Abstract
Hypermethylation of CpG islands in gene promoter regions is an important mechanism of gene inactivation in cancer. Many cellular pathways, including DNA repair, are inactivated by this type of epigenetic lesion, resulting in proposed mutator phenotypes. Promoter hypermethylation of hMLH1 has been implicated in a subset of colorectal cancers that show microsatellite instability (MSI). Transcriptional silencing of O6-methylguanine DNA methyltransferase (MGMT) has also been described in a variety of neoplasms and has been associated with a consequent mutational spectrum. We investigated the relationship between hMLH1 promoter hypermethylation and MGMT promoter hypermethylation in 110 colorectal cancers using methylation-specific polymerase chain reaction. Expression of hMLH1 and MGMT was assessed by immunohistochemistry. MSI testing was performed using the National Cancer Institute consensus panel of five microsatellite markers. Promoter hypermethylation of hMLH1 was detected in 12% of tumors. This was significantly associated with the MSI-high phenotype (P < 0.01) and loss of hMLH1 expression (P < 0.01). Methylation of the MGMT promoter was detected in 43% of tumors, which were mostly microsatellite stable or MSI-low (P = 0.041) and showed loss of MGMT expression (P < 0.01). We demonstrated an inverse relationship between hMLH1 promoter hypermethylation and MGMT promoter hypermethylation (P = 0.041), suggesting that a number of distinct hypermethylation-associated pathways may exist in colorectal cancer.
Tumorigenesis in humans is a multistep process, reflecting an accumulation of genetic changes that lead to the progressive transformation of normal cells into increasingly malignant derivatives.1 The causes of such genetic alterations are multifactorial, with exogenous and endogenous compounds known to induce a variety of genetic alterations, including deletions, insertions, and base substitutions.2 Aberrations in DNA repair, giving rise to proposed mutator phenotypes, are hypothesized to contribute to the accumulation of mutations.3
The first well-characterized mutator pathway in human cancer involves abnormalities in DNA mismatch repair (MMR) proteins.4,5,6 Germline alterations in MMR genes (hMLH1, hMSH2, hMSH6, and hPMS2) are responsible for the majority of hereditary nonpolyposis colorectal cancer cases.7,8,9,10,11 MMR-deficient tumors exhibit increased random point mutation rates and deletions or insertions in microsatellite and repetitive sequences, referred to as microsatellite instability (MSI).12 Because microsatellites and repetitive sequences are frequently present in coding regions, instability can manifest as frameshift mutations that inactivate a variety of genes, including genes that suppress tumor formation, eg, APC, transforming growth factor-βRII, hMSH2, and caspase-5.13,14 This mutator phenotype is also present in 10 to 15% of sporadic cases of colorectal cancer (CRC), in which hMLH1 expression is lost not through mutation but through epigenetic silencing by promoter hypermethylation.15,16
Promoter hypermethylation-associated silencing of O6-methylguanine DNA methyltransferase (MGMT) has been proposed to cause another mutator pathway more prevalent than MSI.17 MGMT is a DNA repair protein that removes promutagenic and cytotoxic adducts from O6-guanine in DNA. It protects cells against potentially deleterious effects of endogenously and exogenously produced O6-alkylating agents, including mutation, sister chromatid exchanges, recombination, and chromosomal aberrations.18 Loss of MGMT function during tumorigenesis has been suggested to result in mutations in key cancer-related genes. Silencing of MGMT has been shown to be strongly linked to the presence of G to A transition mutations in K-ras, the most frequent type of mutation, in colon,19 gastric,20 and gall bladder cancers.21 A similar association has been described with C:G to T:A transitions in p53 of colorectal carcinomas,22 astrocytomas,23 gliomas,24 and non-small-cell lung cancers.25 Although genetic alterations of MGMT are rarely detected in human cancers,26 MGMT silencing associated with promoter hypermethylation has been documented in a variety of different tumor types and is observed in approximately 20 to 40% of CRC cases.27,28,29
The functional interaction between MGMT and DNA mismatch repair has been extensively characterized in transgenic animals and cell lines.30,31,32 These latter studies collectively proposed a model whereby functional mismatch repair over an unrepaired alkylated residue, occurring, for example, in a MGMT-deficient background, results in double-strand breaks (DSBs) and potentially chromosomal alterations. Although the occurrence of each respective mutator phenotype has been described, no specific interrelation has been demonstrated. In this study, we examined the promoter hypermethylation and immunohistochemical status of hMLH1 and MGMT in an unselected series of sporadic colorectal carcinomas and correlated these findings with clinical and pathological data.
Materials and Methods
Tumor Specimens
We randomly selected 110 colorectal cancers from patients attending the SVUH Centre for Colorectal Disease. Histologically normal mucosa, remote from the tumor, was also obtained. Each cancer case selected was sporadic, ie, lacking family history or clinical evidence of familial adenomatous polyposis or hereditary nonpolyposis colorectal cancer. Ethical approval to conduct this study was granted by the St. Vincent’s University Hospital Ethics and Medical Research Committee.
Histopathological Features
Conventional histopathological parameters, including the American Joint Committee on Cancer/International Union Against Cancer tumor-node-metastasis stage, tumor type, and differentiation, were assessed using a standard reporting template. Tumors were typed as adenocarcinomas or mucinous adenocarcinomas according to the criteria of the World Health Organization.33 Differentiation (well/moderate or poor) was determined following the World Health Organization guidelines.33 Additional histological parameters assessed included lymphovascular invasion,34 perineural invasion,35 tumor margin,36 and peritoneal involvement.37,38
Microsatellite Marker Analysis
MSI analysis was performed on DNA from the snap-frozen samples for the paired tumor and normal adjacent mucosa using fluorescently labeled primers for the National Cancer Institute consensus panel of microsatellite markers (D2S123, D5S346, D17S250, BAT25, and BAT26). Size variations were analyzed by capillary gel electrophoresis (ABI 310; Applied BioSystems, Foster City, CA). Tumors were scored as MSI-high (MSI-H) if two or more of the five markers showed instability, MSI-low (MSI-L) if only one of these markers showed instability, or stable (MSS) if none of the markers showed any size variation.
Methylation-Specific Polymerase Chain Reaction (PCR)
DNA methylation patterns in the CpG islands of hMLH1 and MGMT genes were determined using methylation-specific PCR (MSP).39 Briefly, <2 μg of genomic DNA from tumor and paired non-neoplastic tissue was denatured by treatment with NaOH and modified by sodium bisulfite. DNA samples were purified using Wizard DNA purification resin (Promega, Madison, WI), treated with NaOH, precipitated with ethanol, and resuspended in deionized water. The primer sequences used are given in Table 1. PCR products were separated on 2.5% agarose gels and visualized under UV illumination.
Table 1.
Primer Sequences for MSP15
| Gene | Sense | Antisense | Size (bp) | |
|---|---|---|---|---|
| hMLH1 | U | 5′-TTT TGATGTAGATGTTTTATTAGGGTTGT-3′ | 5′-ACCACCTCATCATAACTACCCACA-3′ | 124 |
| M | 5′-ACGTAGACGTTTTATTAGGGTCGC-3′ | 5′-CCTCATCGTAACTACCCGCG-3′ | 115 | |
| MGMT | U | 5′-TTTGTGTTTTGATGTTTGTAGGTTTTTGT-3′ | 5′-AACTCCACACTCTTCCAAAAACAAAACA-3′ | 93 |
| M | 5′-TTTCGACGTTCGTAGGTTTTCGC-3′ | 5′-GCACTCTTCCGAAAACGAAACG-3′ | 81 |
M, methylated; U, unmethylated.
Immunohistochemistry
hMLH1 Expression
Immunostaining for hMLH1 was performed on 5-μm sections of formalin-fixed paraffin-embedded tissues. Sections were dewaxed in xylene and rehydrated through graded alcohol to water. Endogenous peroxidase activity was quenched by incubating the slides in 3% hydrogen peroxide for 7 minutes. Antigen retrieval was performed by immersing sections in 10 mmol/L citrate buffer (pH 6.0) and heating in a domestic microwave oven (800 W) at full power for 5 minutes, followed by boiling in a pressure cooker for 4 minutes. Sections were left for 20 minutes in buffer to cool to room temperature.
Tissues were stained for hMLH1 using the DAKO Envision kit (DAKO A/S, Glostrup, Denmark) according to the manufacturer’s instructions, with the primary antibody against hMLH1 (clone G168-728; PharMingen, San Diego, CA) applied at a dilution of 1:75 for 2 hours. Positive staining was visualized with 3,3-diaminobenzidine substrate solution. Sections were counterstained with Mayer’s hematoxylin. Tumor cells were judged to be negative for hMLH1 only if they lacked nuclear staining in a section in which stromal or inflammatory cell nuclei stained positive (Figure 1D).
Figure 1.
Immunohistochemical staining patterns for MGMT (A–C) and hMLH1. A: Tumor with positive staining; B: tumor showing weak nuclear staining; C: tumor showing absence of nuclear staining; D: tumor showing loss of hMLH1 expression. Intratumoral endothelial cells (arrow) act as an internal positive control. An adjacent area of normal mucosa, staining positive for hMLH1, is present in the upper left corner. Magnification, ×100.
MGMT Expression
Tissue microarrays were assembled from the formalin-fixed paraffin-embedded tissues using a 0.6-mm-diameter punch (Beecher Instruments, Silver Spring, MD). Four cores were taken from the tumor and the paired normal adjacent mucosa for each case. Sections (5 μm) were cut from each tissue microarray block and used for MGMT immunohistochemistry. Sections were dewaxed and endogenous peroxidase quenched as before. Sections were immersed in boiling citrate buffer in a pressure cooker and then boiled for 5 minutes at full pressure (103 kPa). Sections were stained for MGMT using the avidin-biotin method (Vectastain Elite, Vector Laboratories, Burlingame, CA) with the primary antibody against MGMT (clone mT3.1; Neomarkers, Fremont, CA) applied at a dilution of 1:100 overnight at 4°C. Staining was visualized with 3,3-diaminobenzidine and hematoxylin counterstain as before. Only strong nuclear staining was regarded as indicative of MGMT protein expression (Figure 1A). Loss of MGMT expression was defined as either weak nuclear staining (Figure 1B) or complete absence of nuclear staining (Figure 1C).
Statistical Analyses
Fisher’s exact test, χ2 test, and Mann-Whitney U-test were used, as appropriate, to test statistical significance. All calculations were performed using the SPSS 11.0 for Windows statistical software package (SPSS, Inc., Chicago, IL). All reported P values are two tailed.
Results
Promoter Hypermethylation and Relationship to MSI and Immunohistochemical Expression
Tables 2and 3 outline the primary clinicopathological and molecular features of the cohort, respectively. Ten tumors (9%) displayed a MSI-H phenotype and 97 (88%) had no evidence of microsatellite instability. Using the Bethesda panel of microsatellite markers, only three MSI-L tumors (3%) were detected. Immunohistochemical absence of an MMR protein was described in nine cases (8%). Absence of hMLH1 (Figure 1D) was described in eight cases (seven of which were MSI-H), absence of hMLH1 and hMSH2 in one case (an 86-year-old male with a poorly differentiated cecal cancer, MSI-H), and absence of hMSH2 in one case (a 70-year-old male with a moderately differentiated cecal cancer, MSI-H). MSI status was therefore 90% sensitive and 99% specific as an indicator of MMR function characterized immunohistochemically.
Table 2.
Clinicopathological Characteristics of 110 Patients with Colorectal Cancer
| Patient characteristics | |
|---|---|
| Mean age (years [±SD]) | 69 (±11) |
| Gender | |
| Male | 52 (47%) |
| Female | 58 (53%) |
| Tumor site | |
| Rectum/left colon | 74 (66%) |
| Right colon | 36 (33%) |
| Tumor differentiation | |
| Well/moderate | 84 (74%) |
| Poor | 26 (24%) |
| Angiolymphatic invasion | |
| Absent | 62 (56%) |
| Present | 48 (44%) |
| Perineural invasion | |
| Absent | 91 (83%) |
| Present | 19 (17%) |
| Maximum tumor size (cm [±SD]) | 4.7 (±1.9) |
| Tumor stage | |
| A(T2, N0) | 2 |
| B(T3, N0) | 32 |
| (T4, N0) | 11 |
| C(T2, N1) | 2 |
| (T3, N1) | 21 |
| (T4, N1) | 7 |
| (T2, N2) | 7 |
| (T3, N2) | 9 |
| (T4, N2) | 5 |
| D(T1–4, N1–2, M1) | 14 |
Table 3.
Frequencies of Molecular and Immunohistochemical Features in Tumors of 110 Colorectal Cancer Patients
| Variable | Number |
|---|---|
| Microsatellite instability | |
| MSI-H | 10 |
| MSI-L | 3 |
| MSS | 97 |
| Immunohistochemistry | |
| Absent hMLH1 | 9 |
| Absent hMSH2 | 1 |
| Absent MGMT | 37 |
| Promoter hypermethylation | |
| hMLH1 | 13 |
| MGMT | 47 |
Promoter hypermethylation of hMLH1 was detected in 12% (13 of 110) of tumors (Figure 2). This was significantly associated with a MSI-H phenotype (P < 0.001). A positive association was observed between hMLH1 promoter hypermethylation and absence of hMLH1 expression as assayed by immunohistochemistry (P < 0.001). Six of 13 cases exhibiting hMLH1 promoter hypermethylation did not show absence of hMLH1 by immunohistochemistry or any level of microsatellite instability. In one-half of these cases (three of six) hMLH1 promoter hypermethylation was also detected in paired normal adjacent mucosa.
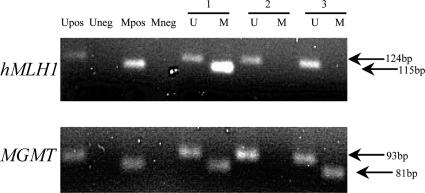
Figure 2.
Methylation analysis of hMLH1 and MGMT promoters. MSP was performed with primers specific for methylated (M) and unmethylated (U) regions. Product sizes: hMLH1 unmethylated, 124 bp; hMLH1 methylated, 115 bp; MGMT unmethylated, 93 bp; and MGMT methylated, 81 bp.
Hypermethylation of the MGMT promoter was detected in 43% (47 of 110) of tumors and was significantly associated with the MSS/MSI-L phenotype (P = 0.041). We found 18% of cases (20 of 110) with complete absence of MGMT expression (Figure 1C), with 18 of 20 displaying MGMT promoter hypermethylation. In 15% of cases (17 of 110), weak nuclear staining was seen (Figure 1B); more than one-half of these cases (9 of 17) showed evidence of MGMT promoter hypermethylation. Combining those cases with complete absence and weak nuclear staining gave 37 cases with loss of MGMT expression. Of these 37, 27 (73%) exhibited promoter hypermethylation at this locus. There was no evidence of promoter hypermethylation of MGMT in 57% of cases (63 of 110). MGMT was expressed in 84% (53 of 63) of these cases, with the majority (7 of 10) of the negative cases in this group showing weak nuclear staining and incomplete absence of MGMT staining. Absence of MGMT was thus significantly associated with MGMT promoter hypermethylation (P < 0.001).
Relationship between Promoter Hypermethylation Status and Clinicopathological Features
hMLH1 promoter hypermethylation was statistically significantly associated with proximal location (P = 0.027), poor differentiation (P = 0.015), lower incidence of lymph node metastasis (P = 0.035), and large tumor size (P = 0.025). Similar trends were described for absence of hMLH1 protein expression: proximal location (P = 0.056), lower incidence of lymph node metastasis (P = 0.078), and large tumor size (P = 0.066). Associations between promoter hypermethylation status and pathological/molecular features are given in Table 4.
Table 4.
Association between Promoter Hypermethylation and Pathological, Molecular, and Immunohistochemical Features in 110 Colorectal Cancers
| Variable |
hMLH1 hypermethylation
|
MGMT hypermethylation
|
||||
|---|---|---|---|---|---|---|
| U (n = 97) | M (n = 13) | P value | U (n = 63) | M (n = 47) | P value | |
| Immunohistochemistry | ||||||
| hMLH1 | ||||||
| Present | 95 | 6 | <0.001 | 56 | 45 | 0.30 |
| Absent | 2 | 7 | 7 | 2 | ||
| MGMT | ||||||
| Present | 61 | 12 | 0.057 | 53 | 20 | <0.001 |
| Absent | 36 | 1 | 10 | 27 | ||
| MSI | ||||||
| MSI-H | 2 | 8 | <0.001 | 9 | 1 | 0.041 |
| MSI-L/MSS | 95 | 5 | 54 | 46 | ||
| Hypermethylation | ||||||
| hMLH1 | ||||||
| Present | 11 | 2 | 0.041 | |||
| Absent | 52 | 45 | ||||
| Tumor site | ||||||
| Rectum/left colon | 69 | 5 | 0.027 | 42 | 32 | 1.0 |
| Right colon | 28 | 8 | 21 | 15 | ||
| Tumor differentiation | ||||||
| Well/moderate | 78 | 6 | 0.015 | 49 | 35 | 0.65 |
| Poor | 19 | 7 | 14 | 12 | ||
| Tumor margin | ||||||
| Expansile | 31 | 6 | 0.35 | 17 | 20 | 0.11 |
| Infiltrative | 66 | 7 | 46 | 27 | ||
| Nodal metastases | ||||||
| N0 | 41 | 10 | 0.035 | 28 | 23 | 0.70 |
| N1/2 | 56 | 3 | 35 | 24 | ||
| Peritoneal involvement | ||||||
| No | 78 | 9 | 0.47 | 52 | 35 | 0.35 |
| Yes | 19 | 4 | 11 | 12 | ||
| Size (cm [±SD]) | 4.6 (±1.7) | 6.1 (±2.5) | 0.025 | 4.8 (±2.0) | 4.6 (1.6) | 0.85 |
M, methylated; U, unmethylated.
Although tumors displaying MGMT promoter hypermethylation were more likely to have an expansile margin (P = 0.105), no associations with the clinical or pathological variables examined were statistically significant. Absence of MGMT protein expression demonstrated similar nonsignificant clinical and pathological trends as MGMT promoter hypermethylation, with only peritoneal involvement (P = 0.047) reaching significance. Associations between immunohistochemical staining patterns and pathological/molecular features are given in Table 5.
Table 5.
Association between Immunohistochemical Staining Patterns of hMLH1 and MGMT and Pathological and Molecular Features in 110 Colorectal Cancers
| Variable | hMLH1 staining
|
MGMT staining
|
||||
|---|---|---|---|---|---|---|
| Present (n = 101) | Absent (n = 9) | P value | Present (n = 73) | Absent (n = 37) | P value | |
| MSI | ||||||
| MSI-H | 2 | 8 | <0.001 | 9 | 1 | 0.16 |
| MSI-L/MSS | 99 | 1 | 64 | 36 | ||
| Tumor site | ||||||
| Rectum/left colon | 71 | 3 | 0.056 | 49 | 25 | 1.0 |
| Right colon | 30 | 6 | 24 | 12 | ||
| Tumor differentiation | ||||||
| Well/moderate | 79 | 5 | 0.21 | 57 | 27 | 0.64 |
| Poor | 22 | 4 | 16 | 10 | ||
| Tumor margin | ||||||
| Expansile | 31 | 6 | 0.059 | 22 | 15 | 0.29 |
| Infiltrative | 70 | 3 | 51 | 22 | ||
| Nodal metastases | ||||||
| N0 | 44 | 7 | 0.078 | 35 | 16 | 0.69 |
| N1/2 | 57 | 2 | 38 | 21 | ||
| Peritoneal involvement | ||||||
| No | 80 | 7 | 1.0 | 62 | 25 | 0.047 |
| Yes | 21 | 2 | 11 | 12 | ||
| Size (cm [±SD]) | 4.6 (±1.7) | 6.3 (±2.9) | 1.0 | 4.8 (±1.9) | 4.6 (1.7) | 0.90 |
Interrelationship between hMLH1 and MGMT Promoter Hypermethylation Statuses
Of this cohort of 110 cases, two tumors displayed promoter hypermethylation at both loci. hMLH1 and MGMT promoter hypermethylation consequently showed a statistically significant inverse association (P = 0.041). This exclusive pattern was reflected at the immunohistochemical level with only one tumor displaying concomitant loss of both proteins.
Discussion
Although the individual instances of hMLH1 and MGMT promoter hypermethylation, at 12 and 43%, respectively, and their association with gene silencing are well described in colorectal cancer,27,29,40 we demonstrate a specific relationship between the patterns of promoter hypermethylation of these two genes.
Tumors characterized on this basis are known to be molecularly and clinicopathologically distinct. Similar to previous descriptions,15,16,19,26,41,42,43,44 tumors in this cohort displaying hMLH1 promoter hypermethylation are more likely to be MSI-H, to be right-sided, to be of poor differentiation, and to have lower levels of nodal metastasis. The phenotypic characteristics of this group are distinct from those tumors displaying MGMT promoter hypermethylation, which tend to be clinically and pathologically somewhat more variable, as determined in this study and by others.29
The interplay between MGMT and MMR functions has been extensively documented in transgenic mice, including MLH1/MGMT−/− double knockout animals, as well as in cell lines.32 The current view regarding the cellular result of an unrepaired O6-meG:C is that replication results in an O6-meG:T mismatch (or possibly an O6-meG:C ambiguous pair). In the next round of replication, this results in an A:T transition mutation and again to an O6-meG:C pair or an O6-meG:T mismatch. The O6-meG:T mismatch is recognized and initiates MMR that creates a gapped duplex after incision of the newly replicated strand. Because O6-meG remains in the template, this process may be repeated in a futile repair loop that eventually results in DSBs that are intermediates in both apoptotic and recombinogenic pathways.31,45 DSBs have been demonstrated to frequently induce various sorts of chromosomal aberrations, including aneuploidy, loss of heterozygosity, and chromosomal translocations—events that are all intimately associated with carcinogenesis46,47,48,49 (Figure 3).
Figure 3.
Schematic of the consequences of attempted mismatch repair over an unrepaired alkylated residue in an MGMT-deficient background. Alkylation of DNA in the absence of MGMT function will result in transition mutations, regardless of mismatch repair proficiency. These lesions, in the presence of proficient mismatch repair, will give rise to futile MMR cycles, resulting in the generation of DSBs in DNA. These DSBs may potentially induce various sorts of chromosomal aberrations, including aneuploidy, loss of heterozygosity, and chromosomal translocations.
Importantly, in the only such study to date in a mammalian MGMT-deficient cell line, the frequency of recombinogenic to apoptotic events resulting from the presence of O6-meG residues is relatively high, on the order of 175:1.50 Thus, the likelihood of cytogenetic change is greater than that of programmed cell death, particularly because the tumor is not specifically being confronted with an alkylating agent. In cells lacking MMR function, DSBs would not be proposed to occur in response to a persistent O6-meG residue. Because the recombinogenic effects of O6-meG requires MMR, the potential for genetic instability in an MGMT-deficient background is potentiated by functional MMR. Consistent with this, DSBs have been demonstrated to occur in MGMT-deficient/MMR-proficient cells after treatment with an O6-alkylating agent but not in MMR-deficient cells, regardless of their MGMT functional status.51 Similarly, the higher relative occurrence of cytogenetic alterations reported in MMR-proficient compared with MMR-deficient colorectal tumors is consistent with this model.52
Thus the largely exclusive relationship of MGMT and hMLH1 hypermethylation and expression that we report here may reflect a positive selective pressure for the retention of MMR function in an MGMT-deficient background. This nominally constitutes the preservation of a tumor suppressor function during tumor progression. In the context of deficient MGMT function, functional MMR, through potentiation of genetic instability, promotes tumorigenesis. We hypothesize that no similar selective pressure would exist against loss of MGMT in an MMR-deficient background and that this may in part explain the occasional occurrence of cases lacking both proteins.
Although statistically significant, promoter hypermethylation status was not an absolute indicator of protein expression as assayed by immunohistochemistry for either hMLH1 or MGMT. This may explain the fact that this inverse relationship has not been specifically noted in previous studies in which promoter hypermethylation but not immunohistochemical status of genes, including hMLH1 and MGMT, have been characterized. In particular, some tumors displaying promoter hypermethylation retained protein expression, possibly because of heterogeneous cell populations in the tumor. Because of the high sensitivity of PCR-based techniques, hypermethylation could potentially be observed, even if only representative of a subset of tumor cells. Also, the inactivation of either hMLH1 or MGMT protein expression secondary to non-hypermethylation-associated mechanisms, although infrequent, has been described.26,53,54,55
The use of alkylating agent-based therapy in the subpopulation of cancer patients exhibiting MGMT promoter hypermethylation has been previously proposed.56,57 The expected sensitivity of MGMT-deficient cancers to alkylating agent-based therapies has been partially confounded by the prediction from experimental data of the existence of a chemo-resistant subpopulation of MGMT−/MMR− patients. The present data suggest that such a subpopulation may, in fact, be minimal.
That hMLH1 and MGMT inactivation in sporadic CRC is usually associated with promoter hypermethylation consequently results in the existence of promoter hypermethylation-associated mutator phenotypes. The observations reported here propose that these phenotypes are distinct and exclusive and that, in an MGMT-deficient background, tumorigenesis may be driven both by chromosomal alterations dependent on intact MMR function as well as by transition mutations in genes such as p53 and K-ras, which are independent of MMR function.
Acknowledgments
We thank Dr. Jacintha O’Sullivan for helpful discussions in reviewing this manuscript.
Footnotes
Supported by the Health Research Board, Ireland.
References
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- Loeb LA, Loeb KR, Anderson JP. Multiple mutations and cancer. Proc Natl Acad Sci USA. 2003;100:776–781. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Powell SM, Jen J, Hamilton SR. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- Peltomaki P, Vasen H. Mutations associated with HNPCC predisposition: update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- Miyaki M, Konishi M, Tanaka K, Kikuchi-Yanoshita R, Muraoka M, Yasuno M, Igari T, Koike M, Chiba M, Mori T. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet. 1997;17:271–272. doi: 10.1038/ng1197-271. [DOI] [PubMed] [Google Scholar]
- Berends MJ, Wu Y, Sijmons RH, Mensink RG, van der Sluis T, Hordijk-Hos JM, de Vries EG, Hollema H, Karrenbeld A, Buys CH, van der Zee AG, Hofstra RM, Kleibeuker JH. Molecular and clinical characteristics of MSH6 variants: an analysis of 25 index carriers of a germline variant. Am J Hum Genet. 2002;70:26–37. doi: 10.1086/337944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- Eshleman JR, Markowitz SD. Mismatch repair defects in human carcinogenesis. Hum Mol Genet. 1996;5:1489–1494. doi: 10.1093/hmg/5.supplement_1.1489. [DOI] [PubMed] [Google Scholar]
- Malkhosyan S, Rampino N, Yamamoto H, Perucho M. Frameshift mutator mutations. Nature. 1996;382:499–500. doi: 10.1038/382499a0. [DOI] [PubMed] [Google Scholar]
- Parsons R, Myeroff LL, Liu B, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res. 1995;55:5548–5550. [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, Thibodeau SN. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58:3455–3460. [PubMed] [Google Scholar]
- Margison GP, Santibanez-Koref MF. O6-alkylguanine-DNA alkyltransferase: role in carcinogenesis and chemotherapy. Bioessays. 2002;24:255–266. doi: 10.1002/bies.10063. [DOI] [PubMed] [Google Scholar]
- Mukai T, Sekiguchi M. Gene silencing in phenomena related to DNA repair. Oncogene. 2002;21:9033–9042. doi: 10.1038/sj.onc.1206095. [DOI] [PubMed] [Google Scholar]
- Esteller M, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Watkins DN, Issa JP, Sidransky D, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000;60:2368–2371. [PubMed] [Google Scholar]
- Park TJ, Han SU, Cho YK, Paik WK, Kim YB, Lim IK. Methylation of O(6)-methylguanine-DNA methyltransferase gene is associated significantly with K-ras mutation, lymph node invasion, tumor staging, and disease free survival in patients with gastric carcinoma. Cancer. 2001;92:2760–2768. doi: 10.1002/1097-0142(20011201)92:11<2760::aid-cncr10123>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Kohya N, Kitajima Y, Kitahara K, Miyazaki K. Mutation analysis of K-ras and beta-catenin genes related to O6-methylguanin-DNA methyltransferase and mismatch repair protein status in human gallbladder carcinoma. Int J Mol Med. 2003;11:65–69. [PubMed] [Google Scholar]
- Esteller M, Risques RA, Toyota M, Capella G, Moreno V, Peinado MA, Baylin SB, Herman JG. Promoter hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res. 2001;61:4689–4692. [PubMed] [Google Scholar]
- Nakamura M, Watanabe T, Yonekawa Y, Kleihues P, Ohgaki H. Promoter methylation of the DNA repair gene MGMT in astrocytomas is frequently associated with G:C → A:T mutations of the TP53 tumor suppressor gene. Carcinogenesis. 2001;22:1715–1719. doi: 10.1093/carcin/22.10.1715. [DOI] [PubMed] [Google Scholar]
- Bello MJ, Alonso ME, Aminoso C, Anselmo NP, Arjona D, Gonzalez-Gomez P, Lopez-Marin I, de Campos JM, Gutierrez M, Isla A, Kusak ME, Lassaletta L, Sarasa JL, Vaquero J, Casartelli C, Rey JA. Hypermethylation of the DNA repair gene MGMT: association with TP53 G:C to A:T transitions in a series of 469 nervous system tumors. Mutat Res. 2004;554:23–32. doi: 10.1016/j.mrfmmm.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Wolf P, Hu YC, Doffek K, Sidransky D, Ahrendt SA. O(6)-Methylguanine-DNA methyltransferase promoter hypermethylation shifts the p53 mutational spectrum in non-small cell lung cancer. Cancer Res. 2001;61:8113–8117. [PubMed] [Google Scholar]
- Halford S, Rowan A, Sawyer E, Talbot I, Tomlinson I. O6-methylguanine methyltransferase in colorectal cancers: detection of mutations, loss of expression, and weak association with G:C>A:T transitions. Gut. 2005;54:797–802. doi: 10.1136/gut.2004.059535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- Herfarth KK, Brent TP, Danam RP, Remack JS, Kodner IJ, Wells SA, Jr, Goodfellow PJ. A specific CpG methylation pattern of the MGMT promoter region associated with reduced MGMT expression in primary colorectal cancers. Mol Carcinog. 1999;24:90–98. doi: 10.1002/(sici)1098-2744(199902)24:2<90::aid-mc3>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Whitehall VL, Walsh MD, Young J, Leggett BA, Jass JR. Methylation of O-6-methylguanine DNA methyltransferase characterizes a subset of colorectal cancer with low-level DNA microsatellite instability. Cancer Res. 2001;61:827–830. [PubMed] [Google Scholar]
- Fedor HL, De Marzo AM. Practical methods for tissue microarray construction. Methods Mol Med. 2004;103:89–102. doi: 10.1385/1-59259-780-7:089. [DOI] [PubMed] [Google Scholar]
- Galloway SM, Greenwood SK, Hill RB, Bradt CI, Bean CL. A role for mismatch repair in production of chromosome aberrations by methylating agents in human cells. Mutat Res. 1995;346:231–245. doi: 10.1016/0165-7992(95)90040-3. [DOI] [PubMed] [Google Scholar]
- Kawate H, Sakumi K, Tsuzuki T, Nakatsuru Y, Ishikawa T, Takahashi S, Takano H, Noda T, Sekiguchi M. Separation of killing and tumorigenic effects of an alkylating agent in mice defective in two of the DNA repair genes. Proc Natl Acad Sci USA. 1998;95:5116–5120. doi: 10.1073/pnas.95.9.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morson BC, Sobin LH. Histological typing of intestinal tumours. Geneva: World Health Organization; International Histological Classification of Tumours. no. 15. 1976 [Google Scholar]
- Talbot IC, Ritchie S, Leighton MH, Hughes AO, Bussey HJ, Morson BC. The clinical significance of invasion of veins by rectal cancer. Br J Surg. 1980;67:439–442. doi: 10.1002/bjs.1800670619. [DOI] [PubMed] [Google Scholar]
- Knudsen JB, Nilsson T, Sprechler M, Johansen A, Christensen N. Venous and nerve invasion as prognostic factors in postoperative survival of patients with resectable cancer of the rectum. Dis Colon Rectum. 1983;26:613–617. doi: 10.1007/BF02552975. [DOI] [PubMed] [Google Scholar]
- Jass JR, Love SB, Northover JM. A new prognostic classification of rectal cancer. Lancet. 1987;1:1303–1306. doi: 10.1016/s0140-6736(87)90552-6. [DOI] [PubMed] [Google Scholar]
- Shepherd NA, Baxter KJ, Love SB. The prognostic importance of peritoneal involvement in colonic cancer: a prospective evaluation. Gastroenterology. 1997;112:1096–1102. doi: 10.1016/s0016-5085(97)70119-7. [DOI] [PubMed] [Google Scholar]
- Lennon AM, Mulcahy HE, Hyland JM, Lowry C, White A, Fennelly D, Murphy JJ, O’Donoghue DP, Sheahan K. Peritoneal involvement in stage II colon cancer. Am J Clin Pathol. 2003;119:108–113. doi: 10.1309/J6BD-TWM2-M792-TN2V. [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Fraga MF, Guo M, Garcia-Foncillas J, Hedenfalk I, Godwin AK, Trojan J, Vaurs-Barriere C, Bignon YJ, Ramus S, Benitez J, Caldes T, Akiyama Y, Yuasa Y, Launonen V, Canal MJ, Rodriguez R, Capella G, Peinado MA, Borg A, Aaltonen LA, Ponder BA, Baylin SB, Herman JG. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum Mol Genet. 2001;10:3001–3007. doi: 10.1093/hmg/10.26.3001. [DOI] [PubMed] [Google Scholar]
- Xu XL, Yu J, Zhang HY, Sun MH, Gu J, Du X, Shi DR, Wang P, Yang ZH, Zhu JD. Methylation profile of the promoter CpG islands of 31 genes that may contribute to colorectal carcinogenesis. World J Gastroenterol. 2004;10:3441–3454. doi: 10.3748/wjg.v10.i23.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind GE, Thorstensen L, Lovig T, Meling GI, Hamelin R, Rognum TO, Esteller M, Lothe RA. A CpG island hypermethylation profile of primary colorectal carcinomas and colon cancer cell lines. Mol Cancer. 2004;3:28. doi: 10.1186/1476-4598-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- Gafa R, Maestri I, Matteuzzi M, Santini A, Ferretti S, Cavazzini L, Lanza G. Sporadic colorectal adenocarcinomas with high-frequency microsatellite instability. Cancer. 2000;89:2025–2037. [PubMed] [Google Scholar]
- Kaina B, Ziouta A, Ochs K, Coquerelle T. Chromosomal instability, reproductive cell death and apoptosis induced by O6-methylguanine in Mex−, Mex+ and methylation-tolerant mismatch repair compromised cells: facts and models. Mutat Res. 1997;381:227–241. doi: 10.1016/s0027-5107(97)00187-5. [DOI] [PubMed] [Google Scholar]
- Coquelle A, Rozier L, Dutrillaux B, Debatisse M. Induction of multiple double-strand breaks within an hsr by meganucleaseI-SceI expression or fragile site activation leads to formation of double minutes and other chromosomal rearrangements. Oncogene. 2002;21:7671–7679. doi: 10.1038/sj.onc.1205880. [DOI] [PubMed] [Google Scholar]
- Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- Pipiras E, Coquelle A, Bieth A, Debatisse M. Interstitial deletions and intrachromosomal amplification initiated from a double-strand break targeted to a mammalian chromosome. EMBO J. 1998;17:325–333. doi: 10.1093/emboj/17.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasouli-Nia A, Sibghat U, Mirzayans R, Paterson MC, Day RS., III On the quantitative relationship between O6-methylguanine residues in genomic DNA and production of sister-chromatid exchanges, mutations and lethal events in a Mer− human tumor cell line. Mutat Res. 1994;314:99–113. doi: 10.1016/0921-8777(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Ochs K, Kaina B. Apoptosis induced by DNA damage O6-methylguanine is Bcl-2 and caspase-9/3 regulated and Fas/caspase-8 independent. Cancer Res. 2000;60:5815–5824. [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Tomlinson IP, Ilyas M, Bodmer WF. Allele loss occurs frequently at hMLH1, but rarely at hMSH2, in sporadic colorectal cancers with microsatellite instability. Br J Cancer. 1996;74:1514–1517. doi: 10.1038/bjc.1996.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herfarth KK, Kodner IJ, Whelan AJ, Ivanovich JL, Bracamontes JR, Wells SA, Jr, Goodfellow PJ. Mutations in MLH1 are more frequent than in MSH2 in sporadic colorectal cancers with microsatellite instability. Genes Chromosomes Cancer. 1997;18:42–49. doi: 10.1002/(sici)1098-2264(199701)18:1<42::aid-gcc5>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Chen FY, Harris LC, Remack JS, Brent TP. Cytoplasmic sequestration of an O6-methylguanine-DNA methyltransferase enhancer binding protein in DNA repair-deficient human cells. Proc Natl Acad Sci USA. 1997;94:4348–4353. doi: 10.1073/pnas.94.9.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Herman JG. Generating mutations but providing chemosensitivity: the role of O6-methylguanine DNA methyltransferase in human cancer. Oncogene. 2004;23:1–8. doi: 10.1038/sj.onc.1207316. [DOI] [PubMed] [Google Scholar]
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]