Abstract
Large expansions in the SCA2 and SCA7 genes (>100 CAG repeats) have been associated with juvenile and infantile forms of cerebellar ataxias that cannot be detected using standard polymerase chain reaction (PCR). Here, we describe a successful application of the fluorescent short tandem repeat-primed PCR method for accurate identification of these expanded repeats. The test is robust, reliable, and inexpensive and can be used to screen large series of patients, although it cannot give a precise evaluation of the size of the expansion. This test may be of practical value in prenatal diagnoses offered to affected or pre-symptomatic at-risk parents, in which a very large expansion inherited from one of the parents can be missed in the fetus by standard PCR.
Large-repeat expansions in SCA2 and SCA7 genes have been associated with infantile- and juvenile-onset forms of spinocerebellar ataxia (SCA).1 SCA2 neonatal- and infantile-onset cases with common (from 57 to 64 CAG repeats) and extreme triplet expansions (from 230 to 500 CAG repeats) have been reported.2,3,4 However, the clinical manifestations of the latter are unusual and include one or more of the following symptoms: severe hypotonia, developmental delay, dysphagia, encephalopathy, chronic seizures, and retinopathy.1,5 SCA7 CAG repeats from 55 to 460 repeats can give a severe disease with onset in childhood and a rapid fatal course.6,7,8,9,10,11
Routine genetic tests for these genes can miss expansions >80 CAG repeats giving a false-negative result. Assays based on the separation of the polymerase chain reaction (PCR) products on agarose gels, blotting, and hybridization with a (CAG)n oligonucleotide have been applied to overcome this limit.1,12,13
Our aim was to develop a simple, robust, and rapid PCR-based test able to detect large SCA2 and SCA7 CAG expansions. Our approach is based on a fluorescent short tandem repeat (STR)-primed PCR, first described for myotonic dystrophy type 1 by Warner et al14 and recently applied by our group to SCA10, SCA12, and FRDA1 genes.15
Materials and Methods
Genomic DNA and Patients
Genomic DNA was extracted from blood samples using the QIAamp DNA mini blood kit (Qiagen, Hilden, Germany) or a standard phenol-chloroform extraction. From our DNA collection of SCA patients and healthy controls, we selected 34 samples with SCA2 repeats in the normal range (22 to 34 CAG repeats) and 12 SCA2 patients carrying the most common repeats within the pathogenic range (37 to 53 CAG repeats). Two patients with very large SCA2 expansions (>200 CAG repeats) were provided by the Molecular Genetics Laboratory of the Mayo Clinic (Rochester, MN) (Table 1). The approximate expansion size of the latter had been determined by PCR-blot assay.1
Table 1.
Comparison of STR-PCR with Standard PCR and PCR-Southern Blot in the Analysis of SCA2 Patients
| Range of expansion | Genotypes (CAG) | N | Standard PCR | PCR- Southern | STR- PCR |
|---|---|---|---|---|---|
| Normal | 22/22 | 25 | + | n.d. | + |
| 22/23 | 4 | + | n.d. | + | |
| 22/25 | 1 | + | n.d. | + | |
| 22/27 | 1 | + | n.d. | + | |
| 22/29 | 1 | + | n.d. | + | |
| 22/32 | 1 | + | n.d. | +* | |
| 22/34 | 1 | + | n.d. | +* | |
| Common | |||||
| mutations | 22/37 | 1 | + | n.d. | +* |
| 22/38 | 1 | + | n.d. | +* | |
| 22/39 | 2 | + | n.d. | +* | |
| 22/40 | 1 | + | n.d. | +* | |
| 22/41 | 1 | + | n.d. | +* | |
| 22/45 | 1 | + | n.d. | + | |
| 22/47 | 1 | + | n.d. | + | |
| 22/49 | 2 | + | n.d. | + | |
| 22/52 | 1 | ± | n.d. | + | |
| 22/53 | 1 | ± | n.d. | + | |
| Rare large | |||||
| mutations | 23/∼200 | 1 | − | + | + |
| 22/∼350 | 1 | − | + | + |
The fluorescent STR-primed PCR could not correctly classify the profile in the normal or expanded range.
PCR results on the three right columns refer to the larger allele. + and − indicate the ability of a technique to identify an expansion. When ± is present, the expansion was not always detected.
N, number of tested subjects. n.d., not done.
For SCA7 analysis, we selected 39 normal subjects with alleles ranging from 9 to 34 CAG repeats and 20 SCA7 patients carrying a 36- to 54-CAG repeat allele. Eight additional patients carried large uncommon alleles of 58 to 306 CAG repeats (Table 2). All normal and expanded alleles had been confirmed by Southern blot-PCR for SCA7.8,12
Table 2.
Comparison of STR-PCR with Standard PCR and PCR-Southern Blot in the Analysis of SCA7 Patients
| Range of expansion | Genotypes (CAG) | N | Standard PCR | PCR- Southern | STR- PCR |
|---|---|---|---|---|---|
| Normal | 9/9 | 5 | + | n.d. | + |
| 9/10 | 1 | + | n.d. | + | |
| 10/10 | 16 | + | n.d. | + | |
| 10/11 | 3 | + | n.d. | + | |
| 10/12 | 4 | + | n.d. | + | |
| 10/13 | 2 | + | n.d. | + | |
| 10/14 | 1 | + | n.d. | + | |
| 10/15 | 1 | + | n.d. | + | |
| 10/16 | 1 | + | n.d. | + | |
| 11/12 | 1 | + | n.d. | + | |
| 11/14 | 1 | + | n.d. | + | |
| 12/12 | 1 | + | n.d. | + | |
| 13/13 | 1 | + | n.d. | + | |
| 12/34 | 1 | + | n.d. | +* | |
| Common | |||||
| mutations | 10/36 | 1 | + | n.d. | +* |
| 11/39 | 1 | + | n.d. | +* | |
| 10/41 | 1 | + | n.d. | + | |
| 7/42 | 1 | + | n.d. | + | |
| 10/42 | 5 | + | n.d. | + | |
| 12/42 | 1 | + | n.d. | + | |
| 10/43 | 2 | + | n.d. | + | |
| 12/43 | 1 | + | n.d. | + | |
| 10/44 | 1 | + | n.d. | + | |
| 10/47 | 2 | + | n.d. | + | |
| 10/48 | 1 | + | n.d. | + | |
| 10/49 | 1 | + | n.d. | + | |
| 10/54 | 2 | + | + | + | |
| Rare large | |||||
| mutations | 10/58 | 1 | + | + | + |
| 12/61 | 1 | + | + | + | |
| 10/65 | 1 | + | + | + | |
| 13/66 | 1 | + | + | + | |
| 10/77 | 1 | + | + | + | |
| 10/90 | 1 | ± | + | + | |
| 13/106 | 1 | ± | + | + | |
| 10/306 | 1 | − | + | + |
The fluorescent STR-primed PCR could not correctly classify the profile in the normal or expanded range.
PCR results on the three right columns refer to the larger allele. + and − indicate the ability of a technique to identify an expansion. When ± is present, the expansion was not always detected.
N, number of tested subjects. n.d., not done.
Standard PCR and Repeat-Primed PCR
Standard PCR analyses of the SCA2 and SCA7 genes were performed following previous reports.12,16,17The fluorescent STR-primed PCR was based on a previously described method14 that we successfully applied to analyze the FRDA1, SCA10, and SCA12 genes.15 Briefly, a fluorescent-labeled primer was designed in a locus-specific region upstream of the unstable repeat of interest. The companion reverse primer (on the complementary strand) consisted of five CTG units and a 5′ tail that was used as an anchor for a second reverse primer, which prevents progressive shortening of the PCR products during subsequent cycles.
PCR reactions were performed on 200 to 1000 ng of genomic DNA with 144 μmol/L dNTPs, 1.3 to1.5 mmol/L MgCl2 (for the SCA7 and SCA2 genes, respectively), 1 mol/L betaine (B0300; Sigma-Aldrich, St. Louis, MO), 1 U of TaqGold in 1× TaqGold buffer (Applied Biosystems, Foster City, CA) in the presence of 0.8 μmol/L of the locus-specific primer (SCA2F, 5′-HEX-ggg ccc ctc acc atg tcg, or SCA7F, 5′-HEX-gcg gtc cca aaa ggg tca gtt gtt aca ttg tag gag cgg aa), 0.08 μmol/L of the repeat-specific oligonucleotide (R1, 5′-tac gca tcc cag ttt gag acg ctg ctg ctg ctg ctg) and 0.8 μmol/L of the “common” flag primer“ (R2, 5′-tac gca tcc cag ttt gag acg).
PCR cycling parameters were as follows. SCA2: initial denaturation of 7 minutes at 95°C, 14 cycles consisting of 30 seconds at 95°C, 30 seconds at 63°C −0.5°C/cycle, 1 minute at 72°C, 35 cycles consisting of 30 seconds at 95°C, 30 seconds at 56°C, 1 minute at 72°C + 10 seconds/cycle, and 10 minutes of final extension at 72°C; SCA7: initial denaturation of 7 minutes at 95°C, 14 cycles consisting of 1 minute at 95°C, 1 minute at 63°C −0.5°C/cycle, 1 minute at 72°C, 35 cycles consisting of 1 minute at 95°C, 1 minute at 56°C, 1 minute at 72°C + 20 seconds/cycle, and 10 minutes of final extension at 72°C.
Analysis of Fluorescent PCR Fragments and Interpretation of Data
Analysis of the fluorescent PCR products was performed using an ABI-Prism 3100 Avant automatic sequencer on a 36-cm capillary array with the POP4 polymer and a ROX-GS500 internal standard marker (Applied Biosystems). Data were examined using the Genescan 3.1 software. Three independent examiners blindly evaluated STR-primed PCR profiles.
Results and Discussion
Fluorescent STR-primed PCR is a robust technique used to detect large-repeat expansions that cannot be amplified using standard PCR.14,15,18,19 Here, we present an application of this technique to SCA2 and SCA7 genes. Their pathogenic CAG expansions are mostly within the PCR detection range, but large expansions have been described in both genes and need Southern-based approaches to be detected.1
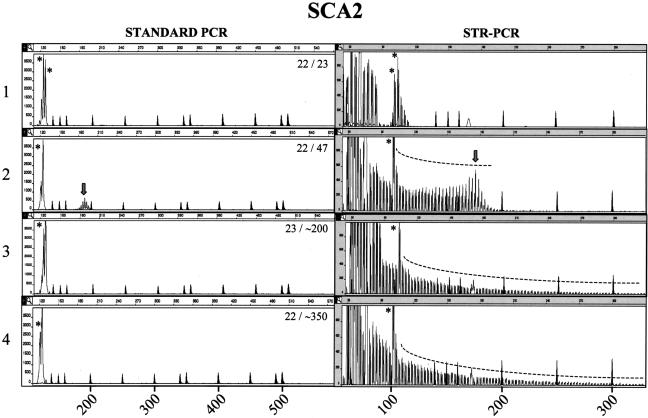
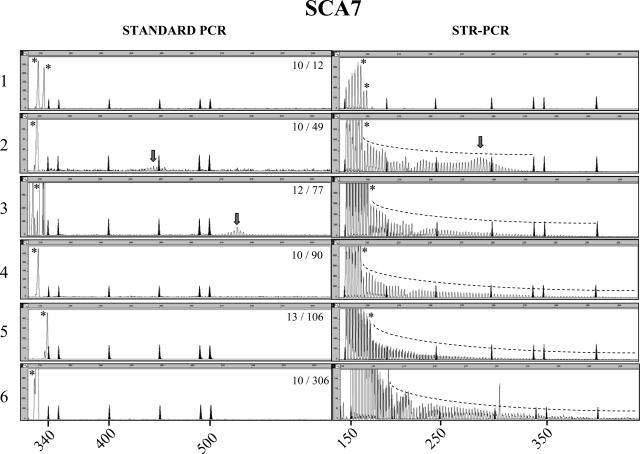
Fluorescent STR-primed PCR showed a short series of discrete peaks with 3-bp periodicity when subjects within the normal range were tested; a few peaks, probably generated by the alternative annealing sites of the reverse primer, always preceded the main allele (Figures 1and 2, panel 1). To evaluate the consistency of STR-primed PCR profiles on expanded alleles, we tested 12 SCA2 and 20 SCA7 patients within the most common mutation ranges of 37 to 53 and 36 to 54 CAG repeats, respectively (Tables 1and 2). Both SCA2 and SCA7 showed a proportionally larger array of peaks compared with controls. For the smallest expansions, the profile assumed a bell shape on the right end of the panel (Figures 1, panel 2, and 2, panel 2), with the highest peak in the bell-shaped array corresponding to the size of the larger allele with a maximum error of ±2 repeats (Figures 1, panel 2 [right], and 2, panel 2 [right]). Large expansions in SCA2 (two patients; ∼200 and ∼350 CAG repeats) and SCA7 (eight patients; range, 58 to 306 CAG repeats) yielded clearly distinguishable profiles, with multiple peaks of progressively lower intensity down to a fluorescence close to zero (Figures 1, panel 3, and 2, panels 3–6). It is notable that SCA7 expansions above 90 repeats were not constantly amplified in our laboratories and that SCA2 mutations >200 CAG repeats and SCA7 mutations >106 were never detected by standard PCR (Figures 1, panels 2 and 3 [left], and 2, panels 4–6 [left]).
Figure 1.
Comparison between standard and STR-primed PCR analysis in the SCA2 locus. Left and right panels represent the analysis of SCA2 CAG repeat in the same subject using standard PCR and STR-primed PCR, respectively. The STR-primed PCR peak profile of a healthy control with two alleles in the normal size range (panel 1) is distinct from the profiles found in a subject carrying a pathogenic allele (panel 2). In the case of an extreme expansion, standard PCR is unable to detect the pathogenic allele, whereas the STR-primed PCR profile clearly shows a gradually descending array of peaks, revealing the presence of a large CAG tract (panels 3 and 4). The genotype of SCA2 CAG triplets is shown in the left panels. Marker peaks are shadowed. On the left panels, their sizes are: 139, 150, 160, 200, 250, 300, 340, 350, 400, 450, 490, and 500 bp. Marker sizes on the right panels are: 75, 100, 139, 150, 160, 200, 250, and 300 bp. Size in base pairs is also reported on the abscissa, whereas the ordinate shows fluorescence intensity as arbitrary unit. An asterisk indicates the peaks corresponding to normal alleles. The arrow points to the central peak of small expansions. SCA2 profiles of normal alleles typically contained one or two major peaks of 101/104 bp corresponding to the most frequent 22- and 23-CAG alleles.
Figure 2.
Comparison between standard PCR and STR-primed PCR analysis in the SCA7 locus. Left and right panels represent the analysis of SCA7 CAG repeat in the same subject using standard PCR and STR-primed PCR, respectively. The genotype of the SCA7 CAG triplets is shown in the left panels. The STR-primed PCR peak profile of a healthy control with two alleles in the normal size range (panel 1) is distinct from the profiles in a subject carrying pathogenic 49 and 77 CAG repeats (panels 2 and 3). In patients carrying >80 CAG repeats, standard PCR often failed to amplify the repeat, whereas STR-primed PCR always detected the pathogenic allele (panels 4–6). Marker peaks are shadowed. Their sizes on the left panels are 340, 350, 400, 450, 490, and 500 bp. Marker sizes on the right panels are 160, 200, 250, 300, 340, 350, and 400 bp. Size in base pairs is also indicated on the abscissa, whereas ordinate shows fluorescence intensity in arbitrary units. An asterisk indicates the peaks corresponding to the normal alleles. The arrow points to the central peak of visible expansions.
These results suggest that fluorescent STR-PCR is reliable for large CAG expansions in the SCA2 and SCA7 genes, although their detection is proved only up to 350 triplets in SCA2 and up to 306 triplets in SCA7. However, it must be considered that this technique was used to detect CAG expansions up to 5 kb in the myotonic dystrophy type 1 gene,14 >1400 GAA in the FRDA1 gene, and >3500 ATTCT in the SCA10 gene.15 Also, considering its rationale, which does not rely on the amplification of the repeat region, we hypothesize that the upper detection limit is virtually absent.
The golden choice for large-repeat expansion detection has been the genomic- or PCR-Southern blot. We believe that STR-PCR has several advantages on these techniques: 1) it uses a small amount of DNA versus standard Southern blot (100 ng versus 10 μg); 2) it is easy to set up, quick to complete (1 to 2 versus 7 to 15 days), and does not need the use of radiochemicals; 3) it is easier to use on screening; 4) in our experience, PCR-Southern blot can give false-positive results on DNA samples extracted from chorionic villi, because of minor maternal contamination and its high sensitivity. On the other hand, STR-PCR requires automated genotyping equipment, which is not available in every laboratory.
A misinterpretation of the expanded profile is possible because of technical reasons: 1) the presence of a very low fluorescent signal (<100 fluorescence units) that, however, must induce to repeat the test trying different DNA dilutions; 2) the presence of a possible somatic mosaicism in which two cell populations co-exist, one with two normal alleles and one heterozygous for an expansion; this event is, however, probably extremely rare.
The lower detection limit of fluorescent STR-PCR was estimated in controls and patients with small CAG expansions. The profiles of normal SCA2 subjects with 32 to 34 CAG repeats could not be precisely distinguished from those of affected patients with small expansions of 37 to 41 repeats (data not shown). In SCA7, a control subject carrying 34 CAG repeats was not distinguished from patients with 36 to 39 repeats (data not shown). A gray zone in which subjects could not clearly be classified as carriers of a mutation exists approximately between 34 and 41 repeats. However, above this range, patients were clearly assigned to the mutated range.
Further data may prove that STR-primed PCR can be routinely used to detect expansions. At present, we propose that routine PCR should precede STR-PCR, which can be applied in case of apparent homozygosity. At this regard, it is noteworthy that, in our experience, common expansions >50 repeats might not be detected by standard PCR. Technical artifacts might affect the results obtained by standard PCR (eg, the phenol used in DNA extractions), disfavoring the amplification of pathological alleles. In such cases, a normal homozygous genotype can be wrongly attributed (Figure 2, panel 2). This was not the case for either gene using the fluorescent STR-PCR technique, providing an alternative method, or a second confirmatory technique and is particularly important in SCA patients with infantile or juvenile onset and an apparently homozygous normal allele when tested with standard techniques.
We also suggest using fluorescent STR-PCR in case a single normal allele is detected in prenatal diagnosis of affected or pre-symptomatic at-risk parents, because in such a case, a very large expansion inherited from one of the parents can be missed in the fetus by standard PCR. Finally, it is also possible to apply our test to routine screening for infantile- or juvenile-onset diseases that suggest the involvement of large expansions in these genes.
Acknowledgments
We thank the Molecular Genetics Laboratory at the Mayo Clinic (Rochester, MN) (Dr. C.A. Kremer), for providing SCA2 samples with very large expansions and Professor Huda Y. Zoghbi (Howard Hughes Medical Institute, Baylor College of Medicine, Houston, TX) for providing the 306-CAG SCA7 patient.
Footnotes
Supported by the Associazione Emma & Ernesto Rulfo per la Genetica Medica, Ministero dell’Università e della Ricerca Scientifica e Tecnologica 60%, Regione Piemonte, Ricerca Sanitaria Finalizzata 2004 (to A. Brusco), the VerUm Foundation (to A. Brice), and the European Community (to the European Integrated Project on Spinocerebellar Ataxias consortium).
References
- Mao R, Aylsworth AS, Potter N, Wilson WG, Breningstall G, Wick MJ, Babovic-Vuksanovic D, Nance M, Patterson MC, Gomez CM, Snow K. Childhood-onset ataxia: testing for large CAG-repeats in SCA2 and SCA7. Am J Med Genet. 2002;110:338–345. doi: 10.1002/ajmg.10467. [DOI] [PubMed] [Google Scholar]
- Cancel G, Durr A, Didierjean O, Imbert G, Burk K, Lezin A, Belal S, Benomar A, Abada-Bendib M, Vial C, Guimaraes J, Chneiweiss H, Stevanin G, Yvert G, Abbas N, Saudou F, Lebre AS, Yahyaoui M, Hentati F, Vernant JC, Klockgether T, Mandel JL, Agid Y, Brice A. Molecular and clinical correlations in spinocerebellar ataxia 2: a study of 32 families. Hum Mol Genet. 1997;6:709–715. doi: 10.1093/hmg/6.5.709. [DOI] [PubMed] [Google Scholar]
- Riess O, Epplen JT, Amoiridis G, Przuntek H, Schols L. Transmission distortion of the mutant alleles in spinocerebellar ataxia. Hum Genet. 1997;99:282–284. doi: 10.1007/s004390050355. [DOI] [PubMed] [Google Scholar]
- Moretti P, Blazo M, Garcia L, Armstrong D, Lewis RA, Roa B, Scaglia F. Spinocerebellar ataxia type 2 (SCA2) presenting with ophthalmoplegia and developmental delay in infancy. Am J Med Genet A. 2004;124:392–396. doi: 10.1002/ajmg.a.20428. [DOI] [PubMed] [Google Scholar]
- Babovic-Vuksanovic D, Snow K, Patterson MC, Michels VV. Spinocerebellar ataxia type 2 (SCA 2) in an infant with extreme CAG repeat expansion. Am J Med Genet. 1998;79:383–387. [PubMed] [Google Scholar]
- Martin J, Van Regemorter N, Del-Favero J, Lofgren A, Van Broeckhoven C. Spinocerebellar ataxia type 7 (SCA7): correlations between phenotype and genotype in one large Belgian family. J Neurol Sci. 1999;168:37–46. doi: 10.1016/s0022-510x(99)00176-8. [DOI] [PubMed] [Google Scholar]
- David G, Durr A, Stevanin G, Cancel G, Abbas N, Benomar A, Belal S, Lebre AS, Abada-Bendib M, Grid D, Holmberg M, Yahyaoui M, Hentati F, Chkili T, Agid Y, Brice A. Molecular and clinical correlations in autosomal dominant cerebellar ataxia with progressive macular dystrophy (SCA7). Hum Mol Genet. 1998;7:165–170. doi: 10.1093/hmg/7.2.165. [DOI] [PubMed] [Google Scholar]
- Benton CS, de Silva R, Rutledge SL, Bohlega S, Ashizawa T, Zoghbi HY. Molecular and clinical studies in SCA-7 define a broad clinical spectrum and the infantile phenotype. Neurology. 1998;51:1081–1086. doi: 10.1212/wnl.51.4.1081. [DOI] [PubMed] [Google Scholar]
- van de Warrenburg BP, Frenken CW, Ausems MG, Kleefstra T, Sinke RJ, Knoers NV, Kremer HP. Striking anticipation in spinocerebellar ataxia type 7: the infantile phenotype. J Neurol. 2001;248:911–914. doi: 10.1007/s004150170082. [DOI] [PubMed] [Google Scholar]
- Ansorge O, Giunti P, Michalik A, Van Broeckhoven C, Harding B, Wood N, Scaravilli F. Ataxin-7 aggregation and ubiquitination in infantile SCA7 with 180 CAG repeats. Ann Neurol. 2004;56:448–452. doi: 10.1002/ana.20230. [DOI] [PubMed] [Google Scholar]
- Johansson J, Forsgren L, Sandgren O, Brice A, Holmgren G, Holmberg M. Expanded CAG repeats in Swedish spinocerebellar ataxia type 7 (SCA7) patients: effect of CAG repeat length on the clinical manifestation. Hum Mol Genet. 1998;7:171–176. doi: 10.1093/hmg/7.2.171. [DOI] [PubMed] [Google Scholar]
- Stevanin G, Giunti P, Belal GD, Durr A, Ruberg M, Wood N, Brice A. De novo expansion of intermediate alleles in spinocerebellar ataxia 7. Hum Mol Genet. 1998;7:1809–1813. doi: 10.1093/hmg/7.11.1809. [DOI] [PubMed] [Google Scholar]
- Snow K, Mao R. Extreme expansion detection in spinocerebellar ataxia type 2 and type 7. Methods Mol Biol. 2003;217:41–50. doi: 10.1385/1-59259-330-5:41. [DOI] [PubMed] [Google Scholar]
- Warner JP, Barron LH, Goudie D, Kelly K, Dow D, Fitzpatrick DR, Brock DJ. A general method for the detection of large CAG repeat expansions by fluorescent PCR. J Med Genet. 1996;33:1022–1026. doi: 10.1136/jmg.33.12.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnoli C, Michielotto C, Matsuura T, Ashizawa T, Margolis RL, Holmes SE, Gellera C, Migone N, Brusco A. Detection of large pathogenic expansions in FRDA1, SCA10, and SCA12 genes using a simple fluorescent repeat-primed PCR assay. J Mol Diagn. 2004;6:96–100. doi: 10.1016/S1525-1578(10)60496-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanpei K, Takano H, Igarashi S, Sato T, Oyake M, Sasaki H, Wakisaka A, Tashiro K, Ishida Y, Ikeuchi T, Koide R, Saito M, Sato A, Tanaka T, Hanyu S, Takiyama Y, Nishizawa M, Shimizu N, Nomura Y, Segawa M, Iwabuchi K, Eguchi I, Tanaka H, Takahashi H, Tsuji S. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat Genet. 1996;14:277–284. doi: 10.1038/ng1196-277. [DOI] [PubMed] [Google Scholar]
- David G, Abbas N, Stevanin G, Durr A, Yvert G, Cancel G, Weber C, Imbert G, Saudou F, Antoniou E, Drabkin H, Gemmill R, Giunti P, Benomar A, Wood N, Ruberg M, Agid Y, Mandel JL, Brice A. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nat Genet. 1997;17:65–70. doi: 10.1038/ng0997-65. [DOI] [PubMed] [Google Scholar]
- Day JW, Ricker K, Jacobsen JF, Rasmussen LJ, Dick KA, Kress W, Schneider C, Koch MC, Beilman GJ, Harrison AR, Dalton JC, Ranum LP. Myotonic dystrophy type 2: molecular, diagnostic and clinical spectrum. Neurology. 2003;60:657–664. doi: 10.1212/01.wnl.0000054481.84978.f9. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Ashizawa T. Polymerase chain reaction amplification of expanded ATTCT repeat in spinocerebellar ataxia type 10. Ann Neurol. 2002;51:271–272. doi: 10.1002/ana.10049. [DOI] [PubMed] [Google Scholar]