Abstract
Apoptotic and necrotic tumor cells release DNA into plasma, providing an accessible tumor biomarker. Tumor-released plasma-circulating DNA can be screened for tumor-specific genetic changes, including mutation, methylation, or allelic imbalance. However, technical problems relating to the quantity and quality of DNA collected from plasma hinder downstream genetic screening and reduce biomarker detection sensitivity. Here, we present a new methodology, blunt-end ligation-mediated whole genome amplification (BL-WGA), that efficiently amplifies small apoptotic fragments (<200 bp) as well as intermediate and large necrotic fragments (>5 kb) and enables reliable high-throughput analysis of plasma-circulating DNA. In a single-tube reaction, purified double-stranded DNA was blunted with T4 DNA polymerase, self-ligated or cross-ligated with T4 DNA ligase and amplified via random primer-initiated multiple displacement amplification. Using plasma DNA from breast cancer patients and normal controls, we demonstrate that BL-WGA amplified the plasma-circulating genome by ∼1000-fold. Of 25 informative polymorphic sites screened via polymerase chain reaction-denaturating high-performance liquid chromatography, 24 (95%) were correctly determined by BL-WGA to be allelic retention or imbalance compared to 44% by multiple displacement amplification. By enabling target magnification and application of high-throughput genome analysis, BL-WGA improves sensitivity for detection of circulating tumor-specific biomarkers from bodily fluids or for recovery of nucleic acids from suboptimally stored specimens.
Solid malignant tumors release a significant amount of genomic DNA into the systemic circulation through cellular necrosis and apoptosis,1,2,3,4,5 and this circulating DNA can be exploited as an accessible tumor-specific biomarker.1,2,3,5,6,7 Tumor-released DNA can be detected as a result of specific genetic changes, including mutations, methylation, translocation, presence of viral genes, or allelic imbalance (AI), a marker for potential deletion of tumor suppressor genes.5,6,7,8,9 For example in breast cancer, tumor-released DNA in plasma provides a useful biomarker for breast cancer diagnosis and for monitoring relapse and metastasis.10,11,12,13,14,15,16,17,18,19,20,21 AI in particular is present in the majority of primary breast tumors and is also encountered in the plasma of breast cancer patients.22,23,24,25,26 In the great majority of cases, AI in plasma corresponds to alterations in the primary tumor of the respective pa-tient.10,12,13,14,16 Plasma-circulating tumor DNA at diagnosis of breast cancer is a predictor of disease-free survival19 and correlates with clinicopathological features and disease stage.13
Although genetic alterations such as AI in plasma-circulating DNA have been shown to have clinical utility as biomarkers for early cancer detection or therapy monitoring, there are technical difficulties limiting their widespread application to cancer screening. Because of the genetic heterogeneity of tumors, no single biomarker is present in 100% of primary tumors.6 Further, plasma-circulating DNA of tumor origin often contains only a fraction of the primary tumor alterations that may reflect the existence of different clones within the same tumor.27,28 In addition, plasma-circulating DNA may contain normal DNA, which complicates detection of tumor-specific genetic changes.7,29 To increase sensitivity and specificity of biomarker-based tumor detection, a panel of biomarkers and techniques must be used for each patient,21 and there is a need for using high-throughput methodologies for detection of DNA alterations in plasma.21 The amount of DNA circulating in the plasma of cancer patients is often low, with a median of ∼59 ng of DNA per ml of blood.7 Because low quantity of DNA template is a source of false-positives (allele dropout) and false-negatives (omission of AI),30,31,32,33,34 at least 5 ng of genomic DNA template is recommended per polymerase chain reaction (PCR).30 Consequently, the number of genes that can be reliably examined for tumor-specific alterations via PCR-based methodologies is limited by the availability of starting material.
Whole genome amplification of plasma-circulating DNA could alleviate problems associated with low-input DNA. Whole genome amplification generates micrograms of DNA when starting from nanogram quantities of material and should enable high-throughput screening for a comprehensive study of genetic abnormalities in plasma-circulating DNA. However, reliable whole genome amplification presents a significant challenge because plasma-circulating DNA consists of a mixture of fragmented apoptotic and necrotic DNA ranging from low (100 to 200 bp) to high (>2 kb) sizes that are difficult to amplify uniformly. Ligation-mediated PCR methodologies35,36 can amplify fragmented DNA;37 however, the genome coverage by PCR is incomplete because only a small fraction (a representation) of the original material is usually amplified.38 Multiple displacement amplification (MDA) provides almost complete genome coverage39,40 but has a low efficiency when used with fragmented DNA,41,42 and thus the lower size DNA fragments of plasma-circulating DNA would not be represented.
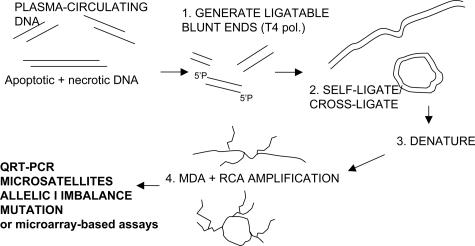
Here we present a new adaptation of MDA developed specifically for efficient amplification of fragmented nucleic acids collected from plasma-circulating DNA or other bodily fluids. The new method utilizes two additional steps before random primer-initiated multiple displacement, ie, conversion of DNA fragments to blunt-ends by T4 DNA polymerase followed by self-ligation or cross-ligation via T4 DNA ligase (Figure 1). The resulting large or circular DNA fragments are then amplified via random-primer initiated MDA. We demonstrate that this blunt-end ligation-mediated whole genome amplification (BL-WGA) enables efficient, unbiased amplification of small (<200 bp), intermediate, and very large (>5 kb) size DNA fragments without production of artifacts in the detection of AI in plasma-circulating DNA. BL-WGA enables high-throughput analysis for sensitive detection, discovery, and evaluation of tumor-specific DNA biomarkers in the blood of cancer patients.
Figure 1.
Outline of whole genome amplification of plasma-circulating DNA via BL-WGA. T4 pol, T4 DNA polymerase; RCA, rolling circle amplification; MDA, multiple displacement amplification.
Materials and Methods
Collection and Extraction of DNA
Human male genomic DNA was obtained from Promega (Madison, WI). Blood samples (plasma, lymphocytes) were obtained from the Dana-Farber/Harvard Cancer Center SPORE Bank, 30 patients with stage II to IV breast cancer and 3 metastatic colon cancer patients, following informed consent and institutional review board approval from the Dana-Farber Cancer Institute. Blood samples were also obtained from 16 healthy individuals. Within 2 to 5 hours of collection, whole blood was centrifuged at 2000 × g for 15 to 30 minutes, and plasma was carefully collected from the top of the supernatant. Plasma-circulating DNA was purified from plasma with QIAamp MinElute virus spin kit (Qiagen, Valencia, CA). Buffy coat was also removed for collection of genomic DNA from white blood cells. Normal and tumor breast tissue surgical specimens from 15 breast cancer patients were obtained from the Massachusetts General Hospital Tumor Bank. The QIAamp DNA mini kit (Qiagen) was used to purify genomic DNA from white blood cells and from normal and tumor tissues. DNA concentrations were measured by PicoGreen (Molecular Probes, Eugene, OR) as well as by a real-time PCR TaqMan assay performed as described.37
Blunt End Ligation-Mediated Whole Genome Amplification (BL-WGA)
A single tube whole genome amplification protocol was developed. Briefly, 2 to 4.5 μl of plasma DNA (∼2 to 5 ng total)) was blunted with 0.3 U of T4 DNA polymerase (New England Biolabs, Beverly, MA) at 12°C for 15 minutes in 5 μl of 1× T4 DNA ligase buffer (New England Biolabs), supplemented with dNTP (Applied Biosystems, Foster City, CA) at a final concentration 100 μmol/L. The T4 DNA polymerase was then heat-inactivated at 75°C for 20 minutes. The blunted DNA was ligated with 0.25 μl of T4 DNA ligase (2000 U/μl, New England Biolabs) in a volume of 5.25 μl at room temperature for 2 hours. Ligase was heat-inactivated at 65°C for 10 minutes. The sample was then amplified using the GenomiPhi (Amersham Biosciences, Buckinghamshire, UK) whole genome amplification kit. Next, 9 μl of random hexamer-containing buffer was added to 5.25 μl of ligated DNA and heated at 95°C for 3 minutes to denature the template followed by rapid cooling on ice. The mixture of 9 μl of reaction buffer plus 1 μl of enzyme mix was added to the cooled sample. The reaction mixture was incubated at 30°C for 16 hours. Finally the reaction was stopped via heating at 65°C for 10 minutes. This BL-WGA protocol can be conducted in a single tube with no intermediate purification steps.
MDA
For comparison to BL-WGA, MDA was performed in parallel on the plasma-circulating DNA samples using the GenomiPhi DNA amplification kit (Amersham Biosciences) per the company’s specifications.
Evaluation of Whole Genome Amplification via TaqMan Real-Time PCR
Real-time PCR TaqMan assays were performed to determine the relative amplification of specific genes after whole genome amplification of plasma-circulating DNA. The assay was done as previously described.37 Briefly, amplification was performed using AmpliTaq Gold (Applied Biosystems) in a smart-Cycler (Cepheid, Sunnyvale, CA). Primers and probes for exonic regions of genes studied were synthesized by Biosearch Technologies (Novato, CA). One μl of amplified plasma-circulating DNA was added to a final volume of 20 μl with a final concentration of 1× ABI TaqMan master mix (Applied Biosciences), 0.2 μmol/L each primer, and 0.1 μmol/L probe. The thermocycling program was as follows: 1 cycle of 50°C for 2 minutes, 1 cycle of 95°C for 10 minutes, 50 cycles of 95°C for 15 seconds, 60°C for 1 minute. Three independent experiments were performed for each gene to generate an average relative copy number and SD. Alternatively, unamplified plasma-circulating DNA was diluted to the same final volume as the BL-WGA/MDA amplification reactions and tested via real-time PCR. The relative gene amplification between unamplified/amplified plasma-circulating DNA was calculated using the comparative threshold (ΔΔCT) method.37,43
Dependence of Amplification on DNA Fragment Size
To estimate the amount of amplification performed by BL-WGA or MDA as a function of DNA fragment size, fragmentation of intact DNA was performed with restriction enzymes that yield predictable DNA fragment sizes ranging from 90 bp to 11,466 bp. DNA digested with restriction enzymes were run on 1% agarose gel to confirm the DNA smear and complete loss of the full-length genomic DNA band. After whole genome amplification, the amplification-fold of specific fragments was assessed via real-time PCR. Specifically, human male genomic DNA (∼1 μg) was digested with TaqI, NlaIII, or DpnII (New England Biolabs) according to the manufacturer’s instructions. Digested DNA was purified with the Qiagen PCR purification kit. DNA concentrations were measured via PicoGreen (Molecular Probes), and 10 ng of purified DNA was amplified via MDA or BL-WGA. An equal amount of DNA was also diluted in GenomiPhi sample/reaction buffer in the absence of the enzyme mix to serve as unamplified control. Real-time TaqMan PCR was performed using unamplified DNA or whole genome-amplified DNA as described previously.37 The amplification-fold for specific genes was calculated by obtaining the relative threshold difference (ΔΔCT) between amplified and unamplified samples.
Detection of AI and Microsatellite Changes via Denaturating High-Performance Liquid Chromatography (dHPLC)
The PCR-dHPLC method for detecting AI44,45 was used to screen amplified and unamplified plasma-circulating DNA at 23 polymorphic (single nucleotide polymorphisms [SNP]), positions described in Table 1. PCR-dHPLC was performed as described.42,46 Briefly, PCR of unamplified or amplified plasma-circulating DNA, or genomic DNA from tissue or white blood cells, was performed using Advantage-HF-2 PCR kit (BD-Clontech Biosciences, Palo Alto, CA). We added 1 to 2 μl of unamplified, or 1:10 diluted amplified plasma-circulating DNA (alternatively, 5 ng genomic DNA from lymphocytes) to a 10-μl reaction with 1× BD-Clonetech HF-2 PCR buffer, 1× dNTP mix, forward and reverse primer each 0.4 μmol/L, and 1× BD Advantage-HF-2 polymerase mix. The PCR cycling was done in a Perkin-Elmer 9600 PCR machine (Perkin-Elmer, Emeryville, CA). Thermocycling conditions were as follows: 94°C for 1 minute (94°C for 20 seconds, 65°C for 20 seconds, 68°C for 1 minute) × 10 cycles, with annealing temperature decreasing 1°C/cycle, touch-down PCR (94°C for 20 seconds, 55°C for 20 seconds, 68°C for 1 minute) × 32 cycles, 68°C for 5 minutes, and 4°C hold. The PCR program was followed by heat-denaturation at 95°C for 2 minutes and slow (1°C/minute) cooling to room temperature to generate heteroduplexes. PCR product (5 μl) was injected into the WAVE dHPLC system (Transgenomic Inc., Omaha, NE) at the corresponding partially denaturing temperature for each PCR product, as described in Table 1. The data were collected and analyzed using Navigator software and finally exported and plotted using Origin software (OrginLab Corp., Northampton, MA). Experiments were repeated at least three independent times, starting from unamplified plasma-circulating DNA each time.
Table 1.
SNPs Used for PCR-dHPLC-Based Detection of Allelic Imbalance
| SNP | dbSNP_ID | Primer | dHPLC temperature (C) |
|---|---|---|---|
| S1 | rs1394437 | forward: AAATCATCTGACTCTGGCTACA | 57.3 |
| reverse: CAGCATCATGCAGTATACCCAT | |||
| S2 | rs950302, rs950303 | forward: CTCCCTGGGTAGCCTCACAACT | 58.6 |
| reverse: GTGAAGTGTGCAAGAGGTCCAA | |||
| S3 | rs1385467, rs1353819 | forward: TCTTGGCCAACTTCTAATCCTA | 55.1 |
| reverse: TTTGTTGGACCAGGTGGATATA | |||
| S4 | rs2134095 | forward: AGGCATCCCAGTTCCGACTTGT | 58.3 |
| reverse: TATATTCCTGGGCATCGTGCTG | |||
| S5 | rs2034558, rs2034557 | forward: GGCGTATATCTTGCTAACCAAA | 58.7 |
| reverse: ATTCTGCCAATGATAGTGTTGA | |||
| S6 | rs2039371, rs2039372 | forward: CACAATGTGTTTCATGCGTCTT | 59.5 |
| reverse: TTCAATATGCAAACCCAATGTC | |||
| S7 | rs1396977, rs1396976 | forward: GACTTGGGTGCTGATAGTTCTT | 58.2 |
| reverse: AAAATGTGGTCCAGCTCATAGA | |||
| S8 | rs720404 | forward: TGCCCTTGAGGAGCTTACAATA | 57.5 |
| reverse: AACCTTCTCCATTGCCAATCCC | |||
| S9 | rs1940436 | forward: GTAGCGCCTAAATCGTTACAAA | 57.5 |
| reverse: TAGCCATCTGTTGTAGGGGATA | |||
| S10 | rs1518790, rs1518789 | forward: CCTCTCACCTTCCTGCACTAGC | 55.3 |
| reverse: ATAGCAAAATCAGACAAGGGTA | |||
| S11 | rs728293 | forward: AGGTCATTCAACAAGGCAATAC | 56.4 |
| reverse: GTAGAGCTGTGGGGCCCAAACT | |||
| S12 | rs1411375, rs1411374 | forward: TAAACAAAACCTGAGGGCGATA | 58.2 |
| reverse: GTCATTGTTGTCTTCAGGACCG | |||
| S13 | rs2115289, rs1947339 | forward: CCCAGCACGATTCTCTACCAT | 57.1 |
| reverse: CCAGTTCTAGAAGGCAAGCATA | |||
| S14 | rs1395547, rs1395548 | forward: ACTGAACACCCTTCCGGTTAAA | 59.5 |
| reverse: GCAGTGCCATGAGGTAGACTCT | |||
| S15 | rs1404843, rs1404841 | forward: AACAACATCTGGGCACTAGGTG | 55.3 |
| reverse: ATGTGCAGTGGTACACGCCTAT | |||
| S16 | rs952084 | forward: GCCCCCAATTAGCTCTTC | 56.6 |
| reverse: GCCAATCTCCCCTTTAGCTTT | |||
| S17 | rs953597 | forward: TGGCCATCTTTTCAAAGTCCT | 54.7 |
| reverse: CTTTCCTGGCGTGCTCATT | |||
| S18 | rs1379981 | forward: CGGCACTCACACTGTTAATG | 57.1 |
| reverse: TCAAGCCCTGAATGTAATCTC | |||
| S19 | rs1080708 | forward: CATTGAGCAGGTCTATTCGT | 56.4 |
| reverse: AACTTAGAGGCCATTGTAGGG | |||
| S20 | rs717796 | forward: TCATGCTTTGGGTCAGAC | 55.6 |
| reverse: TGCTGAGAATATCAGGACCTA | |||
| S21 | rs866435 | forward: GGGATCCATGTCCAGATACTC | 61.3 |
| reverse: CCAATTTTAGCCAACAAGGTC | |||
| S22 | rs4131175 | forward: CTGTTGGTGCCTTGATCT | 56 |
| reverse: TTGGAGGTTTTTGATTACTGA | |||
| S23 | rs718156 | forward: CTCCTCTCTCCTCTTAGC | 58.8 |
| reverse: GTAGGCAACTGAGTCTTAATC |
In addition, the conventional PCR-dHPLC methodology for screening microsatellite changes in unamplified plasma-circulating DNA was performed. Six microsatellite markers, D17S855, D17S654, D16S421, TH2, D10S197, and D9S161, that often display AI in breast tumors and plasma-circulating DNA13,19,20 were screened via PCR-dHPLC. Titanium Taq polymerase (BD-Clontech) was used in a 25-μl PCR reaction volume with 10 ng of genomic DNA. PCR conditions were as follows: 94°C for 30 seconds (94°C for 20 seconds, 65°C for 20 seconds, 68° for 1 minute) × 10 cycles with annealing temperature decreasing 1°C/cycle, touch-down PCR, 25 cycles (94°C for 20 seconds, 55°C for 20 seconds) × 25 cycles, 68°C for 1 minute, and 4°C hold. PCR product (12 μl) was introduced into the mobile phase by the dHPLC autosampler, and the sample was analyzed for microsatellite size changes at 50°C under nondenaturing conditions.
Results
dHPLC Screening of Microsatellite Imbalance in Unamplified Plasma-Circulating DNA
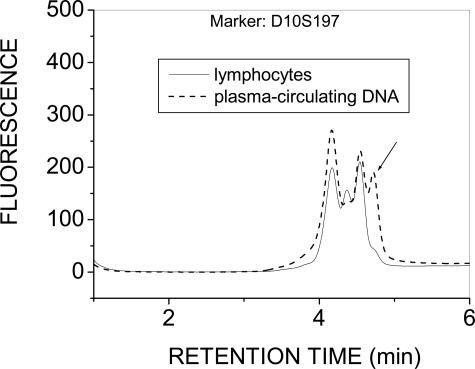
The conventional approach of screening microsatellites in genomic regions frequently presenting loss of heterozygosity in primary breast tumors and in plasma13,19,20 was first applied in samples from five early stage (II/III) breast cancer patients. The PCR-dHPLC methodology44,45 was applied to screen six microsatellite markers for differences in plasma DNA versus the corresponding lymphocyte genomic DNA, as reported by Silva and colleagues.13,19,20 The results demonstrated no microsatellite changes in plasma in any of these samples, except in one sample where microsatellite instability was detected for a single marker (Figure 2). The 1-ml plasma sample available for this study did not allow examination of additional microsatellite markers to increase the probability for detecting AI in at least one marker. Dilution of the starting material was not considered due to the well-documented allele-dropout artifacts associated with limited starting material in plasma-circulating DNA.30,31,32,33,34 Instead, whole genome amplification was applied to a portion of the remaining DNA, followed by AI screening at 23 SNP positions as described below.
Figure 2.
Microsatellite instability detected via PCR-dHPLC directly from unamplified plasma-circulating DNA for one of five patient samples screened using the six microsatellite markers reported by Silva and colleagues.13,19,20 Other markers tested were negative for microsatellite changes in these patients (not shown).
Range of DNA Sizes Amplified via BL-WGA or MDA
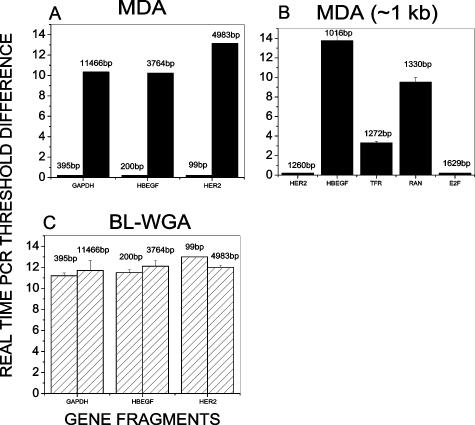
We used a simplified system to study the DNA fragment size dependence of BL-WGA compared to MDA. Using three restriction enzymes to digest intact genomic DNA in separate reactions, distinct DNA fragment sizes ranging from 99 to 11,466 bp were predictably obtained, encompassing a set of genes for which TaqMan real-time PCR probes were available. BL-WGA and MDA were performed in parallel for each restriction enzyme used and real-time PCR was used to quantify the amplification for each gene. Figure 3A demonstrates that MDA successfully amplified (>10 cycles difference, ie, >1000-fold) DNA fragments larger than 3 kb. For fragments ∼1 kb, the MDA results were mixed, ie, MDA substantially amplified 1016-bp (HBEGF) and 1330-bp (RAN) fragments but failed to amplify fragments of 1272 bp (TFR), 1260 bp (HER2), and 1629 bp (E2F) (Figure 3B). As could be expected, MDA consistently did not amplify fragments below 1 kb (Figure 3A). In comparison, BL-WGA amplified all tested DNA fragments with similar amplification efficiency, 2- to 4000-fold (Figure 3C). The data indicate that the method can potentially amplify equally apoptotic (<0.2 to 0.4 kb) and necrotic (>1 kb) DNA fragments from plasma-circulating DNA, resulting in a complete genome coverage similar to that produced when MDA is applied to intact genomic DNA.40
Figure 3.
Range of DNA fragment sizes amplified by MDA and BL-WGA. DNA fragments ranging from 99 to 11,466 bp were obtained after restriction digestion of intact genomic DNA. The amplification-fold after MDA and BL-WGA was evaluated via real-time PCR. A and B: Difference in TaqMan real-time PCR threshold before and after MDA amplification of the designated gene fragments. C: Difference in TaqMan real-time PCR threshold before and after BL-WGA amplification of the designated gene fragments. SD was calculated from three independent BL-WGA and MDA experiments.
Real-Time PCR Evaluation of BL-WGA or MDA-Amplified Plasma-Circulating DNA
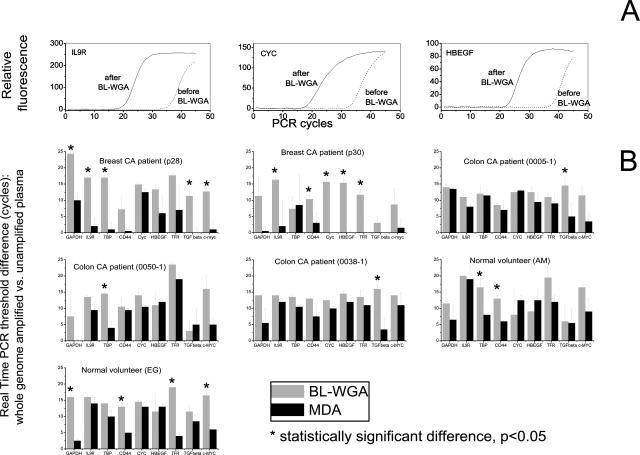
The TaqMan real-time PCR assay was used for an evaluation of amplification efficiency after amplification of plasma-circulating DNA via BL-WGA or MDA. For BL-WGA, representative growth curves obtained for three genes, HBEGF, CYC, and IL9R are depicted in Figure 4A. The difference in PCR threshold (CT) between unamplified and BL-WGA amplified DNA is 10 to 12 cycles, which represents 1000- to 4000-fold amplification. A comparison of the efficiency of the two methods of amplification, BL-WGA and MDA, was then performed for nine genes using real-time PCR primers and probes previously described.37 Results using plasma donated by seven individuals (two breast cancer patients, three colon cancer patients, and two normal volunteers) are depicted in Figure 4B. BL-WGA displays amplification of all nine genes examined in every sample tested. Specifically, BL-WGA generates an amplification of ∼1000- to 4000-fold in most genes examined, although one gene in sample p28 and two genes in p30 were only amplified by 50- to 100-fold. On the other hand, the MDA results are mixed, and amplification varies depending on the individual sample tested. In plasma sample p28 MDA did not amplify five of nine genes (IL9R, TBP, CD44, TGF-β, and C-MYC) while MDA failed to amplify eight of nine genes in sample p30. For the three colon cancer samples and the two normal volunteers, MDA was able to generate a modest degree of amplification in several genes, although BL-WGA consistently produced higher amplification for the same genes. Statistically significant differences (P < 0.05, Student’s t-test) between BL-WGA and MDA are demonstrated with an asterisk above each data point in Figure 4. The data indicate that BL-WGA amplifies plasma-circulating DNA efficiently.
Figure 4.
Real-time PCR-based evaluation of amplification of plasma circulating DNA via BL-WGA or MDA. A: Representative TaqMan real-time PCR growth curves after BL-WGA amplification of plasma-circulating DNA (patient sample 28). Growth curves before and after BL-WGA amplification are demonstrated for three genes. B: Comparison of amplification efficiencies of BL-WGA and MDA throughout nine genes for plasma-circulating DNA from breast and colon cancer patients or normal volunteers. Real-time PCR for each gene was performed before and after whole genome amplification. Amplification efficiency for nine individual genes was expressed as PCR threshold difference. SD was calculated from three independent BL-WGA and MDA experiments.
Evaluation of AI after Amplification of Plasma-Circulating DNA
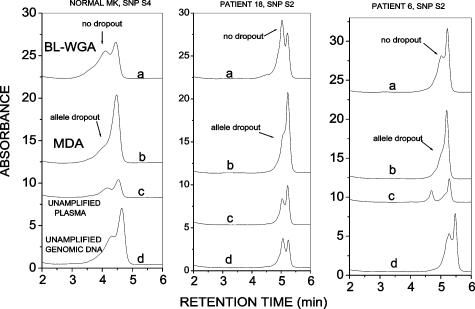
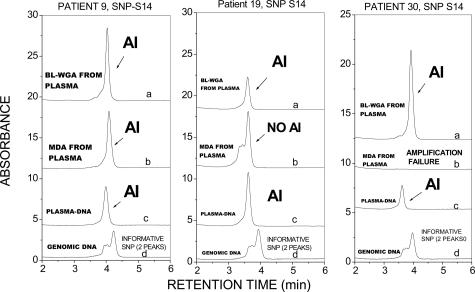
The ability to faithfully amplify both alleles and avoid allele-dropout is one of the most exacting tests for a whole genome amplification method.47 For a thorough evaluation of the fidelity of plasma-circulating DNA amplification, we examined the ability of BL-WGA to reproduce accurately the representation of both alleles, or to correctly diagnose AI in unamplified plasma, for a number of polymorphic (SNP) positions. PCR-dHPLC was used to discriminate between the two alleles in plasma-circulating DNA before and after amplification via BL-WGA or MDA. For each patient sample, matched genomic DNA obtained from lymphocytes was used as a standard against which results from plasma were compared. Twenty-three SNPs were examined in samples obtained from a total of 30 breast cancer patients (stages II to IV) and 16 normal volunteers. Those SNPs that were informative in lymphocyte DNA obtained from the same patients were examined further using unamplified or amplified plasma-circulating DNA. The overall results for the informative SNPs, repeated three independent times, are summarized in Table 2. The data demonstrate that MDA resulted in incorrect calls (allele dropout, amplification failure, or failure to identify AI) in 11 of 25 cases (44%), whereas BL-WGA generated incorrect calls in 1 of 25 cases (5%). Representative PCR-dHPLC chromatograms depicting correct and incorrect allele calls are shown in Figures 5and 6. Data in Figure 5 were obtained from three individuals for SNPs that were informative in the lymphocyte DNA (double peak in curve d). The dual peaks were also present in the unamplified and BL-WGA-amplified plasma-circulating DNA (curve c and a) but converted to single peaks in the MDA-amplified plasma-circulating DNA (curve b) indicating allele dropout. Figure 6 demonstrates results from SNP S14 that demonstrated AI in unamplified plasma-circulating DNA (single peak, curve c) versus the corresponding lymphocyte DNA (double peak, curve d) in three patient samples. Correct detection of AI is seen in BL-WGA-amplified DNA (curve a, single peak) but only in one of three cases in MDA-amplified DNA (curve b).
Table 2.
Evaluation of BL-WGA versus MDA
| A. Evaluation of SNP S14 in 10 patients | |||
|---|---|---|---|
| Patient# | Unamplified | BL-WGA | MDA |
| 5 | R | R | R |
| 6 | R | R | R |
| 9 | AI | AI | AI |
| 13 | R | R | R |
| 18 | R | R | AI |
| 19 | AI | AI | R |
| 22 | R | R | F |
| 23 | R | AI | F |
| 28 | R | R | AI |
| 30 | AI | AI | F |
| B. SNP S1, S7, S11, and S12 for patient 5 | |||
|---|---|---|---|
| SNP | Unamplified | BL-WGA | MDA |
| S1 | R | R | R |
| S7 | R | R | R |
| S11 | R | R | R |
| S12 | R | R | R |
| C. SNP S1, S2, S11, and S12 for patient 6 | |||
|---|---|---|---|
| SNP | Unamplified | BL-WGA | MDA |
| S1 | R | R | R |
| S2 | R | R | AI |
| S11 | R | R | R |
| S12 | R | R | R |
| D. SNP S2, S11 and S12 for patient 18 | |||
|---|---|---|---|
| SNP | Unamplified | BL-WGA | MDA |
| S2 | R | R | AI |
| S11 | R | R | R |
| S12 | R | R | R |
| E. SNP S1, S4, S7 and S12 for normal volunteer M.K. | |||
|---|---|---|---|
| SNP | Unamplified | BL-WGA | MDA |
| S1 | R | R | AI |
| S4 | R | R | AI |
| S7 | AI | AI | AI |
| S12 | R | R | AI |
AI, allelic imbalance; R, Retention; F, persistent PCR failure.
Figure 5.
Confirmation of allelic retention status before and after amplification of plasma-circulating DNA. PCR-dHPLC for the designated polymorphic sites was applied either before (curve c) or after DNA amplification via BL-WGA (curve a) or MDA (curve b) of plasma-circulating DNA. The results are compared with genomic DNA obtained from lymphocytes (curve d). The absence of a second peak in curve b indicates allele dropout artifact by MDA.
Figure 6.
Confirmation of AI status before and after amplification of plasma-circulating DNA. PCR-dHPLC for the designated polymorphic sites was applied either before (curve c) or after DNA amplification of plasma-circulating DNA via BL-WGA (curve a) or MDA (curve b). The results are compared with genomic DNA obtained from lymphocytes (curve d). The loss of a second peak in curve c indicates AI in unamplified plasma-circulating DNA, which is correctly diagnosed by BL-WGA (curve a) but not by MDA (curve b).
In conclusion, although real-time PCR (Figure 4) indicates that MDA may amplify a substantial fraction of plasma-circulating DNA, the data in Table 2 and Figures 5and 6 demonstrate that amplification is often incomplete and results in frequent amplification of only one of two alleles. This is in agreement to published reports.47 On the other hand BL-WGA can reliably (95% accuracy) diagnose the zygosity of the sample after whole genome amplification of plasma-circulating DNA.
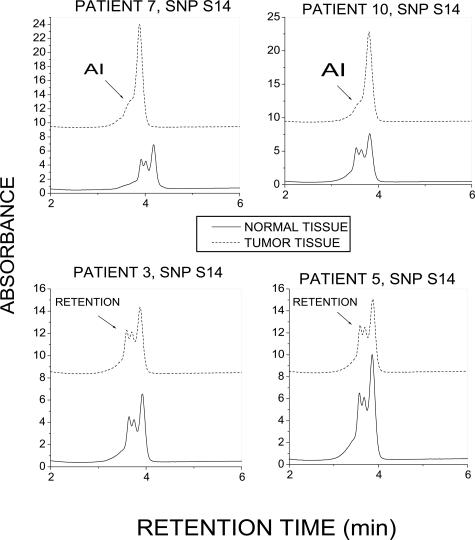
In the process of studying the amplification accuracy of BL-WGA, we identified AI of SNP S14 in the plasma of 3 of 10 informative breast cancer patients (Table 2) but not in the plasma from 7 informative normal volunteers (zero of seven, data not shown). To evaluate if SNP S14 is also encountered in breast tumor tissue, we further screened 20 matched tumor/normal tissue surgical samples donated by patients with infiltrating ductal carcinoma. Nine of twenty samples were informative for SNP S14 and were screened for AI in the corresponding tumor DNA. Two of these nine samples presented AI in tumor tissue for SNP S14, while all matched normal samples demonstrated allelic retention. Figure 7, A and B, depicts chromatograms from the two samples that demonstrated AI for S14 along with two samples that demonstrated allelic retention.
Figure 7.
Identification of AI for SNP S14 in primary breast tumors. PCR-dHPLC indicates allelic loss in tumors from patients 7 and 10 and allelic retention in patients 3 and 5. Matched normal tissue specimens indicated allelic retention for all tissue samples examined.
Discussion
In view of the reported limitations of MDA in amplifying fragmented DNA, such as DNA extracted from formalin-fixed, paraffin-embedded specimens, we recently developed a rolling-circle amplification (RCA)-based whole genome amplification method, RCA-RCA.42 RCA-RCA utilizes a restriction enzyme that digests DNA between successive formalin-induced damage sites and then circularizes the fragment via ligation, followed by rolling circle amplification. RCA-RCA generates a complete genomic amplification in formalin-fixed, paraffin-embedded samples of significant DNA degradation while retaining gene-dosage relations that are present before amplification.42 However, a given DNA fragment must have at least two restriction sites to be amplified via RCA-RCA. Because plasma-circulating apoptotic DNA fragments are, for the most part, smaller than 200 bp,2 it can be expected that a significant fraction of the apoptotic plasma-circulating DNA might not be amplified via RCA-RCA. The blunt-end ligation-mediated amplification described in the present study, BL-WGA, overcomes this problem because no restriction enzyme is required. By obviating the need for restriction sites, BL-WGA achieves efficient amplification of all DNA fragment sizes, including the apoptotic and necrotic DNA fractions that are present in plasma-circulating DNA. Reliable amplification of plasma-circulating DNA was not reported before. BL-WGA efficiently amplified both alleles of the studied genomic loci from plasma-circulating DNA whereas this was not the case for MDA (Figures 4and 5). Quite possibly, samples not amplified by MDA consisted mainly of apoptotic rather than necrotic DNA. By joining these small fragments together, BL-WGA generated DNA template amenable to efficient MDA and downstream analysis in these samples. The present study was aimed at validating reliability of AI detection after whole genome amplification, and therefore a direct comparison with PCR reactions performed from unamplified material was necessary to prove this point. Because the patient-donated plasma was limited (∼1 ml/sample), the study was restricted to examining 23 SNPs. However, it is evident that at least 1000-fold more SNPs could be examined for AI using the BL-WGA product, because the initial ∼5 ng used for whole genome amplification yielded at least 5 μg of DNA. Magnification of the examined target is expected to increase sensitivity in detecting AI or other genetic alterations in plasma-circulating DNA because numerous candidate genomic loci can be examined for alterations.48 After examination of 23 polymorphic sites BL-WGA identified a SNP (db = rs1395548, chromosome 1q23.3) that demonstrates AI in the plasma of 30% (3 of 10) of breast cancer cases examined but not in plasma from normal volunteers. AI in chromosome 1q of primary tumors has been reported49 but was not confirmed in subsequent studies.24 We therefore screened primary breast tumors from a separate set of patients and detected AI for SNP (db = rs1395548) in ∼22% (two of nine) of informative breast cancer cases but not their normal counterparts, suggesting that this chromosomal imbalance can be present in primary tumors as well as in the plasma of breast cancer patients.
Because matched tumor tissue from the patients donating the plasma was not available for a direct comparison, the present study confirms only the technical reliability of whole genome amplification from plasma-circulating DNA, ie, alterations detectable in the unamplified plasma are also present in the BL-WGA-amplified material. Several studies report that in the great majority of cases, plasma AI corresponds to AI in primary tumor.10,12,13,14,16 Therefore, because AI detected in amplified plasma-circulating DNA corresponds faithfully to AI in unamplified plasma, it is likely that this reflects alterations in the primary tumor of the same patients. Further studies are warranted to validate this hypothesis.
In summary, we report on an improved whole genome amplification methodology, BL-WGA, which enables efficient amplification of both apoptotic and necrotic DNA fragments from plasma-circulating DNA and reliable downstream screening for AI or other genetic alterations. The method is simple to perform and enables highly expanded DNA screening that is expected to enhance overall sensitivity in detecting plasma DNA biomarkers. Further, BL-WGA generates adequate amplified material to enable use of microarrays that can examine the entire plasma-circulating genome and aid discovery of cancer fingerprints in plasma or identification of new cancer DNA biomarkers. BL-WGA is expected to have wide application in the comprehensive amplification of apoptotic/necrotic DNA collected from human fluid such as nipple aspirate, saliva, sputum, buccal swabs or dried blood specimens, or potentially in the recovery of suboptimally stored, degraded DNA samples.
Acknowledgments
We thank Mohamet Miri and Frank Haluska, M.D., for assistance in obtaining tissue specimens from the Massachusetts General Hospital Tumor Bank.
Footnotes
Supported in part by the Joint Center for Radiation Therapy Foundation, the Dana-Farber/Harvard Cancer Center (technology award) and SPORE in breast cancer grant, and the National Cancer Institute (grant 1 R21 CA111994-01A1 and R21 CA115439).
References
- Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- Anker P, Mulcahy H, Chen XQ, Stroun M. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev. 1999;18:65–73. doi: 10.1023/a:1006260319913. [DOI] [PubMed] [Google Scholar]
- Mulcahy HE, Lyautey J, Lederrey C, Chen XQ, Lefort F, Vasioukhin V, Anker P, Alstead EM, Farthing MJ, Stroun M. Plasma DNA K-ras mutations in patients with gastrointestinal malignancies. Ann NY Acad Sci. 2000;906:25–28. doi: 10.1111/j.1749-6632.2000.tb06585.x. [DOI] [PubMed] [Google Scholar]
- Wang BG, Huang HY, Chen YC, Bristow RE, Kassauei K, Cheng CC, Roden R, Sokoll LJ, Chan DW, Shih Ie M. Increased plasma DNA integrity in cancer patients. Cancer Res. 2003;63:3966–3968. [PubMed] [Google Scholar]
- Sidransky D. Emerging molecular markers of cancer. Nat Rev Cancer. 2002;2:210–219. doi: 10.1038/nrc755. [DOI] [PubMed] [Google Scholar]
- Chang HW, Lee SM, Goodman SN, Singer G, Cho SK, Sokoll LJ, Montz FJ, Roden R, Zhang Z, Chan DW, Kurman RJ, Shih Ie M. Assessment of plasma DNA levels, allelic imbalance, and CA 125 as diagnostic tests for cancer. J Natl Cancer Inst. 2002;94:1697–1703. doi: 10.1093/jnci/94.22.1697. [DOI] [PubMed] [Google Scholar]
- Chen XQ, Stroun M, Magnenat JL, Nicod LP, Kurt AM, Lyautey J, Lederrey C, Anker P. Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat Med. 1996;2:1033–1035. doi: 10.1038/nm0996-1033. [DOI] [PubMed] [Google Scholar]
- Lo YM, Chan LY, Lo KW, Leung SF, Zhang J, Chan AT, Lee JC, Hjelm NM, Johnson PJ, Huang DP. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 1999;59:1188–1191. [PubMed] [Google Scholar]
- Silva JM, Gonzalez R, Dominguez G, Garcia JM, Espana P, Bonilla F. TP53 gene mutations in plasma DNA of cancer patients. Genes Chromosom Cancer. 1999;24:160–161. [PubMed] [Google Scholar]
- Shao ZM, Wu J, Shen ZZ, Nguyen M. p53 mutation in plasma DNA and its prognostic value in breast cancer patients. Clin Cancer Res. 2001;7:2222–2227. [PubMed] [Google Scholar]
- Silva JM, Dominguez G, Villanueva MJ, Gonzalez R, Garcia JM, Corbacho C, Provencio M, Espana P, Bonilla F. Aberrant DNA methylation of the p16INK4a gene in plasma DNA of breast cancer patients. Br J Cancer. 1999;80:1262–1264. doi: 10.1038/sj.bjc.6690495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JM, Dominguez G, Garcia JM, Gonzalez R, Villanueva MJ, Navarro F, Provencio M, San Martin S, Espana P, Bonilla F. Presence of tumor DNA in plasma of breast cancer patients: clinicopathological correlations. Cancer Res. 1999;59:3251–3256. [PubMed] [Google Scholar]
- Shao ZM, Nguyen M. Tumor-specific DNA in plasma of breast cancer patients. Anticancer Drugs. 2002;13:353–357. doi: 10.1097/00001813-200204000-00003. [DOI] [PubMed] [Google Scholar]
- Kopreski MS, Benko FA, Gocke CD. Circulating RNA as a tumor marker: detection of 5T4 mRNA in breast and lung cancer patient serum. Ann NY Acad Sci. 2001;945:172–178. [PubMed] [Google Scholar]
- Shaw JA, Smith BM, Walsh T, Johnson S, Primrose L, Slade MJ, Walker RA, Coombes RC. Microsatellite alterations in plasma DNA of primary breast cancer patients. Clin Cancer Res. 2000;6:1119–1124. [PubMed] [Google Scholar]
- Hu XC, Wong IH, Chow LW. Tumor-derived aberrant methylation in plasma of invasive ductal breast cancer patients: clinical implications. Oncol Rep. 2003;10:1811–1815. [PubMed] [Google Scholar]
- Kuroi K, Tanaka C, Toi M. Plasma nucleosome levels in node-negative breast cancer patients. Breast Cancer. 1999;6:361–364. doi: 10.1007/BF02966454. [DOI] [PubMed] [Google Scholar]
- Silva JM, Silva J, Sanchez A, Garcia JM, Dominguez G, Provencio M, Sanfrutos L, Jareno E, Colas A, Espana P, Bonilla F. Tumor DNA in plasma at diagnosis of breast cancer patients is a valuable predictor of disease-free survival. Clin Cancer Res. 2002;8:3761–3766. [PubMed] [Google Scholar]
- Silva JM, Garcia JM, Dominguez G, Silva J, Miralles C, Cantos B, Coca S, Provencio M, Espana P, Bonilla F. Persistence of tumor DNA in plasma of breast cancer patients after mastectomy. Ann Surg Oncol. 2002;9:71–76. doi: 10.1245/aso.2002.9.1.71. [DOI] [PubMed] [Google Scholar]
- Wagner PD, Verma M, Srivastava S. Challenges for biomarkers in cancer detection. Ann NY Acad Sci. 2004;1022:9–16. doi: 10.1196/annals.1318.003. [DOI] [PubMed] [Google Scholar]
- Wang ZC, Lin M, Wei L-J, Li C, Miron A, Lodeiro G, Harris L, Ramaswamy S, Tanenbaum DM, Meyerson M, Iglehart JD, Richardson A. Loss of heterozygosity and its correlation with expression profiles in subclasses of invasive breast cancers. Cancer Res. 2004;64:64–71. doi: 10.1158/0008-5472.can-03-2570. [DOI] [PubMed] [Google Scholar]
- Osborne RJ, Hamshere MG. A genome-wide map showing common regions of loss of heterozygosity/allelic imbalance in breast cancer. Cancer Res. 2000;60:3706–3712. [PubMed] [Google Scholar]
- Miller BJ, Wang D, Krahe R, Wright FA. Pooled analysis of loss of heterozygosity in breast cancer: a genome scan provides comparative evidence for multiple tumor suppressors and identifies novel candidate regions. Am J Hum Genet. 2003;73:748–767. doi: 10.1086/378522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002;21:5462–5482. doi: 10.1038/sj.onc.1205606. [DOI] [PubMed] [Google Scholar]
- Szyf M, Pakneshan P, Rabbani SA. DNA methylation and breast cancer. Biochem Pharmacol. 2004;68:1187–1197. doi: 10.1016/j.bcp.2004.04.030. [DOI] [PubMed] [Google Scholar]
- Garcia JM, Silva JM, Dominguez G, Silva J, Bonilla F. Heterogeneous tumor clones as an explanation of discordance between plasma DNA and tumor DNA alterations. Genes Chromosom Cancer. 2001;31:300–301. doi: 10.1002/gcc.1148. [DOI] [PubMed] [Google Scholar]
- Hibi K, Robinson CR, Booker S, Wu L, Hamilton SR, Sidransky D, Jen J. Molecular detection of genetic alterations in the serum of colorectal cancer patients. Cancer Res. 1998;58:1405–1407. [PubMed] [Google Scholar]
- Kolble K, Ullrich OM, Pidde H, Barthel B, Diermann J, Rudolph B, Dietel M, Schlag PM, Scherneck S. Microsatellite alterations in serum DNA of patients with colorectal cancer. Lab Invest. 1999;79:1145–1150. [PubMed] [Google Scholar]
- Sieben NL, ter Haar NT, Cornelisse CJ, Fleuren GJ, Cleton-Jansen AM. PCR artifacts in LOH and MSI analysis of microdissected tumor cells. Hum Pathol. 2000;31:1414–1419. [PubMed] [Google Scholar]
- Coulet F, Blons H, Cabelguenne A, Lecomte T, Lacourreye O, Brasnu D, Beaune P, Zucman J, Laurent-Puig P. Detection of plasma tumor DNA in head and neck squamous cell carcinoma by microsatellite typing and p53 mutation analysis. Cancer Res. 2000;60:707–711. [PubMed] [Google Scholar]
- Silva JM, Bonilla F. Detection of plasma tumor DNA in head and neck squamous cell carcinoma by microsatellite typing and p53 mutation analysis. Cancer Res. 2001;61:8595–8596. [PubMed] [Google Scholar]
- Chen X, Bonnefoi H, Diebold-Berger S, Lyautey J, Lederrey C, Faltin-Traub E, Stroun M, Anker P. Detecting tumor-related alterations in plasma or serum DNA of patients diagnosed with breast cancer. Clin Cancer Res. 1999;5:2297–2303. [PubMed] [Google Scholar]
- Mayall F, Fairweather S, Wilkins R, Chang B, Nicholls R. Microsatellite abnormalities in plasma of patients with breast carcinoma: concordance with the primary tumour. J Clin Pathol. 1999;52:363–366. doi: 10.1136/jcp.52.5.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein CA, Schmidt-Kittler O, Schardt JA, Pantel K, Speicher MR, Riethmuller G. Comparative genomic hybridization, loss of heterozygosity, and DNA sequence analysis of single cells. Proc Natl Acad Sci USA. 1999;96:4494–4499. doi: 10.1073/pnas.96.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrigiorgos GM, Chakrabarti S, Zhang Y, Kaur M, Price BD. A PCR-based amplification method retaining the quantitative difference between two complex genomes. Nat Biotechnol. 2002;20:936–939. doi: 10.1038/nbt724. [DOI] [PubMed] [Google Scholar]
- Wang G, Brennan C, Rook M, Wolfe JL, Leo C, Chin L, Pan H, Liu WH, Price B, Makrigiorgos GM. Balanced-PCR amplification allows unbiased identification of genomic copy changes in minute cell and tissue samples. Nucleic Acids Res. 2004;32:e76. doi: 10.1093/nar/gnh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucito R, Nakimura M, West JA, Han Y, Chin K, Jensen K, McCombie R, Gray JW, Wigler M. Genetic analysis using genomic representations. Proc Natl Acad Sci USA. 1998;95:4487–4492. doi: 10.1073/pnas.95.8.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono S, Faruqi AF, Dean FB, Du Y, Sun Z, Wu X, Du J, Kingsmore SF, Egholm M, Lasken RS. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 2003;13:954–964. doi: 10.1101/gr.816903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, Driscoll M, Song W, Kingsmore SF, Egholm M, Lasken RS. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage JM, Leamon JH, Pejovic T, Hamann S, Lacey M, Dillon D, Segraves R, Vossbrinck B, Gonzalez A, Pinkel D, Albertson DG, Costa J, Lizardi PM. Whole genome analysis of genetic alterations in small DNA samples using hyperbranched strand displacement amplification and array-CGH. Genome Res. 2003;13:294–307. doi: 10.1101/gr.377203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Maher E, Brennan C, Chin L, Leo C, Kaur M, Zhu P, Rook M, Wolfe JL, Makrigiorgos GM. DNA amplification method tolerant to sample degradation. Genome Res. 2004;14:2357–2366. doi: 10.1101/gr.2813404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid C, Stevens J, Livak K, Williams P. Real time quantitative PCR. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; Genome MethodsGenome Research. 1996:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Kleymenova E, Walker CL. Determination of loss of heterozygosity in frozen and paraffin embedded tumors by denaturating high-performance liquid chromatography (DHPLC). J Biochem Biophys Methods. 2001;47:83–90. doi: 10.1016/s0165-022x(00)00154-8. [DOI] [PubMed] [Google Scholar]
- Boettger MB, Sergi C, Meyer P. BRCA1/2 mutation screening and LOH analysis of lung adenocarcinoma tissue in a multiple-cancer patient with a strong family history of breast cancer. J Carcinog. 2003;2:5. doi: 10.1186/1477-3163-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Smith DI, Rechtzigel KJ, Thibodeau SN, James CD. Denaturing high performance liquid chromatography (DHPLC) used in the detection of germline and somatic mutations. Nucleic Acids Res. 1998;26:1396–1400. doi: 10.1093/nar/26.6.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook MS, Delach SM, Deyneko G, Worlock A, Wolfe JL. Whole genome amplification of DNA from laser capture-microdissected tissue for high-throughput single nucleotide polymorphism and short tandem repeat genotyping. Am J Pathol. 2004;164:23–33. doi: 10.1016/S0002-9440(10)63092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorenson GD. A review of studies on the detection of mutated KRAS2 sequences as tumor markers in plasma/serum of patients with gastrointestinal cancer. Ann NY Acad Sci. 2000;906:13–16. doi: 10.1111/j.1749-6632.2000.tb06582.x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Larson PS, Schlechter BL, Zahid N, Finnemore E, de las Morenas A, Blanchard RA, Rosenberg CL. Loss of heterozygosity in serial plasma DNA samples during follow-up of women with breast cancer. Int J Cancer. 2003;106:923–929. doi: 10.1002/ijc.11333. [DOI] [PubMed] [Google Scholar]