Abstract
A 28-year-old Caucasian female with no personal or family history of cystic fibrosis (CF) presented for preconception counseling and screening. Cystic fibrosis transmembrane conductance regulator (CFTR) mutation analysis using the Inno-LiPa CFTR assay revealed lack of hybridization for both the wild-type and mutant oligonucleotides for 3120+1G>A. This region was sequenced, and an apparent homozygous 3120G>A mutation was detected. Additional testing revealed an abnormal sweat chloride (77 mmol/L). Review of systems was essentially unremarkable with an absence of sinus symptoms, occasional nonproductive cough, and no features of malabsorption. Physical examination, chest X-ray, and pulmonary function tests were within normal limits. Only two other patients (siblings) with homozygous 3120G>A mutations have been reported (http://www.genet.sickkids.on.ca/cftr/). Both siblings had pancreatic insufficiency, mild pulmonary symptoms, and abnormal sweat chloride levels. Our findings suggest that a homozygous mutation of a G>A conversion at 3120 is associated with abnormal CFTR function and either a mild form of CF or no overt symptoms of disease, emphasizing the difficulties in assigning genotype/phenotype correlation.
Cystic fibrosis (CF) is one of the most common autosomal recessive genetic disorders among the Caucasian population with a carrier rate of 1 in 29 and a disease frequency of 1 in 3300.1,2,3 CF is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene located on chromosome 7q, resulting in a chloride ion channel defect.4,5,6,7 This defect affects mucus-producing organs including the lungs, pancreas, sweat glands, and reproductive organs.
In 1997, the National Institutes of Health recommended that the United States begin implementing preconception or prenatal carrier screening for CF. Following that recommendation, the American College of Medical Genetics and the American College of Obstetrics and Gynecology published guidelines for carrier CF screening including testing a core panel of the 25 most common mutations among the Caucasian population2,8,9 that has been recently changed to 23 mutations.10 The guidelines recommended that CF mutation screening should be offered to individuals with a family history of CF, reproductive partners of individuals with CF, couples planning a pregnancy or seeking prenatal care when one or both partners are Caucasian or Ashkenazi Jewish, and individuals from other ethnic groups who chose to be screened. The goal of screening for CF gene mutation carriers was to provide appropriate genetic counseling for family planning.
An issue with population screening is the identification of two apparent disease-causing mutations in individuals without a diagnosis of CF. A recent study of 335,204 patients screened for carrier status revealed four individuals with two CF mutations who had not been previously diagnosed with CF.11 This study underscored the lack of definitive genotype/phenotype correlations and discussed the counseling dilemmas associated with such findings. We report a patient who was referred for carrier screening and was found to have two rare mutations but exhibited no overt clinical features of CF.
Patient History
A 28-year-old Caucasian female presented to her gynecologist for preconception counseling. Although there was no family history of CF, the patient chose to have mutation screening. She had a history of bronchitis and chronic cough, which was attributed to smoking, but was an otherwise healthy young adult.
Molecular Studies
Inno-LiPa CFTR33 Assay
The Inno-LiPa CFTR33 Assay (Innogenetics NV, Brussels, Belgium) was used to screen for CF mutations. In brief, the peripheral blood DNA sample underwent multiplex amplification of the CFTR gene using biotinylated primers. Following amplification, the double-stranded DNA was chemically denatured and hybridized to oligonucleotide probes for the wild-type and mutant CF sequences bound to nitrocellulose strips. After hybridization, streptavidin labeled with alkaline phosphatase was applied and bound to biotinylated amplicon-probe hybrids. Incubation with a substrate (chromogen) solution resulted in formation of a purple precipitate. The reaction was stopped by washing, and the reactivity pattern of the probes was analyzed.
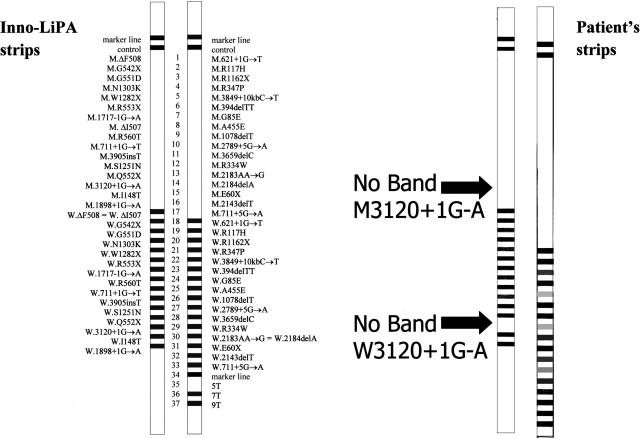
Upon analysis of the patient’s results, absence of both the wild-type and mutant bands for the 3120+1G>A CF mutation was noted (Figure 1). The result was confirmed by performing a second Inno-LiPa test on the patient’s original sample.
Figure 1.
The Inno-LiPa CFTR33 assay results. An example of the assay’s strips is shown on the right. The patient’s results (left) show the absence of both the wild-type and mutant bands (arrows) for 3120+1 G>A.
Sequencing the Region of Interest
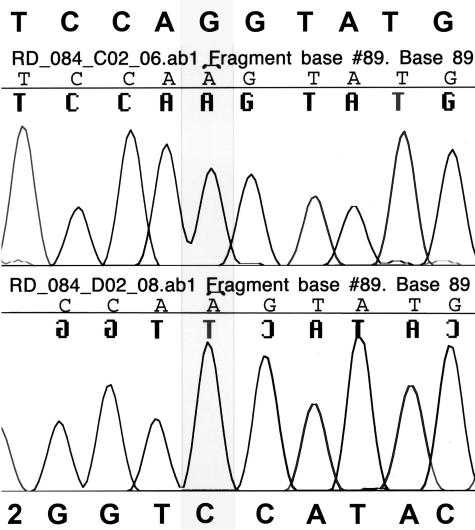
To determine the cause of the unusual result from the CFTR33 mutation assay, sequencing of the region (exon 16) was performed. PCR products were generated using primers that flank exon 16 (including the exonic/intronic boundaries) of the CFTR gene. Products were sequenced bi-directionally using BigDye Terminator sequencing chemistry version 1.0 (Applied Biosystems, Foster City, CA). Unincorporated dye-labeled dideoxy-nucleotides were removed using Sephadex G50 fine (Sigma, St. Louis, MO). Sequencing reactions were electrophoresed using standard sequencing protocols on an AB 3100 capillary genetic analyzer. Sequences were aligned to the CFTR reference sequence. The sequence analysis revealed an apparent homozygous G to A mutation at position 3120 (Figure 2), explaining the Inno-LiPa results. The 3120 mutation apparently interfered with the binding of the PCR product to the wild-type and mutant 3120+1G>A targets on the Inno-LiPa strips (Figure 1).
Figure 2.
Patient’s DNA sequence analysis of the region of interest (exon 16). The electropherogram shows the patient’s results (top-forward sequence; bottom-reverse sequence) and indicates a base change of a G (highlighted box) to an A at the 3120 position.
Patient Clinical Follow-Up
Following the results of our molecular studies, the patient was referred to a pulmonologist. On examination, her height was 5 feet, 4 inches, and her weight was 111 lbs. Chest X-ray and pulmonary function tests were normal. She exhibited no sinus symptoms and no clinical features of malabsorption. She was noted to have an elevated sweat chloride (77 mmol/L) and a history of an occasional nonproductive cough. She declined additional diagnostic evaluation to assess pancreatic function and the presence of bronchiectasis. Attempts to obtain additional information and parental samples for carrier screening studies and/or uniparental disomy studies were declined. Parental consanguinity was denied.
Discussion
The presentation and course of CF is highly variable. Although most cases of CF are diagnosed before the age of 1, some cases do not come to medical attention until adolescence or adulthood.9 The presentation of CF cases can vary from severe pulmonary and/or gastrointestinal symptoms to mild disease9 or, as in this case, no overt symptoms of disease. The currently accepted criteria for the diagnosis of CF include evidence of a CFTR abnormality plus one of the following: one or more characteristic phenotypic features, a history of CF in a sibling, or a positive screening test.12,13 Our patient’s assessment met the criterion based on her abnormal sweat chloride and having two CF mutations. However, if she had not presented for carrier screening, she would not have obtained a diagnosis.
3120G>A is considered a rare CF mutation, having only been reported in three other patients, including two siblings who were homozygous for 3120G>A mutation with pancreatic insufficiency, mild pulmonary symptoms, and abnormal sweat chlorides (>80 mmol/L) and one patient with a severe phenotype carrying the delF508 mutation on the other allele (Cystic Fibrosis Mutation Database, http://www.genet.sickkids.on.ca/cftr/; Hospital for Sick Children, Toronto, Canada). Given the rarity of the 3120G>A mutation, it is unlikely that both of this patient’s parents would be carriers. There are at least two possible mechanisms that could explain the results. One possible mechanism for the apparent homozygosity for this rare CFTR mutation is uniparental disomy for chromosome 7 (UPD7). Several cases of UPD7 and homozygosity of a CFTR mutation have been reported.14,15,16 In addition to having the classical symptoms associated with CF, patients with UPD7 also displayed growth retardation and short stature (∼80% of the cases). Our patient was reported by the referring physician to have a small body frame with a disproportionately large head. These observations along with the homozygous 3120G>A CFTR mutation suggest that she may in fact have UPD7.
A second possible mechanism is that our patient is hemizgyous because of a large deletion of the CFTR gene on the other chromosome that spans the same region as the 3120G>A mutation. Even though few have been characterized, large germline deletions of the CFTR gene have been suggested to account for 1 to 30% of the disease alleles that have not yet been identified.17 Although this mechanism could explain our findings, it seems to be an even more likely explanation for the more severe phenotype observed in the two siblings previously reported. Their phenotypes could be due to the inheritance of a large deletion from one parent, along with inheritance of the 3120G>A mutation from the other parent.
The presence of a modifier affecting the 3120G>A CFTR mutation may also explain the different phenotypes seen in this patient and the other reported homozygotes. Similar to I148T influencing 3199del6 in cis18,19 or R117H being influenced by the 5T variant,20 there could be an additional mutation on the same chromosome causing different phenotypes for those who have the same apparent genotype.
Technical anomalies leading to the incorrect interpretation of the patient’s results cannot be excluded. Although it is feasible to have a separate mutation that interferes with the priming from one allele, this is unlikely because of agreement of the sequencing and Inno-LiPa results along with the fact that the two methods have different primer binding sites (C. Van Loon, Innogenetics NV; A. Alexander, Innogenetics Inc., personal communication).
This case demonstrates that counseling patients who meet laboratory criteria for a CF diagnosis but do not have clinical symptoms of CF is problematic. Given the large numbers of individuals who are undergoing screening for CFTR mutations because of the recommendations made by the American College of Medical Genetics and the American College of Obstetrics and Gynecology, it is likely that other such individuals will be ascertained. The rare mutations detected in these nonsymptomatic patients may not represent independent disease-causing mutations, and care is advised in offering phenotypic predictions.
Acknowledgments
We thank Dr. Denise Quigley for critical reading of this manuscript.
References
- American College of Obstetrics and Gynecology and American College of Medical Genetics Washington, DC: American College of Obstetrics and Gynecology; Preconception and Prenatal Carrier Screening for Cystic Fibrosis, Clinical and Laboratory Guidelines. 2001 [Google Scholar]
- Grody WW, Cutting GR, Klinger KW, Richard CS, Watson MS, Desnick RJ. Laboratory standards and guidelines for population-based cystic fibrosis carrier screening. Genet Med. 2001;3:149–154. doi: 10.1097/00125817-200103000-00010. [DOI] [PubMed] [Google Scholar]
- Palomaki GE, Haddow JE, Bradley LA, FitzSimmons SC. Updated assessment of cystic fibrosis mutation frequencies in non-Hispanic Caucasians. Genet Med. 2002;4:90–94. doi: 10.1097/00125817-200203000-00007. [DOI] [PubMed] [Google Scholar]
- Rommens JM, Iannuzzi MC, Kerem B-S, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- Riordan JR, Rommens JM, Kerem B-S, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Kerem B-S, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- Tsui LC. The cystic fibrosis transmembrane regulator gene. Am J Respir Crit Care Med. 1995;151:S47–S53. doi: 10.1164/ajrccm/151.3_Pt_2.S47. [DOI] [PubMed] [Google Scholar]
- Genetic testing for cystic fibrosis. NIH Consens Statement. 1997;15:1–37. [PubMed] [Google Scholar]
- National Institutes of Health Consensus Development Conference Statement. Genetic testing for cystic fibrosis. Arch Intern Med. 1999;159:1529–1539. [PubMed] [Google Scholar]
- Watson MS, Cutting GR, Desnick RJ, Driscoll DA, Klinger K, Mennuti M, Palomaki GE, Popovich BW, Pratt VM, Rohlfs EM, Strom CM, Richards CS, Witt DR, Grody WW. Cystic fibrosis population carrier screening: 2004 revision of American College of Medical Genetics mutation panel. Genet Med. 2004;6:387–391. doi: 10.1097/01.GIM.0000139506.11694.7C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom CM, Crossley B, Redman JB, Buller A, Quan F, Peng M, McGinnis M, Sun W. Cystic fibrosis screening: lessons learned from the first 320,000 patients. Genet Med. 2004;6:136–140. doi: 10.1097/01.gim.0000127268.65149.69. [DOI] [PubMed] [Google Scholar]
- Rosenstein BJ, Cutting GR. The diagnosis of cystic fibrosis: a consensus statement. Cystic Fibrosis Foundation Consensus Panel. J Pediatr. 1998;132:589–596. doi: 10.1016/s0022-3476(98)70344-0. [DOI] [PubMed] [Google Scholar]
- Lotem Y, Barak A, Mussaffi H, Shohat M, Wilschanski M, Sivan Y, Blau H. Reaching the diagnosis of cystic fibrosis: the limits of the spectrum. Isr Med Assoc J. 2000;2:94–98. [PubMed] [Google Scholar]
- Spence JE, Perciaccante RG, Greig GM, Willard HF, Ledbetter DH, Hejtmancik JF, Pollack MS, O’Brien WE, Beaudet AL. Uniparental disomy as a mechanism for human genetic disease. Am J Hum Genet. 1988;42:217–226. [PMC free article] [PubMed] [Google Scholar]
- Voss R, Ben-Simon E, Avital A, Godfrey S, Zlotogora J, Dagan J, Tikochinski Y, Hillel J. Isodisomy of chromosome 7 in a patient with cystic fibrosis: could uniparental disomy be common in humans? Am J Hum Genet. 1989;45:373–380. [PMC free article] [PubMed] [Google Scholar]
- Spotila LD, Sereda L, Prockop DJ. Partial isodisomy for maternal chromosome 7 and short stature in an individual with a mutation at the COL1A2 locus. Am J Hum Genet. 1992;51:1396–1405. [PMC free article] [PubMed] [Google Scholar]
- Audrezet MP, Chen JM, Raguenes O, Chuzhanova N, Giteau K, Le Marechal C, Quere I, Cooper DN, Ferec C. Genomic rearrangements in the CFTR gene: extensive allelic heterogeneity and diverse mutational mechanisms. Hum Mutat. 2004;23:343–357. doi: 10.1002/humu.20009. [DOI] [PubMed] [Google Scholar]
- Strom CM, Huang D, Buller A, Redman J, Crossley B, Anderson B, Entwistle T, Sun W. Cystic fibrosis screening using the college panel: platform comparison and lessons learned from the first 20,000 samples. Genet Med. 2002;4:289–296. doi: 10.1097/00125817-200207000-00007. [DOI] [PubMed] [Google Scholar]
- Rohlfs EM, Zhou Z, Sugarman EA, Heim RA, Pace RG, Knowles MR, Silverman LM, Allitto BA. The I148T allele occurs on multiple haplotypes: a complex allele is associated with cystic fibrosis. Genet Med. 2002;4:319–323. doi: 10.1097/00125817-200209000-00001. [DOI] [PubMed] [Google Scholar]
- Kiesewetter S, Macek M, Jr, Davis C, Curristin SM, Chu CS, Graham C, Shrimpton AE, Cashman SM, Tsui LC, Mickle J. A mutation in CFTR produces different phenotypes depending on chromosomal background. Nat Genet. 1993;5:274–278. doi: 10.1038/ng1193-274. [DOI] [PubMed] [Google Scholar]