJ. Sentinel-lymph-node biopsy for breast cancer: not yet the standard of care. N Engl J Med. 1998;339:990–995. doi: 10.1056/NEJM199810013391410. [DOI] [PubMed] [Google Scholar]
- Ivens D, Hoe AL, Podd TJ, Hamilton CR, Taylor I, Royle GT. Assessment of arm morbidity from complete axillary dissection. Br J Cancer. 1992;66:1368–1373. doi: 10.1038/bjc.1992.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissin MW, Querci della Rovere G, Easton D, Westbury G. Risk of lymphoedema following the treatment of breast cancer. Br J Surg. 1986;73:580–584. doi: 10.1002/bjs.1800730723. [DOI] [PubMed] [Google Scholar]
- Turner RR, Hansen NM, Stern SL, Giuliano AE. Intraoperative examination of the sentinel lymph node for breast carcinoma staging. Am J Clin Pathol. 1999;112:627–634. doi: 10.1093/ajcp/112.5.627. [DOI] [PubMed] [Google Scholar]
- Van Diest PJ, Torrenga H, Borgstein PJ, Pijpers R, Bleichrodt RP, Rahusen FD, Meijer S. Reliability of intraoperative frozen section and imprint cytological investigation of the sentinel lymph nodes in breast cancer. Histopathology. 1999;35:14–18. doi: 10.1046/j.1365-2559.1999.00667.x. [DOI] [PubMed] [Google Scholar]
- Weiser MR, Montgomery LL, Susnik B, Tan LK, Borgen PI, Cody HS. Is routine intraoperative frozen-section examination of sentinel lymph nodes in breast cancer worthwhile? Ann Surg Oncol. 2000;7:651–655. doi: 10.1007/s10434-000-0651-3. [DOI] [PubMed] [Google Scholar]
- Watson MA, Dintzis S, Darrow CM, Voss LE, DiPersio J, Jensen R, Fleming TP. Mammaglobin expression in primary, metastatic, and occult breast cancer. Cancer Res. 1999;59:3028–3031. [PubMed] [Google Scholar]
- Fleming TP, Watson MA. Mammaglobin, a breast-specific gene and its utility as a marker for breast cancer. Ann NY Acad Sci. 2000;923:78–89. doi: 10.1111/j.1749-6632.2000.tb05521.x. [DOI] [PubMed] [Google Scholar]
- Zehentner BK, Carter D. Mammaglobin: a candidate diagnostic marker for breast cancer. Clin Biochem. 2004;37:249–257. doi: 10.1016/j.clinbiochem.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Noguchi S, Aihara T, Motomura K, Inaji H, Imaoka S, Koyama H. Detection of breast cancer micrometastases in axillary lymph nodes by means of reverse transcriptase-polymerase chain reaction: comparison between MUC1 mRNA and keratin 19 mRNA amplification. Am J Pathol. 1996;148:649–656. [PMC free article] [PubMed] [Google Scholar]
- Houghton RL, Dillon DC, Molesh DA, Zehentner BK, Xu J, Jiang J, Schmidt C, Frudakis A, Repasky E, Maltez Filho A, Nolasco M, Badaro R, Zhang X, Roche PC, Persing DH, Reed SG. Transcriptional complementarity in breast cancer: application to detection of circulating tumor cells. Mol Diagn. 2001;6:79–91. doi: 10.1007/BF03262038. [DOI] [PubMed] [Google Scholar]
- Zehentner BK, Dillon DC, Jiang Y, Xu J, Bennington A, Molesh DA, Zhang X, Reed SG, Persing D, Houghton RL. Application of a multigene reverse transcription-PCR assay for detection of mammaglobin and complementary transcribed genes in breast cancer lymph nodes. Clin Chem. 2002;48:1225–1231. [PMC free article] [PubMed] [Google Scholar]
- Ghadersohi A, Sood AK. Prostate epithelium-derived Ets transcription factor mRNA is overexpressed in human breast tumors and is a candidate breast tumor marker and a breast tumor antigen. Clin Cancer Res. 2001;7:2731–2738. [PubMed] [Google Scholar]
- Mitas M, Mikhitarian K, Hoover L, Lockett MA, Kelley L, Hill A, Gillanders WE, Cole DJ. Prostate-specific Ets (PSE) factor: a novel marker for detection of metastatic breast cancer in axillary lymph nodes. Br J Cancer. 2002;86:899–904. doi: 10.1038/sj.bjc.6600190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink L, Seeger W, Ermert L, Hanze J, Stahl U, Grimminger F, Kummer W, Bohle RM. Real-time quantitative RT-PCR after laser-assisted cell picking. Nat Med. 1998;4:1329–1333. doi: 10.1038/3327. [DOI] [PubMed] [Google Scholar]
- Mitas M, Mikhitarian K, Walters C, Baron PL, Elliott BM, Brothers TE, Robison JG, Metcalf JS, Palesch YY, Zhang Z, Gillanders WE, Cole DJ. Quantitative real-time RT-PCR detection of breast cancer micrometastasis using a multigene marker panel. Int J Cancer. 2001;93:162–171. doi: 10.1002/ijc.1312. [DOI] [PubMed] [Google Scholar]
- Manzotti M, Dell’Orto P, Maisonneuve P, Zurrida S, Mazzarol G, Viale G. Reverse transcription-polymerase chain reaction assay for multiple mRNA markers in the detection of breast cancer metastases in sentinel lymph nodes. Int J Cancer. 2001;95:307–312. doi: 10.1002/1097-0215(20010920)95:5<307::aid-ijc0153>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Bostick PJ, Chatterjee S, Chi DD, Huynh KT, Giuliano AE, Cote R, Hoon DS. Limitations of specific reverse-transcriptase polymerase chain reaction in the detection of metastases in the lymph nodes and blood of breast cancer patients. J Clin Oncol. 1998;16:2632–2640. doi: 10.1200/JCO.1998.16.8.2632. [DOI] [PubMed] [Google Scholar]
- Merrie AE, Yun K, Gunn J, Phillips LV, McCall JL. Analysis of potential markers for detection of submicroscopic lymph node metastases in breast cancer. Br J Cancer. 1999;80:2019–2024. doi: 10.1038/sj.bjc.6690636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed W, Bohler PJ, Sandstad B, Nesland JM. Occult metastases in axillary lymph nodes as a predictor of survival in node-negative breast carcinoma with long-tern follow-up. Breast J. 2004;10:174–180. doi: 10.1111/j.1075-122X.2004.21328.x. [DOI] [PubMed] [Google Scholar]
- Dowlatshahi K, Fan M, Anderson JM, Bloom KJ. Occult metastases in sentinel nodes of 200 patients with operable breast cancer. Ann Surg Oncol. 2001;8:675–681. doi: 10.1007/s10434-001-0675-3. [DOI] [PubMed] [Google Scholar]