Abstract
Heterogeneous clinical expression of mitochondrial DNA (mtDNA) disorders depends on both qualitative and quantitative changes in mtDNA. We developed a sensitive and effective method that simultaneously detects mtDNA deletion(s) and quantifies total mtDNA content. The percentage of deletions and mtDNA content of 19 patients with single or multiple deletions were analyzed by real-time quantitative polymerase chain reaction (real-time qPCR) using TaqMan probes specific for mtDNA (tRNA leuUUR, ND4, ATPase8, and D-loop regions) and nuclear DNA (AIB1, β-2-microglobulin, and β-actin). The proportion of deletion mutants determined by real-time qPCR was consistent with that determined by Southern analysis. Most patients with mtDNA deletions also demonstrated compensatory mtDNA over-replication. Multiple mtDNA deletions that were not detectable by Southern analysis due to low percentage of each deletion molecule were readily detected and quantified by real-time qPCR. Furthermore, 12 patients with clinical features and abnormal biochemical/histopathological results consistent with mitochondrial respiratory chain disorders without identified mtDNA mutations had either substantially depleted or significantly over-replicated mtDNA content, supporting the diagnosis of mitochondrial disease. Our results demonstrate that both qualitative and quantitative analyses are important in molecular diagnosis of mitochondrial diseases. The presence of deletion(s) and mtDNA depletion or compensatory over-replication can be determined simultaneously by real-time qPCR.
Mitochondrial disorders have a dual genome complex etiology. They are not only clinically diverse but also genetically heterogeneous. The mitochondrial genome is a circular double-stranded 16,569-bp DNA molecule that encodes 13 respiratory chain protein subunits. Mitochondrial protein synthesis requires mitochondrial encoded ribosomal RNAs and 22 mitochondrial transfer RNAs1 (http://www.mitomap.org). The majority of mitochondrial proteins are nuclear encoded, including most of the respiratory chain enzyme subunits. The biogenesis of mitochondria depends on nuclear genes to fulfill essential functions such as replication, transcription, and translation. Therefore, a mitochondrial disorder can be caused by mutations in the nuclear DNA or the mitochondrial DNA (mtDNA).2,3
To provide accurate genetic counseling, diagnosis of a mitochondrial disorder through DNA-based molecular analysis is usually quite helpful. Molecular diagnosis of mitochondrial disorders has been focused on mutational analysis of mtDNA due to the high mutation rate and the relatively small-size mitochondrial genome. More than 100 mutations in the mtDNA have been identified4 (http://www.mitomap.org). Most pathogenic mtDNA mutations are heteroplasmic, meaning that there is coexistence of the wild-type and mutant mtDNA. Clinical manifestations of mitochondrial diseases are heterogeneous because of the differences in the tissue threshold effect for different mutations and the distribution of these mutant mtDNA molecules among different tissues. Comprehensive analysis of mtDNA in patients with a mitochondrial disease demonstrates that the molecular defects in the majority of patients remain unidentified.5,6,7 MtDNA depletion due to molecular defects in nuclear genes that are responsible for either maintaining the deoxynucleotide pools or mtDNA replication may influence mtDNA content.8,9,10 Thus, in the absence of mtDNA point mutations and deletions, the abnormal amount of mtDNA may be explained by molecular defects in nuclear genes.
Mitochondrial proliferation can be a cellular response to mitochondrial dysfunction. This is evidenced by the presence of ragged red fibers in the affected muscle specimens of patients with mtDNA mutations in tRNA genes.11,12 Compensatory amplification of mtDNA has also been reported in a Kearns Sayre syndrome patient who had mild and delayed clinical manifestations but a high proportion (92%) of a deletion mutant.13 Thus, the prediction of phenotypic expression of mitochondrial diseases cannot be solely based on the proportion of mutant load in the affected tissue. The total amount of mtDNA is also a determining factor.
Detection of mtDNA deletions is usually done by Southern blot analysis. However, this method requires a large amount of DNA, and it does not detect mutant molecules present at low percentage of heteroplasmy. Although high levels of mtDNA deletions in Kearns Sayre syndrome or Pearson syndrome can be detected by Southern analysis, multiple deletions and mtDNA depletion caused by various nuclear genes such as polymerase γ, DNA helicase, and thymidine phosphorylase may not be readily detectable, even though the percentage of total deletion molecules may be high. Recently, real-time quantitative polymerase chain reaction (qPCR) method has been used to detect and quantify the common point mutations; 3243A>G14,15 and 8344A>G16,17 and the common 5-kb deletion.18 More recently, this method was applied to the study of deletion, depletion, and over-replication.13,19,20 In this study, we developed a comprehensive real-time qPCR method using various mtDNA and nuclear DNA probes to simultaneously detect and quantify mtDNA deletion, depletion, and over-replication in muscle specimens of patients with known deletions and in patients without identified mutations who showed mtDNA depletion and over-replication.
Materials and Methods
Patients, DNA Samples, and DNA Isolation
Patients’ specimens were sent to the Molecular Genetics Laboratory at the Institute for Molecular and Human Genetics, Georgetown University Medical Center for the mutational evaluation of mitochondrial disorders. Total DNA was isolated from peripheral blood leukocytes using salting-out method21 or from muscle using proteinase K digestion followed by standard phenol/chloroform extraction and ethanol precipitation.22
The presence of common point mutations in mtDNA was assessed by multiplex PCR/allele-specific oligonucleotide hybridization analysis.23,24 Large deletions and DNA rearrangements were detected using Southern blot analysis.13,25 Fourteen patients with Southern blot-detectable single deletions and five patients with PCR detectable multiple deletions were further evaluated by real-time quantitative PCR (real-time qPCR) for the detection and quantification of deletion mutant molecules and the total mtDNA content. In addition, 18 patients that had clinical manifestations consistent with a mitochondrial disorder but without identified mtDNA mutations were studied for the multiple deletions and mtDNA content using the real-time qPCR method.
Probes for Southern Blot Analysis
The probes used for Southern analysis of mtDNA were a mixture of eight PCR fragments generated with eight pairs of overlapping primers covering the entire mitochondrial genome (np1351-3135, np3085-4927, np4881-6656, np6606-9169, np9104--11757, np11688-13738, np13695-16543, and np16411-1424). A probe for the 18S rRNA gene was generated by PCR using a forward primer, 18S-641F (5′-TTTCGAGGCCCTGTAATTGG-3′), and a reverse primer, 18S-1650R (5′-CGCTGAGCCAGTCAGTGTA-3′) (GenBank GI: 337376). The concentration of the PCR products was quantified using DyNA Quant 200 fluorometer with Hoechst dye 33258 and diluted to 6 ng/40 μl. The probes were mixed and labeled with [α-32P]dCTP using Rediprime II random prime labeling system (Amersham Biosciences. Inc., Piscataway, NJ) before use.
Primers and TaqMan Probes
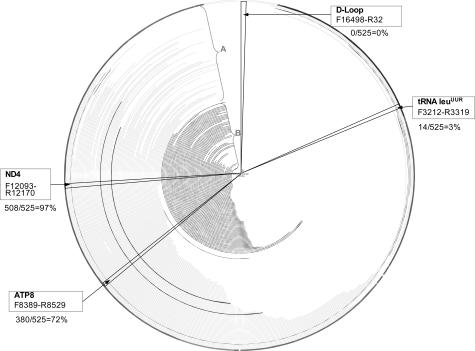
The mitochondrial DNA content was determined by real-time PCR using TaqMan probes specific for the tRNA leuUUR, ND4, ATP8, and D-loop regions. These four regions were amplified with primers listed in Table 1. The probe in tRNA leuUUR was a sequence-specific probe targeted to the most common mutation site np3243A. This region is deleted in only about 3% of deletion molecules reported (http://www.mitomap.org; accessed on March 8, 2003). If the mtDNA contains the mutation, the relative copy number calculated from using this probe and another probe in nondeleted region of mtDNA will show reduced copy number for wild-type mtDNA. Large deletions including the D-loop region have never been reported, although this region is highly polymorphic, and the primers and probes were carefully designed from the mtDNA sequences that do not contain reported mutations or polymorphisms occurring at >1.6% in the population (http://www.genpat.uu.se/mtDB/Polysites). ND4 region is found deleted in 97% of all deletion molecules reported (http://www.mitomap.org). Another region is the ATP8 gene, which is found deleted in 72% of the deletion mtDNA molecules reported (http://www.mitomap.org) (Figure 1).
Table 1.
Primers and Probes Used for Real-Time qPCR
| Genes | Sequence used | Region | Characteristic |
|---|---|---|---|
| mtDNA-tRNAleu | http://www.mitomap.org/mitomap/mitoseq.html | 3212–3319 | Usually not deleted; contains common A3243G mutation |
| mtDNA-ND4 | http://www.mitomap.org/mitomap/mitoseq.html | 12093–12170 | In common deletion region; 97% of the time deleted |
| mtDNA-D loop | http://www.mitomap.org/mitomap/mitoseq.html | 16498–32 | Always not deleted |
| mtDNA-ATP8 | http://www.mitomap.org/mitomap/mitoseq.html | 8389–8529 | 72% of the time deleted |
| AIB1 | GI:2331249 | 739–872 | Single copy nuclear gene |
| β2-Microglobulin | GI:37704380 | 589–674 | Single copy nuclear gene |
| β-Actin | GI:177967 | 2279–2394 | Several (∼5) copies) |
Table 1.
Continued
| Primer sequence (5′→3′) | Amplicon size (bp) | Probe sequence (5′→3′) |
|---|---|---|
| F: CACCCAAGAACAGGGTTTGT | 108 | 6FAM-TTACCGGGCTCTGCCATCT-TAMRA |
| R: TGGCCATGGGTATGTTGTTA | (np3252–3234) | |
| F: TCCTCCTATCCCTCAACCCC | 78 | 6FAM-CATCATTACCGGGTTTTCCTCTTGTA-TAMRA (np12115–12140) |
| R: CACAATCTGATGTTTTGGTTAAAC | ||
| F: CATCTGGTTCCTACTTCAGGG | 104 | 6FAM-CTTAAATAAGACATCACGATGGATCAC-TAMRA (np16549–6) |
| R: TGAGTGGTTAATAGGGTGATAGA | ||
| F: ATGGCCCACCATAATTACCC | 141 | 6FAM-TACACTATTCCTCATCACCCA-AMRA |
| R: CATTTTGGTTCTCAGGGTTTG | (np8419–8439) | |
| F: GAGTTTCCTGGACAAATGAG | 134 | 6FAM-GCCGTATGTTGATGAAAACACCACA-TAMRA (790–814) |
| R: CATTGTTTCATATCTCTGGCG | ||
| F: TGCTGTCTCCATGTTTGATGTATCT | 86 | VIC-TTG CTC CAC AGG TAG CTC TAG GAG G-TAMRA (621–645) |
| R: TCTCTGCTCCCCACCTCTAAGT | ||
| F: AGCGGGAAATCGTGCGTGAC | 116 | 6FAM-GCTACGTCGCCCTGGACTTCGAGCA-TAMRA (2315–2339) |
| R: AGGCAGCTCGTAGCTCTTCTC |
Figure 1.
Location of mtDNA probes for the real-time qPCR analysis of mtDNA deletion and depletion. The first nucleotide is at the 12 o’clock position. Clockwise are tRNA leuUUR, ATP8, ND4, and D-Loop. The arcs in the circle represent the deleted regions in reported cases, from the starting nucleotide position (np) of the deletion to the ending np of the deletion, based on mitomap database (http://www.mitomap.org, accessed on March 8, 2003). A: Reported large-scale [≥264 bp] single deletions. B: Reported multiple deletions. The two bolded arcs in A represent the two most common large-scale single deletions reported.
Nuclear genes used for normalization are 18S rRNA, β-actin, β-2-microglobulin (β2M), and AIB1 (amplified in breast cancer 1). The 18S rRNA gene is present in hundreds of copies, but its exact copy number is not defined among different individuals and among different tissues. Recent reports have suggested that the copy number of 18S rRNA gene is polymorphic.26,27 In this study, the 18S rRNA gene is used as a probe for Southern analysis but not for real-time qPCR studies. β-actin has several homologs, whereas β2M and AIB1 genes are single copy. These three genes are used as nuclear gene normalizers for the measurement of mtDNA content by real-time qPCR.
Age-Matched Control Specimens
A mixture of 10 to 20 matched muscle tissue DNA samples from age-matched individuals who do not carry deleterious mtDNA point mutations or deletions and whose mtDNA contents are within one SD of age-matched mean is always included in each run. The muscle specimens are divided into 10 age groups: 0 to 0.3, 0.4 to 0.6, 0.7 to 1.0, 1.1 to 2.0, 2.1 to 5, 5.1 to 10, 11 to 20, 21 to 40, 41 to 60, and 60 years up.20 The SD of mtDNA content of each age group is about 30% of the age-matched mean.
Real-Time Quantitative PCR
The real-time PCR reaction was performed in triplicate for each reaction. The 20-μl PCR reaction contained 1× Platinum qPCR SuperMix-UDG Master Mix (Invitrogen, Carlsbad, CA), 300 nmol/L of each primer, 100 nmol/L TaqMan probe, 0.4 μl of Rox dye (supplied by the manufacturer), and 4 ng of total genomic DNA extract. Real-time PCR conditions were 2 minutes at 50°C and 10 minutes at 95°C, followed by 45 cycles of 15 seconds of denaturation at 95°C and 60 seconds of annealing/extension at 60°C. Alternatively, sequence-specific TaqMan probe can be replaced by SYBR Green fluorescence dye for detection of amplification. Fluorescent signal intensity of PCR products was recorded and analyzed on Sequence Detection System ABI-Prism 7700 (Applied Biosystems, Foster City, CA) using SDS v1.9 software. The threshold cycle or CT value within the linear exponential phase was used to construct the standard curve and to measure the original copy number of DNA template. If a sample had a measurement above 10,000,000 or below 1,000 copies of mtDNA, or above 1,000,000 or below 100 copies of β2M or AIB1 gene, respectively, the assay was repeated at lower or higher dilutions of the DNA extract so that the measurement would fall within a linear range of DNA copy number.
Preparation of Standard DNA
Standard DNA for each target sequence was generated by cloning the PCR products of the corresponding target sequence into pCR2.1-TOPO vector. The PCR products of mtDNA-ND4, mtDNA-D-loop, mtDNA-ATP8, β-actin, and β2M gene are listed in Table 1. The mtDNA-tRNA leuUUR was amplified using mtF3212 and mtR3471 primers, producing a 260-bp fragment. The exon 5 of AIB1 gene was amplified using the forward primer 5′-CAAGCGATCAAATGAGGGTAG-3′ and the reverse primer 5′-CATTGTTTCATATCTCTGGCG-3′, giving a 439-bp PCR product. The copy number was calculated based on the molecular weight of the plasmid DNA. Serial dilutions were made, and the real-time qPCR reactions were performed to construct the standard curve for each individual gene.
Measurement of Mutant Heteroplasmy and mtDNA Copy Number
The standard curve for each target sequence is always included in each run. The copy number of the target sequence in the sample is calculated from the corresponding standard curve. The proportion of deletion mutant is calculated from the copy number of the deleted region and the nondeleted region. The mtDNA content is equivalent to the mtDNA copy number divided by the copy number of the single-copy nuclear gene. mtDNA depletion or over-replication is determined by comparing the total mtDNA content of the patient to that of the age-matched mean.
Results
DNA samples from 14 patients were analyzed by Southern blot and restrictive fragment length polymorphism (RFLP) analysis (Figure 2A). The percentage of deletion was calculated from the signal intensities of the deletion mutant and the full-length molecules using densitometry. The same samples were analyzed by real-time qPCR using probes in the regions listed in Table 1. Figure 2B shows that results obtained from these two methods are consistent with a R2 of 0.94. The clinical and laboratory findings of the patients are listed in Table 2. Nine patients presented typical Kearns Sayre syndrome, others had atypical Kearns Sayre Syndrome or Pearson syndrome with a multisystemic disease expression. The percentages of deletion mutants obtained using the probes for ND4 (np12093-12170) and ATP8 (np8389-8529) are consistent except for patients 12, 13, 14, 16, and 24, whose deletions do not cover the ATP8 probe region, and patient 19 (both blood and muscle), whose deletion does not cover ND4 probe region (Table 2). When normalized to a single-copy nuclear gene, β2M gene, the total amount of mtDNA in all patients but one (patient 17), with an mtDNA deletion demonstrated mtDNA over-replication in muscle specimens when compared with the age-matched mean. The degree of mtDNA over-replication does not appear to correlate with the size and location of the deletion. Patient 21 was found to have multiple deletions within regions between np8295 and np13921 as judged by PCR (data not shown) in addition to the major deletion listed in Table 2, which covers ND4 but not ATP8 region. This may explain the discrepancy between the percentage of deletions calculated from using the ND4 or the ATP8 probe (43 and 23%, respectively).
Figure 2.
The heteroplasmic mtDNA deletions determined by Southern analysis and real-time qPCR. A:Southern blot analysis. The number on the top is patient’s number. The number at the bottom is the percentage of deletion molecules. The percentage of deletion heteroplasmy is the intensity of mtDNA with deletion (bottom band) divided by the sum of the intensity of mtDNA with deletion and the intact mtDNA (top band). The intensity of the DNA bands is measured using Scion Image for Windows (β 4.02) software (http://www.scioncorp.com). B: The correlation between the percentages of deletion mutant measured by real-time qPCR and Southern blot. The calculation of percent of mtDNA deletion by real-time qPCR was described in the footnote of Table 2. Only the data points from the deleted region(s) were included in the plot: ATP8 (9 filled circles) and ND4 (12 open circles). Patient 21 in Table 2, which has multiple deletions, was not included in the plot. The linear regression analysis was performed using SigmaStat for Windows Version 2.03, and the graph was plotted using SigmaPlot 2000 for Windows Version 6.00. The P value is <0.001.
Table 2.
Clinical and Molecular Features of the Patients Harboring Single mtDNA Deletion
| Patient | Sex | Clinical features | Age (years) | Characterization of mtDNA deletion
|
mtDNA content (mtDNA/B2M ratio)
|
% of mtDNA deletion by Southern blot and real-time qPCR
|
|||
|---|---|---|---|---|---|---|---|---|---|
| Size (kb) | Breakpoints | % of age matched mean | Southern blot | Real-time QPCR (ND4) | Real-time QPCR (ATP8) | ||||
| 11m | F | KSS | 33 | Common, 5 | (8469–8482):(13447–13460) | 129 | 51 | 60 | 65 |
| 12m | F | Atypical Pearson/multisystemic/KSS | 8 | 4.5 | 10559:14981 | 174 | 55 | 57 | 0 |
| 13b | M | Pearson/multisystemic | 0.8 | 2.3 | 12113:14421 | 152 | 90 | 86 | 0 |
| 14b | F | Multisystemic | 1.3 | 7.1 | 8536:15642 | 105 | 65 | 74 | 0 |
| 15m | F | KSS | 39 | Common, 5 | (8469–8482):(13447–13460) | 259 | 64 | 67 | 66 |
| 16b | M | Pearson/multisystemic | 1.2 | 5.2 | 10418:15570 | 173 | 80 | 84 | 0 |
| 17m | F | KSS | 79 | Common, 5 | (8469–8482):(13447–13460) | 95 | 27 | 24 | 29 |
| 18m | M | KSS | 20 | Common, 5 | (8469–8482):(13447–13460) | 181 | 88 | 90 | 90 |
| 19m | F | Atypical KSS (mild) | 40 | del-dupl, 3.1 | (8420–8424):(11498–11502) | 391 | 92 | 0 | 96 |
| 19b | F | KSS | 40 | Blood of 19 | (8420–8424):(11498–11502) | ND | 0 | 0 | 12 |
| 20m | F | KSS/ multisystemic | 40 | 7.7 | (6331–6341):(13994–14004) | 165 | 24 | 41 | 42 |
| 21m | F | KSS | 48 | 5 | (9256–9265):(13625–13634) | 152 | 23 | 43 | 23 |
| 22b | M | KSS/multisystemic | 9 | 7.7 | (6331–6341):(13995–14004) | 155 | 44 | 53 | 50 |
| 23b | M | KSS | 9 | 5 | (8469–8482):(13447–13460) | 240 | 53 | 61 | 63 |
| 24m | F | KSS | 25 | 4 | (10951–10957):(13989–13995) | 150 | 48 | 44 | 0 |
b, blood; m, muscle; KSS, Kearns Sayre Syndrome; ND, not done; mtDNA content, the ratio of mtDNA copy number to B2M gene copy number. Usually the results from the two non-deleted mtDNA probes (tRNA leuUUR and D-loop region) were consistent with each other (coefficient of variance <3%), the mean value of the two nondeleted regions was used to calculate the mtDNA content. Percentages of mtDNA deletion by real-time qPCR were calculated from the copy number of the deleted region (ND4 or ATP8 region depending on the region of deletion) and the copy number of the nondeleted region (mean of tRNA leuUUR and D-loop region) using the formula: [(copy number of nondeleted region − copy number of deleted region)/copy number of nondeleted region] × 100%. Percentage of mtDNA deletion by Southern blot was calculated as described in the legend of Fig 2A. The mtDNA content obtained from a pool of muscle DNA samples from 10∼20 age-matched individuals was used as age-matched mean.
Several patients with multiple mtDNA deletions that were not readily detected by Southern analysis (Figure 3A) showed multiple deletions on PCR (Figure 3B). It would be difficult to determine the percentage of total mtDNA deletion mutant by Southern blot because each deletion molecule was present at a low percentage. The design of the probes at regions (ND4 and ATP8, backup each other) involved in almost all deletion mutants and probes at nondeleted regions (tRNA leuUUR and D-loop backup each other) (Table 1 and Figure 1) for real-time qPCR analysis facilitated the detection and quantification of the overall level of multiple mtDNA deletions in five samples listed in Table 3. Lower levels of mtDNA contents were also found in three patients with multiple deletions (patients 102, 103, and 105). All five patients fulfilled the diagnostic criteria for having a definite mitochondrial disease based on the results from enzymatic, histochemical, or clinical studies.28 Unlike the patients with a single deletion, only two patients with multiple mtDNA deletions demonstrated a slight increase in mtDNA content. Multiple deletions in muscle are observed in older patients (Table 3) but are rarely observed in young children (Bai and Wong, unpublished data) unless the patients carry nuclear gene mutations that affect mtDNA biosynthesis.
Figure 3.
Detection of mtDNA multiple deletions by Southern analysis (A) and PCR (B). Lanes 1 to 6 are normal control and patients 101 to 105, respectively. Lane 7 in B is no template control in PCR. A: Southern blot analysis. Total DNA was digested with HindIII followed by electrophoresis in 0.8% agarose gel and Southern analysis. The blot was hybridized with mtDNA probes (see Materials and Methods). B: PCR using primers mtF7234-R16133 (I), mtF8295-14499 (II), and mtF5681-R14686 (III). M, 100-bp DNA marker.
Table 3.
Clinical and Laboratory Findings of the Patients Harboring Multiple mtDNA Deletions
| Patient | Sex | Age (years) | Clinical features | mtDNA content (% of age matched mean) | % of mtDNA deletion
|
|
|---|---|---|---|---|---|---|
| Southern blot | Real-time qPCR | |||||
| 101 | M | 67 | Muscle cramps, peripheral sensory neuropathy, gait ataxia, demyelination of peripheral nerve axons, mitochondrial proliferation | 131 | Not visible | 16 |
| 102 | F | 43 | Not available | 49 | Visible multiple bands | 41 |
| 103 | M | 40 | Exercise intolerance, easy fatigability, short staure | 71 | Not visible | 11 |
| 104 | M | 49 | Not available | 161 | Not visible | 11 |
| 105 | F | 79 | Ptosis, CPEO, encephalomyopathy, ragged-red fibers | 74 | Not visible | 22 |
CPEO, chronic progressive external ophthalmoplegia. mtDNA content, the ratio of mtDNA copy number to B2M gene copy number. Percentages of mtDNA deletion by real-time qPCR were calculated as described in the footnote of Table 2, except the copy number of the commonly deleted region was ND4 or ATP8, whichever is lower. Percent of mtDNA deletion by Southern blot was calculated as described in the legend of Fig 2A. Age-matched mean, the same as described in the footnote of Table 2.
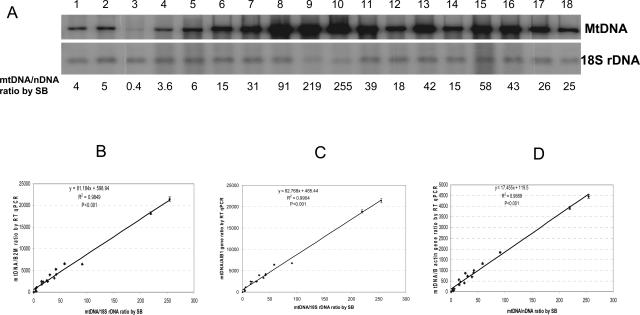
To assess the utility of the real-time qPCR method in the diagnosis of mitochondrial diseases, we evaluated 300 muscle specimens from patients suspected of having a mitochondrial disorder. We arbitrarily chose 18 samples, 6 each with high, medium, and low levels of mtDNA determined by Southern blot analysis (Figure 4A) to be used for further analysis with real-time qPCR (Figure 4B). Four mtDNA probes and three nuclear genes listed in Table 1 were used for the real-time qPCR analysis. mtDNA deletions were not observed, consistent with the regular PCR results (data not shown). The mtDNA content measured using mtDNA probes in the deleted and nondeleted regions were essentially the same with the difference less than 3.2% (Table 4). Thus, the minimum level of deletion heteroplasmy that can be reliably detected by this method should be higher than 3.2%. In Table 3, an 11% mtDNA multiple deletion mutant load was detected in two patients. Therefore, the sensitivity of detection of unknown, multiple mtDNA deletion is estimated to be between 3 and 11%. However, if the DNA sequence characterizing the junction fragment where the deletion occurred is known, a specific TaqMan probe targeted to this junction can be designed resulting in much greater detection sensitivity as low as to 0.01% mutant load (Bai and Wong, unpublished data).
Figure 4.
Measurement of mtDNA content. A: Southern analysis. The upper blot was hybridized with mtDNA probe, and the lower blot was hybridized with 18S rDNA probe. The numbers on the top (1 to 18) are DNA samples from patients 201 to 218, respectively. The numbers at the bottom are ratios of signal intensities of mtDNA to 18S rDNA. Total muscle DNA (0.15 μg) was digested with EagI followed by Southern analysis. B through D: MtDNA content determined by real-time qPCR using β2M (B), AIB1(C), or β-actin gene (D) as nuclear gene reference and their correlation with the mtDNA content determined by Southern blot analysis using 18S rDNA as the reference nuclear gene. mtDNA is the mean of copy number of mtDNA from tRNA leuUUR and D-loop region determined by standard curves.
Table 4.
MtDNA Contents and Clinical and Laboratory Findings of the Patients Analyzed by Real-Time qPCR
| Patient | Sex | Clinical and laboratory findings | Age (years) | % of age matched mean | mtDNA content (mtDNA-to-B2M ratio)
|
||
|---|---|---|---|---|---|---|---|
| Del | Non-Del | Del/(Non-Del) | |||||
| 201 | F | Hypotonia, seizures, muscle weakness, developmental delay, failure to thrive | 3.3 | 10.2 | 208 | 203 | 1.0233 |
| 202 | M | Complex I,II/III, and IV deficiency, hypotonia, areflexic, myopathy | 6 | 7.8 | 259 | 260 | 0.9992 |
| 203 | M | Developmental delay, hypotonia, ketosis, lactic acidosis, ragged-red fibers | 0.3 | 1.6 | 24 | 25 | 0.9770 |
| 204 | F | Cardiomyopathy | 2.7 | 52 | 1017 | 1029 | 0.9884 |
| 205 | M | Developmental delay, seizures, hypotonia, ataxia, abnormal MRI, muscle weakness, lactic acidosis, CEPO, DM, SNHL | 2.7 | 53 | 1085 | 1051 | 1.0321 |
| 206 | F | Developmental delay, hypotonia, ataxia, cardiomyopathy | 4 | 121 | 2435 | 2408 | 1.0111 |
| 207 | F | Lactic acidosis, ophthalmoparesis, lethargy, MELAS | 3 | 205 | 4119 | 4071 | 1.0118 |
| 208 | M | Congenital muscular dystrophy, failure to thrive, hypotonia | 0.5 | 414 | 6409 | 6465 | 0.9913 |
| 209 | F | Mitochondrial proliferation | 36 | 549 | 17966 | 18147 | 0.9900 |
| 210 | F | Developmental delay, hypotonia, dementia, migraine, exercise intolerance, abnormal brain MRI | 17 | 672 | 21795 | 21884 | 0.9959 |
| 211 | F | Developmental delay, perinatal insult, abnormal brain MRI | 2 | 161 | 3237 | 3183 | 1.0169 |
| 212 | M | Developmental delay, seizures, abnormal brain MRI, lactic acidosis | 1.2 | 154 | 2420 | 2404 | 1.0066 |
| 213 | F | Hearing loss, hypoglycemia, abnormal brain MRI | 30 | 160 | 5274 | 5321 | 0.9911 |
| 214 | F | Ataxia, myoclonus, muscle weakness, exercise intolerance, diarrhea | 34 | 56 | 1828 | 1827 | 1.0006 |
| 215 | F | Muscle weakness, exercise intolerance | 57 | 192 | 6625 | 6649 | 0.9964 |
| 216 | F | Muscle weakness, ragged-red fibers, mitochondrial proliferation | 50 | 119 | 4130 | 4112 | 1.0044 |
| 217 | M | Neuropathy, muscle weakness, retinitis pigmentosa, hearing loss | 87 | 73 | 2450 | 2493 | 0.9830 |
| 218 | F | Ragged-red fibers, increased lipid | 62 | 110 | 3724 | 3759 | 0.9908 |
CEPO, chronic progressive external ophthalmoplegia; DM, diabetes mellitus; SNHL, sensorineural hearing loss; MELAS, mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes; Non-Del, nondeleted region (mean of tRNA leuUUR and D-loop region); Del, commonly deleted region (ND4 or ATP8, whichever is lower); Age-matched mean, the same as described in the footnote of Table 2.
The mtDNA content obtained from real-time qPCR analysis using different nuclear genes as references correlates well with the results from Southern blot analysis using 18S rRNA gene as the normalizer (Figure , 4B4C4D). The mtDNA content of each patient was compared with that of the age-matched mean (Table 4). Three patients (patients 201, 202, and 203) had extremely low levels of mtDNA (3 to 10% of the age-matched mean), whereas patients 207 through 210 had a 2- to 6.4-fold amplification of mtDNA. None of these patients had PCR detectable deletions. Analysis of the entire mitochondrial genome of these 18 patients revealed numerous reported polymorphisms but deleterious mtDNA mutations were not found (data not shown). Because these patients had clinical findings (Table 4) consistent with a mitochondrial respiratory chain disorder, the results suggest that mtDNA depletion and over-replication in these patients could be secondary because of primary molecular defects in nuclear genes.
To evaluate the real-time qPCR method using SYBR green for detection instead of TaqMan probe, the measurement of percentage of deletion in patients 11, 15, 16, and 17 were repeated using SYBR green. The results (not shown) revealed a 54, 67, 80, and 20% deletion mutant load for the above mentioned patients, respectively, using primers for the ND4 region, and a 56, 66, 0, and 24%, respectively, using primers for the ATP8 region. These results are similar to the results shown in Table 2 (TaqMan method), suggesting that these two detection methods are consistent. The deletion region in patient 16 spans from np10418 to np15570 and does not cover the probe region in ATP8 gene. The total mtDNA content in these patients determined using SYBR green was similar to those listed in Table 2 (TaqMan method). We also selected three patients with 73, 75, and 83% A3243G mutant heteroplasmy, respectively, for real-time qPCR analysis using both TaqMan and SYBR green detection methods. These results showed a consistent percentage of mutant heteroplasmy with a 69, 69, and 79% mutant load, respectively, when the sequence-specific TaqMan probe was used. However, SYBR green could not detect the presence of A3243G mutation, because it only detected the total PCR products. These data indicated that real-time qPCR with TaqMan probes appropriately selected can detect and measure percentage of mutant load in patient mtDNA samples that have the most common mtDNA point mutation, single or multiple deletions, depletion, or over-replication, all at the same time.
Discussion
The main focus of molecular diagnosis of mitochondrial disorders has been on the detection of mtDNA point mutations and large deletions. However, identification of a mutation is often not sufficient to explain the clinical phenotype. The proportion of mutant heteroplasmy and the total mtDNA level also play an important role in determining the disease expression. Without considering mutant load in affected tissues, patients with A3243G common mutation may be totally asymptomatic or affected with diabetes, retinopathy, hearing loss, or manifest the full spectrum of MELAS.29 Patients with high proportions of deletion mutant load may manifest only mild mitochondrial myopathy due to a compensatory mtDNA over-replication.13 Thus, in a comprehensive molecular diagnosis of mitochondrial disorders, it is highly desirable to include quantitative analysis of the mutant heteroplasmy and the total mtDNA content. Quantification of point mutations has usually been based on PCR/RFLP. Recently, the one-step detection and quantification of heteroplasmic point mutation using real-time qPCR was reported.15 For quantitative assessment of large mtDNA deletions and alterations in mtDNA content due to depletion or over-replication, Southern blot analysis has been the gold standard.25 However, this approach requires a large amount of DNA and is a tedious, time-consuming procedure that involves the use of radioactive material and serial steps. Thus, errors can potentially be introduced in each step resulting in variability and reduced sensitivity. The real-time qPCR method described here allows the simultaneous detection and quantification of a single deletion or multiple deletions combined with the determination of mtDNA depletion or over-replication relative to a group of age-matched controls. In addition, because the amount of DNA required for analysis is very small, it is feasible to study mtDNA from noninvasive tissues such as hair follicles, buccal mucosal cells, or urine sediment. Detection and quantification of mtDNA deletion and mtDNA content is important in providing accurate genetic counseling to family members. The alteration in mtDNA content may also suggest molecular defects in nuclear genes that are involved in mtDNA biogenesis.
During the development of real-time qPCR approach, we have noticed several potential problems that have been taken into consideration. The quantity of PCR products depends on the efficiency of PCR. Because mtDNA is highly polymorphic among individuals, nucleotide mismatches in the sequences in which the primer or TaqMan probe binds will reduce the PCR efficiency. To avoid this problem, all primers are designed in regions where mtDNA polymorphisms are rarely found, and at least two regions were analyzed. Choosing two regions in the nondeleted regions (tRNA leuUUR and D-loop regions) and two regions in the commonly deleted regions (ND4 and ATP8 regions) allows detection of the presence, the proportion, and the range of deletion. For example, the location of the deletion in patients 12, 13, 14, 16, and 24, was confirmed to not include the ATP8 region, whereas the deletion in patient 19 did not include ND4 region. The other patients in Table 2 had deletions including both ND4 and ATP8 regions, because they gave consistent mtDNA copy numbers with both probes. Although the tRNA leuUUR gene region in the minor arc was rarely deleted, it was found that the deletion of the minor arc occurred in 73% of skin cancer specimens in a recent report.30 Thus, using a second probe in the D-loop region provides a backup for the measurement of the nondeleted molecules. The primers and TaqMan probes are located in regions where mtDNA polymorphisms are reported to be present in <1.6% of individuals (http://www.genpat.uu.se/mtDB/Polysites). If these probes give inconsistent mtDNA content, the possibility of polymorphisms can be confirmed by sequencing the regions.31 Because many nuclear genes display copy number variation,26 the use of two to three single-copy nuclear genes minimizes the problem of copy number polymorphism, such as in the case of 18S rRNA and β-actin genes.
One of the complications in making the diagnosis of mitochondrial disease based on mtDNA content is the high variability among normal individuals. Even within the same individual, mitochondrial biogenesis may fluctuate in response to changes in physiological conditions. This problem will always exist because true controls are difficult to define and in many cases difficult to obtain. In addition, different tissues have different mtDNA copy number. For example, we have studied mtDNA content in 300 muscle and 200 blood specimens from patients of all ages; mtDNA content in muscle is at least 10 times higher than that in blood, and in new-born muscle, it is only about 20 to 30% of that in adult muscle.20 In this study, pooled tissue-matched DNA samples from age-matched individuals were always used for comparison. Because the mtDNA contents vary among different individuals, only when mtDNA contents are three times SD below or above the age-matched mean is the mtDNA depletion or over-replication considered significant.
Two different detection methods were used for comparison in this study. Both use the same primers for PCR. The TaqMan probes were designed for the detection of specific target sequence, whereas the SYBR green detected all double-stranded PCR products. In this study, TaqMan probes were oligonucleotides within the amplified DNA fragments. If a particular deletion is known, the specificity and sensitivity of detection can be enhanced by designing a TaqMan oligonucleotide probe complementary to the specific junction sequence of the breakpoints.18 We have used this approach to detect the common 4977-bp deletion and found that a heteroplasmic deletion as low as 0.01% can be detected (data not shown). The application of this method in clinical diagnosis is significant if different probes consistently show more than 10% deletion, an estimated reliable lower limit of detection sensitivity for clinical diagnosis. Our recommendation is that to detect and quantify deletion and mtDNA content simultaneously, all four mitochondrial probes (two in commonly deleted region, two in always not deleted region) and at least one single nuclear gene should always be analyzed to obtain reliable results. If there is a discrepancy, then the reasons will be determined. For example, polymorphisms in D-Loop region causing lower amplification, or deletion region does not include ATP8 region, etc. Using the probe at the major deletion arc allows the detection of multiple deletions undetectable with Southern blot analysis. Three of five patients with multiple deletions also showed decreased amount of mtDNA (Table 3). On the contrary, most patients with single deletions, regardless of age, showed a certain degree of mtDNA over-replication. Patients with mtDNA depletion (Table 4, patients 201 to 205) do not necessarily have PCR detectable mtDNA multiple deletions. Thymidine phosphorylase (TP), polymerase γ (POLG), thymidine kinase 2 (TK2), and DNA helicase (Twinkle) in patients 201 to 205 were sequenced. Deleterious mutations in these genes were not found except that patient 202 was found to be homozygous for p.T150M polymorphism in TK2 gene and heterozygous p.E662K for POLG gene. Patient 204 was heterozygous for p.V170I in exon 1 of Twinkle gene, and patient 205 was heterozygous for p.V170I of Twinkle gene and p.S471L of TP gene. We believe the observed mtDNA depletion is secondary to yet unidentified primary nuclear gene defect. Alternatively, the mechanism of synergistic heterozygosity may play a role. The patients with multiple deletions listed in Table 3 are all older than 40 years of age. All five patients with mtDNA depletion are young patients with an average age of 3 years. It is not clear whether the multiple deletions are due to the aging process or to an intrinsic primary molecular defect in one or more of the nuclear genes.
During the study of mtDNA content, it is noticed that an increased number or size of mitochondria does not always correlate with mtDNA over-replication. Mitochondrial biogenesis relies on intergenomic communication. In response to mitochondrial dysfunction, there may be disproportional synthesis of nuclear encoded proteins that make up the mitochondrial mass, but the mitochondria may be devoid of mtDNA because of a mtDNA replication defect. On the other hand, to compensate for defective mitochondrial gene expression, there may be over-replication of the mitochondrial genome causing mitochondrial proliferation. Therefore, an abnormal amount of mtDNA content may have implications in primary molecular defects.
In conclusion, our results demonstrated that both qualitative and quantitative analyses are important in molecular diagnosis of mitochondrial diseases. Using the appropriate probes from mitochondrial and nuclear genes, the presence of deletion(s) and mtDNA depletion or compensatory over-replication can be determined simultaneously by real-time qPCR. Because the analysis requires only a small amount of DNA, the assay can be applied to noninvasive tissues and can be used for carrier analysis. The data obtained will be valuable in genetic counseling, clinical prognosis, phenotype correlation, and patient management.
Acknowledgments
We thank the physicians and patients who contributed DNA samples for mitochondrial mutation analysis and Dr. Li Guo for technical assistance in allele-specific oligonucleotide analysis.
Footnotes
Supported in part by a grant from Muscular Dystrophy Association (to L.-J.C.W.).
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Shoffner JM, Wallace DC. Oxidative phosphorylation diseases. Scriver CR, Beaudet AL, Sly WS, Valle D, editors. New York: McGraw-Hill,; The Metabolic and Molecular Bases of Inherited Disease. 1995:1535–1629. [Google Scholar]
- Wallace DC. Disease of mitochondrial DNA. Annu Rev Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- Servidei S. Mitochondrial encephalomyopathies: gene mutation. Neuromusc Disord. 2003;13:685–690. doi: 10.1016/s0960-8966(01)00265-6. [DOI] [PubMed] [Google Scholar]
- Wong L-JC, Liang M-H, Kwon H, Bai RK, Alper O, Gropman A. A CF patient with two novel mutations in mitochondrial DNA: mild disease led to delayed diagnosis of both disorders. Am J Med Genet. 2002;113:59–64. doi: 10.1002/ajmg.10767. [DOI] [PubMed] [Google Scholar]
- Wong L-JC, Liang M-H, Kwon H, Park J, Bai R, Tan D. Comprehensive scanning of the whole mitochondrial genome for mutations. Clin Chem. 2002;48:1901–1912. [PubMed] [Google Scholar]
- Chen TJ, Boles RG, Wong LJ. Detection of mitochondrial DNA mutations by temporal temperature gradient gel electrophoresis. Clin Chem. 1999;45:1162–1167. [PubMed] [Google Scholar]
- Nishino I, Spinazzola A, Hirano M. Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science. 1999;283:689–692. doi: 10.1126/science.283.5402.689. [DOI] [PubMed] [Google Scholar]
- Lamantea E, Tiranti V, Bordoni A, Toscano A, Bono F, Servidei S, Papadimitriou A, Spelbrink H, Silvestri L, Casari G, Comi GP, Zeviani M. Mutations of mitochondrial DNA polymerase gammaA are a frequent cause of autosomal dominant or recessive progressive external ophthalmoplegia. Ann Neurol. 2002;52:211–219. doi: 10.1002/ana.10278. [DOI] [PubMed] [Google Scholar]
- Saada A, Shaag A, Mandel H, Nevo Y, Eriksson S, Elpeleg O. Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat Genet. 2001;29:342–344. doi: 10.1038/ng751. [DOI] [PubMed] [Google Scholar]
- Moslemi AR, Tulinius M, Holme E, Oldfors A. Threshold expression of the tRNA(Lys) A8344G mutation in single muscle fibres. Neuromuscul Disord. 1998;8:345–349. doi: 10.1016/s0960-8966(98)00029-7. [DOI] [PubMed] [Google Scholar]
- Tokunaga M, Mita S, Sakuta R, Nonaka I, Araki S. Increased mitochondrial DNA in blood vessels and ragged-red fibers in mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS). Ann Neurol. 1993;33:275–280. doi: 10.1002/ana.410330308. [DOI] [PubMed] [Google Scholar]
- Wong LJ, Perng CL, Hsu CH, Bai RK, Schelley S, Vladutiu GD, Vogel H, Enns GM. Compensatory amplification of mtDNA in a patient with a novel deletion/duplication and high mutant load. J Med Genet. 2003;40:e125. doi: 10.1136/jmg.40.11.e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo K, Yamano A, Nagashima M, Mori Y, Anzai K, Akehi Y, Nomiyama R, Asano T, Urae A, Ono J. Mitochondrial gene mutations in the tRNA(Leu(UUR)) region and diabetes: prevalence and clinical phenotypes in Japan. Clin Chem. 2001;47:1641–1648. [PubMed] [Google Scholar]
- Bai RK, Wong LJ. Detection and quantification of heteroplasmic mutant mitochondrial DNA by real-time amplification refractory mutation system quantitative PCR analysis: a single-step approach. Clin Chem. 2004;50:996–1001. doi: 10.1373/clinchem.2004.031153. [DOI] [PubMed] [Google Scholar]
- Szuhai K, Ouweland J, Dirks R, Lemaitre M, Truffert J, Janssen G, Tanke H, Holme E, Maassen J, Raap A. Simultaneous A8344G heteroplasmy and mitochondrial DNA copy number quantification in myoclonus epilepsy and ragged-red fibers (MERRF) syndrome by a multiplex molecular beacon based real-time fluorescence PCR. Nucleic Acids Res. 2001;29:E13. doi: 10.1093/nar/29.3.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Chinnery PF, Durham SE, Blakely EL, Wardell TM, Borthwick GM, Taylor RW, Turnbull DM. Detection and quantification of mitochondrial DNA deletions in individual cells by real-time PCR. Nucleic Acids Res. 2002;30:e68. doi: 10.1093/nar/gnf067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wurmb-Schwark N, Higuchi R, Fenech AP, Elfstroem C, Meissner C, Oehmichen M, Cortopassi GA. Quantification of human mitochondrial DNA in a real time PCR. Forensic Sci Int. 2002;126:34–39. doi: 10.1016/s0379-0738(02)00026-9. [DOI] [PubMed] [Google Scholar]
- Chabi B, Mousson de, Camaret B, Duborjal H, Issartel JP, Stepien G. Quantification of mitochondrial DNA deletion, depletion, and over-replication: application to diagnosis. Clin Chem. 2003;49:1309–1317. doi: 10.1373/49.8.1309. [DOI] [PubMed] [Google Scholar]
- Bai R, Perng C-L, Hsu C-H, Wong LJC. Quantitative PCR analysis of mitochondrial DNA content in patients with mitochondrial disease. Ann NY Acad Sci. 2004;1011:304–309. doi: 10.1007/978-3-662-41088-2_29. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Nurnberger JI., Jr A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res. 1991;19:5444. doi: 10.1093/nar/19.19.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L-JC, Lam C. Alternative, noninvasive tissues for quantitative screening of mutant mitochondrial DNA. Clin Chem. 1997;43:1241–1243. [PubMed] [Google Scholar]
- Wong L-JC, Senadheera D. Direct detection of multiple point mutations in mitochondrial DNA. Clin Chem. 1997;43:1857–1861. [PubMed] [Google Scholar]
- Liang MH, Wong L-JC. Yield of mtDNA mutation analysis in 2000 patients. Am J Med Genet. 1998;77:385–400. [PubMed] [Google Scholar]
- Shanske S, Wong LJC. Molecular analysis for mitochondrial DNA disorders. Mitochondrion. 2004;4:403–415. doi: 10.1016/j.mito.2004.07.026. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Delany ME, Krupkin AB. Molecular characterization of ribosomal gene variation within and among NORs segregating in specialized populations of chicken. Genome. 1999;42:60–71. doi: 10.1139/g98-110. [DOI] [PubMed] [Google Scholar]
- Bernier FP, Boneh A, Dennett X, Chow CW, Cleary MA, Thorburn DR. Diagnostic criteria for respiratory chain disorders in adults and children. Neurology. 2002;59:1406–1411. doi: 10.1212/01.wnl.0000033795.17156.00. [DOI] [PubMed] [Google Scholar]
- Chae JH, Hwang H, Lim BC, Cheong HI, Hwang YS, Kim KJ. Clinical features of A3243G mitochondrial tRNA mutation. Brain Dev. 2004;26:459–462. doi: 10.1016/j.braindev.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Harbottle A, Krishnan KJ, Birch-Machin MA. Implications of using the ND1 gene as a control region for real-time PCR analysis of mitochondrial DNA deletions in human skin. J Invest Dermatol. 2004;122:1518–1521. doi: 10.1111/j.0022-202X.2004.22608.x. [DOI] [PubMed] [Google Scholar]
- McComsey G, Bai RK, Maa JF, Seekins D, Wong LJ. Extensive investigations of mitochondrial DNA genome in treated HIV-infected subjects: beyond mitochondrial DNA depletion. J Acquir Immune Defic Syndr. 2005;39:181–188. [PubMed] [Google Scholar]