Abstract
Speed is of the essence when evaluating an infant with symptoms consistent with sepsis. Because of the high morbidity and mortality associated with neonatal sepsis, infants receive multiple, broad-spectrum antibiotics before receiving finalized blood culture results. Incorporating an additional, reliable, yet rapid assay to detect bacteria directly from blood would facilitate timely diagnosis and appropriate care. To this end, we designed a real-time polymerase chain reaction (PCR) assay targeting the highly conserved 380 bases of 16S rDNA. DNA was extracted from whole-blood samples using a Qiagen column. The limit of detection for the TaqMan-based assay, using a Smartcycler instrument, was 40, 50, or 2000 colony-forming units per milliliter of blood (CFU/ml) of Escherichia coli, group B Streptococcus, and Listeria monocytogenes, respectively, when white blood cell counts were below 39,000/μl. Implementing this approach requires less than 4 hours for both sample preparation and real-time PCR compared with 1 to 2 days to detect growth in culture or 5 days to finalize no-growth culture results. There was an overall agreement between the results of culture and real-time PCR of 94.1% (80 of 85) in this study. These results suggest that molecular techniques can augment culture-based methods for diagnosing neonatal sepsis, especially in infants whose mothers have received intrapartum antibiotic prophylaxis.
Neonatal sepsis is a high-risk, low-incidence disease, in which many evaluations are undertaken to discover one case. In addition, accurate diagnosis of neonatal sepsis is difficult because signs and symptoms in the infant may be subtle, often mimicking other medical conditions that frequently occur in the newborn such as hypoglycemia, delayed transition, or transient tachypnea.1,2,3,4 Infants admitted to the neonatal intensive care unit (NICU) for sepsis evaluation are usually managed clinically with bacterial cultures, a complete blood count (CBC), and combination antimicrobial therapy for presumed sepsis; for example, the antibiotics given to the term infant might include ampicillin and gentamicin, or ampicillin and cefotaxime.5,6,7,8,9 Traditionally, infants are provided with antibiotic therapy for 72 hours pending negative culture results.10,11,12
Currently, there is no definitive test for diagnosing neonatal sepsis because even blood culturing techniques have unacceptably low sensitivities.2 Some of the reasons for this include intermittent seeding of low numbers of bacteria within the blood stream, the small volumes of blood obtained from infants for culture, and the increasingly common practice of providing intrapartum antibiotic prophylaxis to mothers of high-risk deliveries.13,14,15 It is well known that the smaller the blood volume obtained for culture, the lower the chances of recovering organisms.16,17 Although infants tend to have a somewhat greater magnitude of bacteremia compared with adults—10 to 300 CFU/ml versus 1 to 30 CFU/ml—the amount of blood sampled for culturing from neonates is significantly less than that taken from adults: <1 ml versus 10 to 30 ml.18
Most approaches to blood culturing use automated instrumentation, like the BACTEC (Becton Dickinson, Sparks, MD), BacT/Alert (Biomérieux, Durham, NC), or ESP (Trek Diagnostic Systems, Inc., Westlake, OH) system, which use sensors that either continuously monitors the CO2 being produced by actively metabolizing microorganisms or sense changes in pressure within the blood culture bottle. Detecting bacterial growth in blood culture bottles requires approximately 12 to 24 hours of incubation for gram-negative organisms or 24 to 48 hours for gram-positive organisms, with additional time needed to isolate the organism contained within the blood culture bottle for identification and antimicrobial susceptibility testing. A study by Kurlat et al19 illustrated positive and negative predictive values of blood cultures at days 2 and 4 of 92 and 99%, respectively. However, speed is of the essence when evaluating an infant who has been admitted to a NICU with symptoms of sepsis. Incorporating an additional, reliable, yet rapid assay that could detect bacteria directly from blood samples would facilitate the timely diagnosis and appropriate care for these infants.
Nucleic acid amplification testing such as the polymerase chain reaction (PCR) assay is rapid to perform, and has been used with success to diagnose a wide range of infectious diseases, including bacterial, yeast/fungal, and protozoan infections.20 In fact, nucleic acid amplification testing is now considered the gold standard for diagnosing herpes simplex virus meningitis in the neonate.21 PCR detection of blood-borne bacteria using targets within 16S rDNA has been described previously.22,23,24 Jordan et al22 (J.A. Jordan, M.B. Durso, A.R. Butchko, J. Jones, B. Brozanski, unpublished data) published the first two large-scale studies for detecting sepsis in the neonate. These studies compared the results of culture (Cx) to conventional PCR in which whole blood was first pre-enriched in tryptic soy broth (TSB) for between 5 and 10 hours at 35°C before sample preparation and testing occurred.
The objective of this current research project was to design a sample preparation protocol that would eliminate the tryptic soy broth pre-enrichment step, saving 5 to 10 hours, and to convert the conventional PCR assay to a more rapid, real-time platform. If successful, this approach would significantly reduce the time needed to obtain laboratory results on infants being evaluated for bacterial sepsis; a real-time PCR approach analyzing the sample directly can be completed in less than 4 hours, whereas conventional PCR amplification using agarose gel-based electrophoresis detection along with TSB pre-enrichment of the sample would require more than 1 full day to complete, whereas bacterial growth in culture requires 1 to 2 days to detect, or 5 days to finalize a no-growth blood culture result.
Materials and Methods
Patient Selection and Sample Rejection Criteria
Our study received hospital institutional review board approval before its initiation. To be eligible, an infant had to be admitted to the neonatal intensive care unit and have blood drawn for both a blood Cx and CBC analyses. The study was designed to use the unused portion of the CBC for 16S rDNA real-time PCR analysis, which eliminated the need for an additional blood draw. Sample rejection for PCR analysis included blood that was either grossly hemolyzed and/or clotted.
Blood Culturing and Phenotypic Identification
Whole blood (0.5–1.0 ml) was collected for culture by either a venous or arterial draw and inoculated directly into a single, pediatric sample-sized resin-containing blood culture bottle (Peds Plus; Becton Dickinson). Each bottle was loaded into the Bactec 9240 automated blood culture instrument within 1 hour from the time of receipt in the laboratory in accordance with the manufacturer’s recommendation.
Bottles flagged by the Bactec instrument for detectable levels of CO2 production had fluid withdrawn for gram stain and subculture on the appropriate agar-based culture plates. Purified bacterial colonies were analyzed either by an automated identification system (MicroScan Dade, West Sacramento, CA) or with the appropriate biochemical reagents for phenotypic identification.
Agar Dilution Method for Quantitating Bacterial Colony-Forming Units Added to Whole-Blood Samples
Individual tryptic soy broth cultures, inoculated with single colony of a pure isolate, were incubated overnight at 37°C. The density of these broth cultures were adjusted to equal that of a 0.5 McFarland standard, representing 2.4 × 108 CFU/ml. Ten-fold dilutions of each 0.5 McFarland standard were prepared, and 10 μl of each dilution was inoculated onto sheep blood agar plates (Becton Dickinson) and incubated overnight at 37°C. The number of colonies found growing on the plates was counted, and the exact number of colony-forming units per milliliter was calculated. The Escherichia coli and Listeria monocytogenes used in the limit of detection studies were purchased from the American Type Culture Collection and consisted of E. coli (ATCC no. 11775) and L. monocytogenes (ATCC no. 15313). The group B Streptococcus (Streptococcus agalactiae) (GBS) used to determine limit of detection and as the positive control included in each real-time PCR assay was isolated in our laboratory from a neonatal blood culture and identified as a β-hemolytic, catalase-negative, gram-positive cocci in chains that reacted with GBS antisera in a latex agglutination reaction (Oxoid Typing kit; Fisher Scientific, Pittsburgh, PA).
Whole-Blood Collection for CBC Analysis
Whole blood (0.3–1.0 ml) was collected for CBC analysis by either venous or arterial draw or by a heel-stick method into a pediatric sample-sized ethylenediamine tetraacetic acid-containing Vacutainer tube (Becton Dickinson). The unused portion of the CBC specimen was made available for real-time 16S rDNA PCR analysis once the CBC result was finalized.
Whole-Blood Sample Processing for PCR
The entire content of the discarded CBC sample was used; sample volumes ranged from 10 to 500 μl (average and median volumes were 154 and 125 μl, respectively). Mutanolysin (Sigma, St. Louis, MO) was added directly to the whole-blood sample; 10 μl of 10 U/μl enzyme was added for every 200 μl of blood being processed, and the sample was incubated for 30 minutes at 37°C. A modified protocol using the QIAamp DNA Mini kit (Qiagen Inc., Valencia, CA) was performed as follows: 20 μl of QIAGEN Proteinase K (20 mg/ml) was added for every 200 μl of whole blood processed along with an equal volume (200 μl) of buffer AL, and the sample was incubated for 30 minutes at 57°C. After incubation, an equal volume (200 μl) of 100% ethanol was added, and the resulting lysate was loaded onto the QIAamp DNA Mini kit column (Qiagen). The column was centrifuged at 6,000 × g for 1 minute, washed with 500 μl of wash buffer AW1 (Qiagen), then centrifuged at 6,000 × g for 1 minute, washed with 500 μl of wash buffer AW2, and centrifuged at 13,000 × g for 3 minutes. A volume of 100 μl of buffer AE (0.2-μm filter sterilized) was added to the column and left at room temperature for 5 minutes before being centrifuged at 6,000 × g for 1 minute. The resulting eluate from the column was reapplied for a second time, allowed to remain at room temperature for 5 minutes, and centrifuged as previously described. A 0.2-μm filter was used to filter the following reagents before use: PCR primers, distilled, deionized water, Mutanolysin, Proteinase K, ethanol, and Qiagen buffers AE, AW1, and AW2.
Controls Included with Each Sample Processed
Each real-time 16S rDNA PCR assay included a positive control, containing 1000 CFU of GBS/reaction. The other controls incorporated with each real-time PCR 16S rDNA assay included a no template control (NTC), which was used to monitor potential contamination in the master mix, DNA adequacy of the sample was assessed using β-globin PCR as described previously,25 and specimen inhibition was assessed by spiking 1000 CFU of GBS into each of two separate 25-μl 16S rDNA PCR reactions that contained master mix either with or without the prepared blood specimen. The cycle threshold (CT) value of the spiked master mix, containing specimen, was compared with the CT value of the spiked master mix alone lacking specimen. A specimen was considered to have significant inhibition if the spiked master mix containing specimen had a CT value >1.0 above the spiked master mix without specimen.
Real-Time 16S rDNA PCR Assay
The real-time PCR assay was developed using a SmartCycler (SC) (Cepheid, Sunnyvale, CA). The primers and TaqMan probe used in the real-time 16S rDNA PCR assay were the well-characterized RW01 and DG74 primers and RDR 245 universal probe that have been described previously.22,26 The sequences were as follows: forward primers, 5′-AAC TGG AGG AAG GTG GGG AT3′; reverse primer, 5′-AGG AGG TGA TCC AAC CGC A-3′; and the TaqMan probe, 5′-(6-FAM)-TA CAA GGC CCG GGA ACG TAT TCA CCG-(TAMRA)-3′. The TaqMan probe was synthesized at Applied Biosystems (Foster City, CA). The 380-bp amplicon resides within the highly conserved C8 and C9 regions of the bacterial 16S rDNA target. Each 100-μl PCR reaction contained 30 μl of Qiamp column-purified DNA, and 70 μl of reaction mixture consisting of 50 μl of TaKaRa 2× Premix Ex Taq PCR buffer (catalog no. RR039A; TaKaRa Bio Inc., Otsu, Shiga, Japan), especially formulated for real-time PCR containing 2.5 U of TaKaRa Ex (hot start) Taq, undisclosed concentrations of MgCl2 and dNTPs, and 0.25 μm of each primer and 0.1 μm of probe. Cycling conditions included a 2-minute denaturation step at 95°C, followed by 35 cycles of 30 seconds at 95°C, 60 seconds at 60°C, and 30 seconds at 72°C. Fluorescence measurements on the SC were collected during the extension phase of each cycle, and a CT value for each sample and control was calculated by determining the point at which the fluorescence exceeded the threshold limit, which was set at 30.0 U.
Results
Real-Time 16S rDNA PCR Assay Consistently Detected 40, 50, or 2000 CFU/ml of E. coli, GBS, or L. monocytogenes, Respectively, Spiked into Neonatal Whole-Blood Samples
The limit of detection for the 16S rDNA PCR assay was determined using whole-blood samples spiked with known amounts of GBS, E. coli, or L. monocytogenes, which was calculated using an agar dilution method. The amount of bacteria added to the whole-blood samples ranged from approximately 5 to 50,000 CFU/ml.
The spiked whole-blood samples used to determine the assay’s limit of detection were prepared and amplified under exactly the same conditions as those described for those used for the clinical specimens. Using this approach, as few as 40, 50, or 2000 CFU/ml E. coli, GBS, or L. monocytogenes, respectively, could be detected directly in whole-blood samples (data shown for GBS).
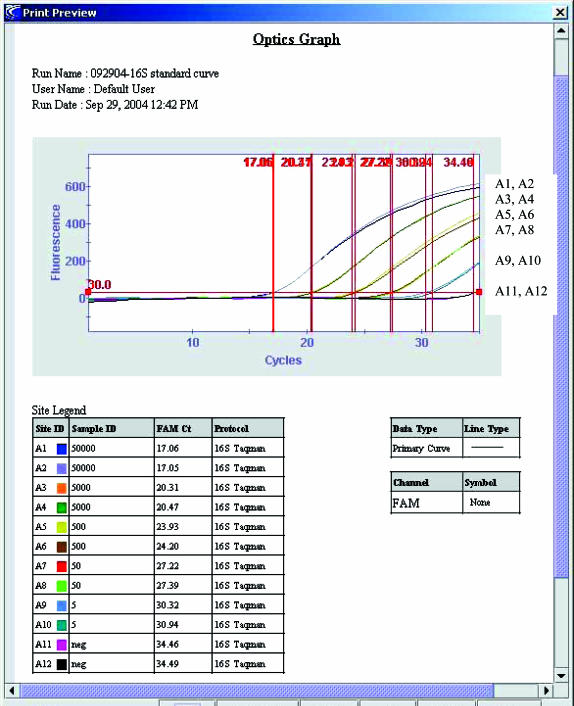
Figure 1 illustrates the data from a typical 10-fold dilution series using GBS. Each 10-fold dilution was run in duplicate within an experiment, and the experiment repeated a total of four times. The slope and r value of the line generated for a typical 10-fold dilution series was −3.65 and 0.995, respectively. The CT values obtained from the four experiments in which whole-blood specimen were spiked with 50 CFU/ml GBS ranged from 27.22 to 28.64; the calculated mean, SD, and %CV for these four experiments were 27.98, 0.5805, and 2.1, respectively.
Figure 1.
Fluorescence measurements derived using the SmartCycler for 10-fold serial dilutions of GBS (5 to 50,000 CFU/ml) spiked into neonatal whole-blood samples and analyzed by 16S rDNA PCR. Site ID numbers represent samples tested in duplicate at the following concentrations: A1 to 2, 5 × 104 CFU/ml; A3 to 4, 5 × 103 CFU/ml; A5 to 6, 5 × 102 CFU/ml; A7 to 8, 50 CFU/ml; A9 to 10, 5 CFU/ml; and A11 to 12, negative controls lacking target DNA.
Real-Time 16S rDNA PCR Assay Successfully Detected Bacteria in Whole-Blood Samples Collected from Neonates with Culture-Proven Sepsis
Fifty-three cases of culture-positive sepsis were analyzed using the real-time PCR assay. The bacteria isolated from these blood cultures were typical of those commonly associated with neonatal sepsis and included 26 coagulase-negative Staphylococcus species (sp.) (CoNS), 8 GBS, 5 E. coli, 4 Staphylococcus aureus, 2 Enterococcus sp., 2 Klebsiella pneumoniae, and 1 each of Haemophilus influenzae, Pseudomonas aeruginosa, Enterobacter ag-glomerans, Serratia marcescens, Bacillus sp., and a diptheroid (Table 1). One infant had two positive blood cultures, an earlier one contained GBS and a later one had S. aureus.
Table 1.
A Summary of the 53 Culture-Positive Samples and the Corresponding PCR Results
| Culture identification | n | PCR results* |
|---|---|---|
| CoNS | 26 | 26 of 26 Positive |
| GBS | 8 | 8 of 8 Positive |
| E. coli | 5 | 5 of 5 Positive |
| S. aureus | 4 | 3 of 4 Positive, 1 of 4 equivocal |
| Enterococcus sp. | 2 | 2 of 2 Positive |
| K. pneumoniae | 2 | 1 of 2 Positive, 1 of 2 equivocal |
| H. influenzae | 1 | 1 of 1 Equivocal |
| P. aeruginosa | 1 | 1 of 1 Equivocal |
| E. agglomerans | 1 | 1 of 1 Equivocal |
| S. marcescens | 1 | 1 of 1 Positive |
| Bacillus sp. | 1 | 1 of 1 Positive |
| Diptheroid | 1 | 1 of 1 Equivocal |
Those results having ≥2.5 cycles differences between specimen and its NTC were considered to be PCR positive. Those results having <2.5 cycle differences between the specimen and its NTC were considered to be PCR equivocal.
The time required for growth to become detectable within a blood culture ranged from 7.8 hours for GBS to 44.1 hour for CoNS, with an average time of 16.7 hours, and a median time of 16 hours. The volume of whole-blood processed for the 16S rDNA PCR assay ranged from 10 to 500 μl, with an average volume of 154 μl and a median volume of 125 μl.
Fifty-one of the 53 (96.2%) whole-blood samples prepared from the discarded CBC specimens of the 52 infants with culture-proven sepsis were positive by the real-time PCR assay for the 16S rDNA target. Two (2 of 53, 3.8%) samples from culture-proven cases of sepsis failed to be detected the 16S rDNA target by real-time assay; one blood culture had H. influenzae and the other Enterococcus sp. The CT values of the 51 culture-positive/PCR-positive (Cx+/PCR+) specimens ranged from 12.27 to 33.54, with an average and median CT value of 25.68 and 25.87, respectively. The CT values of the positive controls ranged from 22.13 to 26.63, with both the average and median CT values being 24.68.
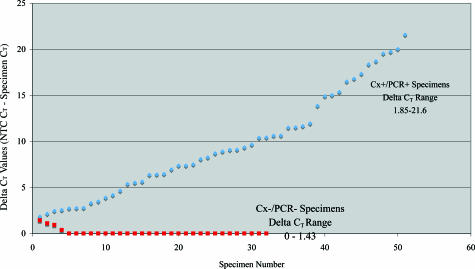
The NTC remained undetected, not crossing the established threshold of 30.0 U for 35 cycles in 21 of the 32 runs. In the remaining 11 runs, the NTC crossed the threshold between 33.41 and 34.79 cycles. Figure 2 illustrates a graph comparing the delta CT values for the 51 Cx+/PCR+ specimens, plotted in ascending order; the delta CT was generated by subtracting the CT value of the PCR+ specimen away from the CT value of its corresponding NTC. Those NTC CT values that did not cross the cut-off threshold after 35 cycles were arbitrarily given a CT value of 36. For the Cx+/PCR+ specimens, the average difference between the CT values of the specimen and its corresponding NTC was 8.92 cycles, with a median of 9.61 cycles and a range of 1.85 to 21.6 cycles.
Figure 2.
A plot comparing the ΔCT values of the Cx+/PCR+ specimens, in ascending order, with those of the Cx−/PCR− specimens, in descending order.
All of the whole-blood specimens were analyzed for DNA adequacy using β-globin PCR22,25 and found to be positive. The whole-blood specimens were also tested for inhibitory substances by performing “spiking” experiments. Briefly, the CT value of the spiked master mix containing the specimen was compared with the CT value of the spiked master mix that lacked specimen. The difference in the two CT values ranged from 0.01 to 1.03, with the average difference being 0.34 and a median difference being 0.23 (data not shown); only one of the 53 samples had a difference in CT values that was >1.0. This sample was PCR negative but culture positive and had an elevated white blood cell (WBC) count (39,900 WBC/μl).
If one required a minimum 2.5-cycle difference to exist between the CT values of the specimen and its corresponding NTC, then three additional Cx+ specimens from this study would have been considered equivocal. The corresponding blood cultures on these specimens grew an E. agglomerans, S. aureus, or K. pneumoniae, respectively. The real-time PCR results from these three specimens revealed ΔCT values of 1.85, 2.14, and 2.46, respectively, with raw CT values of 32.94, 32.65, and 33.54, respectively, with corresponding NTC CT values of 34.79, 34.79, and >35, respectively. Using this cutoff, there was an agreement between the culture-positive and real-time PCR-positive results of 88.7% (47 of 53).
Real-Time 16S rDNA PCR Assay Showed Excellent Agreement with Culture-Negative Whole-Blood Samples Collected from Neonates
Thirty-two culture-negative whole-blood samples were analyzed by real-time PCR for the presence of the 380-bp 16S rDNA target, for DNA adequacy using β-globin PCR22,25 and for inhibitory substances by performing “spiking” experiments as described previously. All 32 samples were negative for the 16S rDNA target, positive for β-globin and negative for inhibitory substances. Figure 2 illustrates the ΔCT values of these specimens analyzed for 16S rDNA, which were plotted in descending order, making it easier to visualize any overlap with the values from the PCR-positive specimens, which were plotted in ascending order. As with the PCR-positive samples, the ΔCT was generated by subtracting the CT value of the specimen from the CT value of its corresponding NTC. Those NTC CT values that did not reach the cut-off threshold after 35 cycles were arbitrarily given a CT value of 36. The CT values for the culture-negative samples ranged from 34.94 to >35, with all but two specimens having a CT value of >35.
Discussion
Each year about 10% of the infants born in the United States are admitted to a neonatal intensive care unit (NICU).27 At Magee Women’s Hospital, 70% of our NICU admissions represent near term/term infants who are at risk for systemic infection, whereas the remaining 30% represent preterm infants (data not shown). Standard of care for these infants includes antimicrobial therapy with at least two antibiotics (usually ampicillin and gentamicin in the case of the term infant) for 48 to 72 hours while awaiting culture results. If an infant is truly septic, such an admission is warranted, because early-onset sepsis is a life-threatening medical emergency. However, most near term/term infants are not infected, but will have symptoms consistent with other medical conditions that mimic sepsis, such as hypoglycemia, delayed transition, or transient tachypnea.
The results of current culture-based techniques are not available soon enough to impact on the initial antibiotic management. The no-growth blood cultures are finalized on day 5, whereas on average, culture-positive bloods require 12 to 24 hours to detect gram-positive bacteria or 24 to 48 hours to detect gram-negative bacteria. This diagnostic uncertainty leads to a greater use of broad-spectrum antibiotics, a practice known to promote antibiotic drug resistance. Changing this practice will require newer diagnostic tests that are both rapid and accurate, which can provide results in a timely manner that can impact on the care of the patient.
Recently, the rapidity of the real-time PCR approach in molecular testing has become more appreciated and has been used successfully to detect a variety of bacterial agents from sterile body sites.28,29 However, most of these approaches use primers that target specific bacterial species, so one must know in advance what bacteria you are suspecting. In contrast, we chose a set of universal primers that recognize a highly conserved region within the 16S rDNA target that successfully detects the bacterial pathogens relevant to neonatal sepsis.22,26
The specificity of these two primers, RW01 and DG-74, and probe, RDR 245, have been demonstrated previously in conventional PCR assay using agarose gel electrophoresis and DNA dot blot conformation.22,26 Representative isolates of the bacterial pathogens associated with neonatal sepsis were retested using the real-time PCR platform, which uses the same primers and probe as the conventional PCR assay, to ensure a similar specificity; all of the isolates were successfully detected using the real-time PCR platform as described here (data not shown). The initial approach used incorporated a pre-enrichment step, in which the whole-blood sample was added to tryptic soy broth (TSB) and incubated for 5 to 10 hours before a crude cell lysate was produced for PCR analysis and limited the number of amplification cycles to 25. That assay demonstrates a sensitivity of approximately 250 CFU/ml for GBS and E. coli.22
This study had two objectives: 1) eliminate the pre-enrichment step so the blood could be processed directly, thus saving time, and 2) design a real-time 16S rDNA PCR assay that could be used to more rapidly analyze the sample. We realized the first objective by replacing the TSB pre-enrichment with a readily available, manual DNA extraction and purification method (Qiagen). Time saving was also appreciated when accomplishing the second objective; developing a real-time 16S rDNA PCR assay that would rapidly and reliably detect bacterial DNA from whole-blood samples from neonates with culture-proven sepsis.
The data generated from this study suggest that the real-time 16S rDNA PCR assay performed using the SmartCycler instrument would be useful for rapidly detecting bacterial DNA directly from whole-blood samples. In contrast to the 16 hours on average that was required for a blood culture bottle to turn positive, DNA extraction and real-time PCR could be accomplished in under 4 hours; this calculation requires that molecular testing is a 24/7 operation, which may not be the case. However, even if molecular testing occurred only on first shift, this approach would detect the slower growing organisms like CoNS, in which the average time for culture detection was 19.7 hours. This argument would also hold true for the no-growth blood cultures that are not finalized until day 5, which is the case with the majority of blood cultures drawn on term infants being evaluated for sepsis.
The limit of detection for the real-time 16S rDNA PCR assay was approximately 40, 50, or 2000 CFU/ml E. coli, GBS, and L. monocytogenes, respectively, which compared favorably with other real-time PCR assays.28,29 With an automated blood culturing system, under ideal conditions, 100 CFU/ml GBS will require 10 to 24 hours of incubation before the instrument will detect the bacterial growth.30 This is in comparison to less than 4 hours when using a real-time PCR assay.
In this study, 2 of 53 culture-proven cases were not detected by real-time PCR. One isolate was a H. influenzae and the other an Enterococcus sp. Both isolates from culture were successfully amplified by PCR using the 16S rDNA assay, ruling out lack of primer recognition. Rapid DNA sequencing using pyrosequencing was also performed, and the typical bacterial fingerprint was obtained on these two isolates, ruling out lack of probe recognition. The sample that did not detect H. influenzae by PCR may have represented transient bacteremia that was present at collection of blood cultures but not when whole blood was collected for CBC and subsequent PCR. However, it may also reflect a limitation of this approach for detecting bacteremia, which often are present at very low CFU of bacteria per milliliter. The sample that failed to detect the Enterococcus sp. had a high WBC count (39.9 × 103/μl), which may explain why the bacterial DNA was not detected. More specifically, the high levels of human genomic DNA that exist in samples with high WBC counts can interfere with the ability of the relatively lower level of bacterial DNA within a sample to bind to the column. The binding capacity of the QIAmp column is approximately 50 μg—the amount of DNA represented in ∼5 × 106 WBCs (verbal communications with Qiagen). In this study, the WBC count of whole blood in clinical samples ranged from 8,000 to 39,900/μl (data not shown). Therefore, bacterial DNA present within whole-blood samples with high WBC counts may be susceptible to competition from human genomic DNA for binding to the column and thus affect assay sensitivity. It will be important to continue to monitor WBC counts from neonatal whole-blood samples being tested for bacterial sepsis. Perhaps a dilution of the sample before sample purification would be helpful in these cases. Except for the issue of high WBC counts in clinical specimens, there do not appear to be problems with carryover of inhibition when using Qiagen column chromatography for sample preparation for this real-time assay.
For low-level or questionable positives, ie, those specimens with high CT values, we found it useful to calculate the specimen’s ΔCT value, which represents the difference between the NTC CT value and sample CT value. Although no overlap existed between the lowest ΔCT value (1.85) for the Cx+/PCR+ results and the highest CT value (1.43) for the Cx−/PCR− results, the two numbers were close. Using a ΔCT value of 2.5 cycles for the cutoff, there was an overall agreement between the results of culture and real-time PCR of 94.1% (80 of 85) in this study.
In implementing a real-time PCR assay, one must also determine whether or not to include an equivocal range, which would necessitate repeat testing of samples. Taking these steps will allow us to detect as many true positives as possible, while minimizing the number of false-positives. One of the most difficult issues to face is the reliable detection of bacteria within a specimen that is at or near the assay’s limit of detection.
Molecular techniques incorporating consensus primers for universal bacterial PCR must take into account the risk of contamination from the environment and from the reagents used in the making of the master mix or that which can occur during sample processing. We routinely filter many of the reagents used in sample processing and in the PCR assay using 0.2-μm filters. We also use UV light to irradiate pipettes routinely, and we use a bleach solution to disinfect all working surfaces after each run.
We also recommend avoiding blood samples obtained via heel stick, because one runs the risk of detecting a skin contaminant and not a true pathogen. In future studies, we plan to use a dedicated venous or arterial blood sample that has been collected at the same time as the blood for culture for use with the real-time 16S rDNA PCR assay. Real-time PCR will not eliminate the need for obtaining blood cultures, because purified isolates are still needed for antimicrobial susceptibility testing. However, combining real-time 16S rDNA PCR with rapid, short-read sequencing methodology (pyrosequencing) may provide important information to the neonatologist as to the identity to the bacteria31 in less than 8 hours and may impact on the choice of antibiotic(s) used to treat the infected infant or, alternatively, may save on antibiotic doses given to the uninfected, asymptomatic infants. These changes in practice have the potential to help reduce the risks of promoting bacterial drug resistance and the side effects associated with antibiotic usage.
In conclusion, this study represents a limited assessment of the real-time PCR assay’s performance on specimens from infants being evaluated for sepsis. To properly assess the clinical sensitivity and positive predictive value of the real-time PCR assay requires a prospective study with a large, well-defined population of infants.
Footnotes
Supported by National Institutes of Health National Institute of Child Health and Development grant R01 HD038559.
References
- Cerase PA. Neonatal sepsis. J Perinat Neonat Nurs. 1989;3:48–57. doi: 10.1097/00005237-198910000-00007. [DOI] [PubMed] [Google Scholar]
- Gerdes JS. Clinicopathologic approach to the diagnosis of neonatal sepsis. Clin Perinatol. 1991;18:361–381. [PubMed] [Google Scholar]
- Klein JO. Philadelphia: W.B. Saunders,; Bacterial sepsis and meningitis. Infectious Diseases of the Fetus and Newborn. 2001:943–998. [Google Scholar]
- Witek-Janusek L, Cusack C. Neonatal sepsis: confronting the challenge. Crit Care Nurs Clin North Am. 1994;6:405–419. [PubMed] [Google Scholar]
- Chadwick EG, Yogev R, Shulman ST. Combination antibiotic therapy in pediatrics. Am J Med. 1986;80:166–171. doi: 10.1016/0002-9343(86)90496-1. [DOI] [PubMed] [Google Scholar]
- de Louvois J, Harvey D. Strategies for the treatment of bacterial infections in the newborn. Ann Acad Med Singapore. 1985;14:631–641. [PubMed] [Google Scholar]
- Fanos V, Dall’Agnola A. Antibiotics in neonatal infections: a review. Drugs. 1999;58:405–427. doi: 10.2165/00003495-199958030-00003. [DOI] [PubMed] [Google Scholar]
- Klein JO, Dashefsky B, Norton CR, Mayer J. Selection of antimicrobial agents for treatment of neonatal sepsis. Rev Infect Dis. 1983;5(Suppl 1):S55–S64. doi: 10.1093/clinids/5.supplement_1.s55. [DOI] [PubMed] [Google Scholar]
- Remington JS, Klein JO. Philadelphia: W.B. Saunders,; Infectious Diseases of the Fetus Newborn Infant. (ed 4) 1995 [Google Scholar]
- Garcia-Prats JA, Cooper TR, Schneider VF, Stager CE, Hansen TN. Rapid detection of microorganisms in blood cultures of newborn infants utilizing an automated blood culture system. Pediatrics. 2000;105:523–527. doi: 10.1542/peds.105.3.523. [DOI] [PubMed] [Google Scholar]
- Mustafa MM, McCracken GHJ. Philadelphia: W.B. Saunders,; Perinatal bacterial diseases. Textbook of Pediatric Infectious Diseases. 2004:929–966. [Google Scholar]
- Pichichero ME, Todd JK. Detection of neonatal bacteremia. J Pediatr. 1979;94:958–960. doi: 10.1016/s0022-3476(79)80233-4. [DOI] [PubMed] [Google Scholar]
- Alfa M, Sanche S, Roman S, Fiola Y, Lenton P, Harding G. Continuous quality improvement for introduction of automated blood culture instrument. J Clin Microbiol. 1995;33:1185–1191. doi: 10.1128/jcm.33.5.1185-1191.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaliere TA. Pharmacologic treatment of neonatal sepsis: antimicrobial agents and immunotherapy. J Obstet Gynecol Neonatal Nurs. 1995;24:647–658. doi: 10.1111/j.1552-6909.1995.tb02547.x. [DOI] [PubMed] [Google Scholar]
- Schuchat A, Whitney C, Zangwill K. Prevention of perinatal group B streptococcal disease: a public health perspective. MMWR Recomm Rep. 1996;45:1–24. [PubMed] [Google Scholar]
- Brown DR, Kutler D, Rai B, Chan T, Cohen M. Perinatal/neonatal clinical presentation: bacterial concentration and blood volume required for a positive blood culture. J Perinatol. 1995;15:157–159. [PubMed] [Google Scholar]
- Dietzman DE, Fischer GW, Schoenknecht FD. Neonatal Escherichia coli septicemia: bacterial counts in blood. J Pediatr. 1974;85:128–130. doi: 10.1016/s0022-3476(74)80308-2. [DOI] [PubMed] [Google Scholar]
- Yagupsky P, Nolte FS. Quantitative aspects of septicemia. Clin Microbiol. 1990;3:269–279. doi: 10.1128/cmr.3.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurlat I, Stoll BJ, McGowan JE., Jr Time to positivity for detection of bacteremia in neonates. J Clin Microbiol. 1989;27:1068–1071. doi: 10.1128/jcm.27.5.1068-1071.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich GD, Greenberg SJ. Boston: Blackwell Scientific Publications,; PCR-Based Diagnostics in Infectious Diseases. 1994 [Google Scholar]
- Kimberlin D. Herpes simplex virus, meningitis and encephalitis in neonates. Herpes. 2004;11(Suppl 2):65A–76A. [PubMed] [Google Scholar]
- Jordan JA, Durso MB. Comparison of 16S rRNA gene PCR and BACTEC 9240 for detection of neonatal bacteremia. J Clin Microbiol. 2000;38:2574–2578. doi: 10.1128/jcm.38.7.2574-2578.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforgia N, Coppola B, Carbone R, Grassi A, Mautone A, Iolascon A. Rapid detection of neonatal sepsis using polymerase chain reaction. Acta Paediatr. 1997;86:1097–1099. doi: 10.1111/j.1651-2227.1997.tb14815.x. [DOI] [PubMed] [Google Scholar]
- McCabe KM, Khan G, Zhang YH, Mason EO, McCabe ER. Amplification of bacterial DNA using highly conserved sequences: automated analysis and potential for molecular triage of sepsis. Pediatrics. 1995;95:165–169. [PubMed] [Google Scholar]
- Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statistics NCFH2001 Natality Data Set Hyattsville, MD: U.S. Department of Health and Human Services CDC, National Center for Health Statistics; Vital and Health Statistics. 2003 [Google Scholar]
- Golden SM, Stamilio DM, Faux BM, dela Cruz WP, Shoemaker CT, Blackmon CL, Stassen SD, Clark VM, Smith JW, Johnson OL. Evaluation of a real-time fluorescent PCR assay for rapid detection of group B Streptococci in neonatal blood. Diag Microbiol Infect Dis. 2004;50:7–13. doi: 10.1016/j.diagmicrobio.2004.04.021. [DOI] [PubMed] [Google Scholar]
- van Haeften R, Palladino S, Kay I, Keil T, Heath C, Waterer GW. A quantitative LightCycler PCR to detect Streptococcus pneumoniae in blood and CSF. Diag Microbiol Infect Dis. 2003;47:407–414. doi: 10.1016/s0732-8893(03)00129-9. [DOI] [PubMed] [Google Scholar]
- Riest G, Linde HJ, Shah PM. Comparison of BacT/Alert and BACTEC NR 860 blood culture systems in a laboratory not continually staffed. Clin Microbiol Infect. 1997;3:345–351. doi: 10.1111/j.1469-0691.1997.tb00624.x. [DOI] [PubMed] [Google Scholar]
- Jordan J, Butchko AR, Durso MB. Use of pyrosequencing of 16S rRNA fragments to differentiate between bacteria responsible for neonatal sepsis. J Mol Diagn. 2005;7:105–110. doi: 10.1016/s1525-1578(10)60015-3. [DOI] [PMC free article] [PubMed] [Google Scholar]