Abstract
A disposable 0.2-ml polymerase chain reaction (PCR) tube modified with an aluminum oxide membrane (AOM) has been developed for the extraction, amplification, and detection of nucleic acids. To assess the dynamic range of AOM tubes for real-time PCR, quantified herpes simplex virus (HSV) DNA was used to compare AOM tubes to standard PCR tubes. AOM PCR tubes used for amplification and detection of quantified HSV-1 displayed a crossing threshold (CT) shift 0.1 cycles greater than PCR tube controls. Experiments with HSV-1-positive cerebrospinal fluid (CSF) examined the extraction, amplification, and detection properties of the AOM tubes compared to the Qiagen DNA blood mini kit. The AOM extraction, amplification, and detection of HSV-1 in CSF displayed differences of less than one CT when compared to Qiagen-extracted samples. Experiments testing the AOM method using clinical CSF samples displayed 100% concordance with reported results. AOM tubes have no adverse effects on amplification or fluorescence acquisition by real-time PCR and can be effectively used for the extraction, amplification, and detection of HSV from CSF. The AOM single tube method is a fast, reliable, and reproducible technique for the extraction, amplification, and detection of HSV in CSF.
A majority of molecular diagnostic assays use a three-phase process of nucleic acid extraction, amplification, and detection of amplification products. For many polymerase chain reaction (PCR)-based assays, amplification and detection have been combined into a real-time, single-tube format using fluorescent oligonucleotides, probes, or DNA-binding dyes.1,2,3 The integration of nucleic acid extraction, amplification, and detection into a single PCR tube format is a significant challenge and would be a notable improvement to current molecular diagnostic assays.
Extraction of DNA or RNA for molecular testing typically requires lysis and/or proteolytic digestion, often in the presence of a chaotropic salt.4,5 The most common extraction methods use adsorption of nucleic acids onto silica matrices.4 The nucleic acid adsorption is performed in high-salt concentrations and eluted in a low-salt buffer or water. The development of a single chamber extraction, amplification, and detection device based on silica poses technical challenges because the high volume of elutent may be greater than desired for most PCR reactions, and any contaminating silica may inhibit the PCR reaction.6
As an alternative to silica, we have been investigating porous aluminum oxide membranes (AOMs) as a matrix for nucleic acid extraction. Previous reports have described the use of 200-nm pore size AOMs for sample filtration and nucleic acid capture,7,8 and as a matrix for the extraction and amplification of human genomic target sequences from whole blood.9 This article describes the development of a custom 0.2-ml polypropylene PCR tube fitted with an AOM filter for the extraction, amplification, and detection of nucleic acids from biological samples. Experiments characterizing the performance of AOM tubes for the detection of herpes simplex virus (HSV) in cerebrospinal fluid (CSF) are presented. HSV was chosen as the model infectious agent based on availability of clinical samples, a quantified DNA target control, and an established reference assay that could be used to compare AOM methods versus reported clinical results. CSF was chosen as the biological medium to test the AOM methodology because of its low viscosity.
Materials and Methods
Aluminum Oxide Custom Tube Assembly
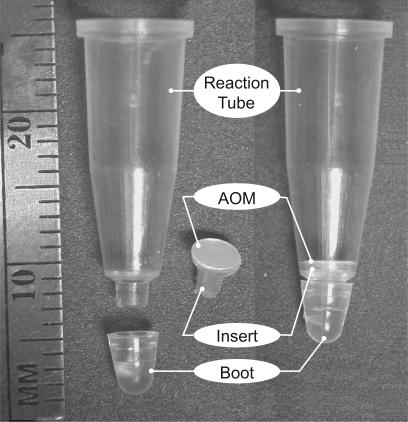
AOM PCR tubes were designed for use in top-reading real-time PCR machines such as the Bio-Rad iCycler (Hercules, CA) or Applied Biosystems 7000 series (Foster City, CA). Each AOM tube consisted of a 0.2-ml injection-molded polypropylene reaction tube, a 200-nm pore diameter Anopore membrane filter insert (Whatman, Middlesex, UK), and a boot (Figure 1). The 0.2-ml AOM tube resembles a standard PCR tube with the bottom removed. The Anopore membrane was thermally sealed to a polypropylene insert using a modified air-actuated ABgene thermal sealer (MarshBio, Rochester, NY) at 150°C, 3 seconds at 49 psi. The polypropylene segments were manufactured by Product Development Manufacturing, Packaging, Inc. (Leesburg, VA), and all subsequent process and assembly steps were performed at ARUP Laboratories (Salt Lake City, UT). Before use, all inserts and tubes were visually inspected for defects such as membrane cracks.
Figure 1.
AOM disposables exploded view (left), and assembled (right). The AOM tubes are designed to work in standard top-reading real-time PCR machines. Scale is 1 mm (ARUP photo by Mark Herrmann).
Clinical Samples and Reference Materials
The clinical samples in the current studies were residual and deidentified following the Health Insurance Portability and Accountability Act of 1996. They were used in accordance with University of Utah institutional review board protocol number 7275, which covers research conducted by ARUP Laboratories. HSV-positive CSF samples were used to test AOM extraction, amplification, and detection using real-time PCR. HSV-negative CSF samples were pooled in sets of 10 and used for negative controls. A DNA standard purified from the HSV-1 virus (1 × 104 DNA copies/μl, Macintyre strain; Advanced Biotechnologies, Inc. (ABI), Columbia, MD) was used as a quantified control standard.
AOM Extraction of Nucleic Acids
Five μl of proteinase K (20 mg/ml; Qiagen, Valencia, CA), 100 μl of 50% AL buffer (Qiagen), and 100 μl of CSF were added to a 1.5-ml microfuge tube and briefly vortexed. Samples were incubated at 56°C for 10 minutes and centrifuged at 5,000 × g for 30 seconds to pellet any particulates. After digestion, 200 μl of the samples were transferred to AOM tubes seated in a custom 48-well filtration block (ARUP), and filtered by vacuum. The CSF sample solutions required 30 to 60 seconds to filter at 400 to 450 mmHg. The AOM was washed once with 200 μl of 100 mmol/L NaCl, and once with 200 μl of nanopure water. After the final wash, the boot was applied to the AOM tubes, master mix was added, and the top capped with optically clear PCR strips or film (ABgene, Surrey, UK).
All experiments described in this study used Qiagen AL buffer, a proprietary lysis buffer containing 25 to 50% guanidium chloride according to the manufacturer’s material safety data sheet. In a separate set of experiments, a modified nonproprietary α-casein lysis buffer (5.25 mol/L guanidine HCl, 50 mmol/L Tris-HCl, pH 6.4, 20 mmol/L ethylenediaminetetraacetic acid, 1.3% w/v Triton X-100, 1 mg/ml α-casein)6 was evaluated. The α-casein lysis buffer and the Qiagen AL buffer performed equally in parallel extraction, amplification, and detection experiments using AOM (data not shown).
PCR Amplification of HSV
Primers to a 192-bp conserved region of the HSV DNA polymerase gene (GenBank accession number X04771) were used for all AOM HSV amplification experiments (HSVdpolF1015-1034, 5′-AGCGACGTCGAGTTTAACTG-3′; HSVdpolR1206-1187, 5′-GGACAGGTCGTAGAGCAGAC-3′ (Integrated DNA Technologies, Coralville, IA). The HSVdpolF1015-1034/HSVdpolR1206-1187 primer set amplifies both HSV-1 and HSV-2 with equal efficiency, and the amplicon cannot be discriminated by melting analysis on an iCycler. A master mix containing Bio-Rad IQ SYBR Green Supermix (50 mmol/L KCl, 20 mmol/L Tris-HCl, pH 8.4, 0.2 mmol/L dNTPs, 1.25 U iTaq DNA polymerase, 3 mmol/L MgCl2, SYBR Green I, 20 nmol/L fluorescein), 500 nmol/L HSVdpolF1015-1034 and HSVdpolR1206-1187 primers, 0.5 mg/ml bovine serum albumin, 5 mmol/L sodium phosphate buffer, pH 8.4, and an additional 0.32× SYBR Green I (Molecular Probes, Eugene, OR) was used for AOM experiments. Sodium phosphate buffer was not added to PCR tube controls because high concentrations of phosphate inhibit PCR (data not shown). All AOM and PCR tube control experiments were run in 50-μl volumes on a Bio-Rad iCycler real-time PCR machine using the following cycling conditions: 95°C for 3:00 minutes and (95°C for 0:15 seconds and 58°C for 0:15 seconds and 72°C for 0:30 seconds) × 50 cycles for amplification (detection at 72°C using SYBR 490-nm filter). Melting was performed after amplification using the following protocol: 95°C for 0:30 minutes and 75°C for 0:10 minutes and (0.5°C for 0:10 minutes/cycle up to 95°C).
HSV Reference Assay
The reference method used for comparison was an Epoch minor groove binding probe-based assay (Epoch Biosciences, Bothell, WA). The Epoch minor groove binding probe possesses 100% identity to a region of the glycoprotein D gene in HSV-1 and HSV-2 and cannot discriminate genotype. Samples were extracted with the Qiagen 96-well blood kit (Qiagen) and run on an ABI 7900 real-time PCR instrument as described by Stevenson and colleagues.10
Determination of HSV DNA Amplification Efficiency in AOM Tubes
The efficiency of PCR amplification in AOM tubes compared to standard PCR tubes was evaluated by real-time PCR. Quantified HSV-1 DNA (ABI), 1000 copies per reaction, was used for this study. Four different reactions were compared: 1) standard PCR tubes with HSV-1 DNA added to master mix; 2) AOM PCR tubes with HSV-1 DNA added to master mix supplemented with 5 mmol/L phosphate buffer; 3) AOM PCR tubes with HSV-1 DNA in 50% AL buffer filtered onto AOM, washed, and master mix with 5 mmol/L phosphate buffer added; and 4) AOM PCR tubes with HSV-1 DNA in 50% AL buffer filtered onto AOM, washed, and master mix with 0.75 mmol/L phosphate buffer added. A no DNA control using AOM tubes containing master mix supplemented with 5 mmol/L phosphate buffer was run as a negative control. All reactions were run in triplicate.
Determination of HSV Dynamic Range Using AOM Method
HSV-negative CSF (pooled from 10 HSV-negative samples) was spiked with high-titer HSV-1 (determined by glycoprotein D sequencing, data not shown) from a clinical swab to create a homogenous, HSV-positive control (designated HSV1-PC+). To estimate the viral concentration of the HSV1-PC+, a serial dilution was made in HSV-negative CSF and purified using a QIAamp DNA blood mini kit (QDM). A second control dilution series using quantified HSV-1 DNA (ABI) spiked into HSV-negative CSF was extracted by the QDM method. Both the HSV1-PC+ and the quantified HSV-1 DNA (ABI) QDM extracted serial dilutions were run in standard PCR tubes on a real-time PCR machine. Crossing thresholds of the dilution series were compared to obtain an estimate of the relative HSV viral concentration in the HSV1-PC+ CSF (data not shown). After establishing HSV1-PC+ viral concentration, a new dilution series of HSV1-PC+ was prepared that corresponded to 100, 1000, 10,000, and 100,000 copies of HSV per ml of CSF and aliquots were processed using the AOM method. This HSV1-PC+ dilution series was analyzed in parallel to a dilution series of HSV-1 DNA (ABI) prepared in 50% AL buffer consisting of 100, 1000, 10,000, and 100,000 copies of HSV-1 DNA per ml and filtered in AOM tubes. All reactions were run in triplicate.
Comparison of AOM and Qiagen Extraction
Nucleic acid extraction using the AOM method was compared to the QDM extraction method using HSV-positive CSF. A direct comparison of the extraction efficiencies of the AOM and QDM methods was complicated by the fact that the QDM procedure is based on the extraction and elution of nucleic acids whereas the AOM method is based on nucleic acid capture. The following approach was used to compare the two procedures. HSV1-PC+ was diluted 1:100 and 1:100,000 in HSV-negative CSF and split into two 500-μl stocks. Two hundred μl of neat HSV1-PC+, the two HSV1-PC+ dilutions, and an HSV-negative CSF control were processed in duplicate by the QDM method and resuspended in 80 μl of elution buffer (EB) (Qiagen). Ten μl of extracted DNA, or 12.5% of each 80-μl QDM extraction, was used as template for real-time PCR in AOM tubes. The extracted DNA was not filtered onto the AOM but added to the master mix supplemented with 5 mmol/L phosphate.
The same HSV1-PC+, HSV1-PC+ dilutions, and HSV-negative CSF control were processed in parallel using the AOM method. In an effort to extract similar quantities of nucleic acid using the QDM and AOM methods, 25 μl of each CSF sample, or 12.5% of 200 μl, was used for AOM extractions. Twenty-five μl of sample, 5 μl of proteinase K, and 175 μl of 50% AL buffer were combined in a 1.5-ml microfuge tube, and incubated at 56°C for 10 minutes. The extraction solutions were filtered through AOM tubes and washed. Master mix supplemented with 5 mmol/L sodium phosphate buffer was added to the AOM tubes and real-time PCR was performed. All AOM extractions were performed in quadruplicate. The crossing thresholds from AOM extracted samples were compared to the crossing thresholds from QDM extracted, eluted, amplified, and detected samples (Figure 4).
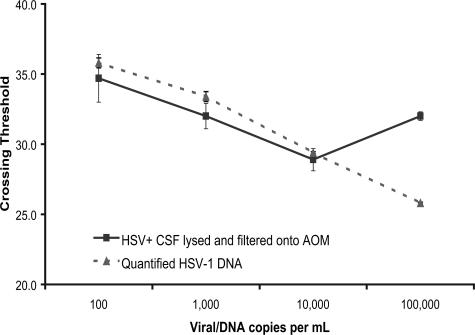
Figure 4.
Comparison of AOM and Qiagen extraction efficiencies. Nucleic acid extraction using the AOM method was compared to the QDM extraction method using HSV-positive CSF. Neat HSV1-PC+ was diluted 1:100 and 1:100,000 and extracted using the AOM and QDM methods as described in Materials and Methods. Equal amounts of input nucleic acid were extracted, amplified, and detected by real-time PCR. The AOM extracted, amplified, and detected HSV dilution series is represented by solid lines. The QDM extracted HSV-1 DNA from the serial dilutions was amplified and detected in AOM tubes (dashed lines). HSV-negative CSF was extracted by the AOM method and represented by the light solid line. AOM extractions were performed in quadruplicate and the QDM extractions were performed in parallel.
AOM Clinical Comparison
Previously characterized HSV-positive clinical samples were tested using the AOM method. Samples were randomly chosen and prepared as previously described. Reactions were run in duplicate on a Bio-Rad iCycler real-time PCR machine. The CSF-negative control was pooled from 10 HSV clinical samples reported as negative by ARUP.
Results
The custom AOM-based reaction tube used in this study is shown in Figure 1. Initial characterization experiments with AOM tubes showed that the addition of phosphate buffer was necessary for optimum PCR efficiency (Figure 2). All experiments described in the current study comparing AOM tubes to standard PCR tubes used 5 mmol/L sodium phosphate buffer supplemented into the AOM master mix. It was observed that sodium phosphate buffer inhibited real-time PCR amplification in standard PCR tubes; therefore, no additional phosphate was added to the master mix for PCR tube controls.
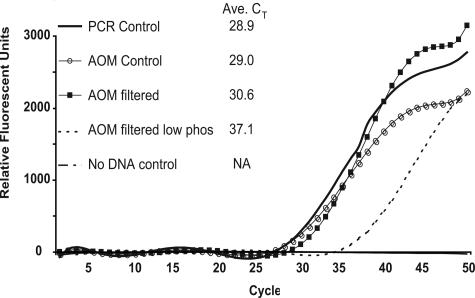
Figure 2.
Real-time PCR amplification of HSV-1 DNA in AOM tubes. One thousand copies of quantified HSV-1 DNA (ABI) were used to compare crossing thresholds between standard PCR and AOM tubes. PCR control: PCR tube control reaction. AOM control: AOM tube reaction with 5 mmol/L phosphate in master mix. AOM filtered: AOM tube reaction with DNA filtered onto the membrane with 5 mmol/L phosphate in master mix. AOM filtered (low phos): AOM tube reaction with DNA filtered onto the membrane with 0.75 mmol/L phosphate in master mix. No DNA control: AOM tube with no DNA with 5 mmol/L phosphate in master mix. All reactions run in triplicate.
HSV DNA Amplification Efficiency in AOM Tubes
To determine whether the assembled AOM tube adversely impacted PCR, a quantified HSV-1 DNA standard (ABI) was used to examine real-time PCR amplification efficiency in AOM tubes compared to standard PCR tubes. A master mix containing 1000 copies of HSV-1 DNA (ABI) was added to AOM and standard PCR tubes. The AOM tubes were supplemented with 5 mmol/L phosphate before capping. Each AOM and standard PCR amplification and detection was performed in triplicate using real-time PCR. The average crossing thresholds (CT) were 29.0 cycles for AOM tubes and 28.9 cycles for PCR tube controls, indicating that AOM tubes did not compromise PCR amplification or fluorescence detection (Figure 2).
Dynamic Range in AOM Tubes
To study the amplification efficiency of HSV-1 DNA filtered onto AOM, two experiments using HSV-1 DNA (ABI) filtered in AOM tubes were performed. One master mix containing the optimized concentration of 5 mmol/L sodium phosphate; the second used 0.75 mmol/L sodium phosphate. The CT obtained with the 5 mmol/L phosphate master mix was 30.6 cycles compared to 37.1 cycles observed with the 0.75 mmol/L phosphate master mix. A CT shift of 1.7 cycles between HSV DNA filtered onto an AOM membrane and amplified in the presence of 5 mmol/L phosphate (CT, 30.6) versus a PCR tube control (CT, 28.9) indicated a compromise in amplification when DNA was filtered onto AOM (Figure 2).
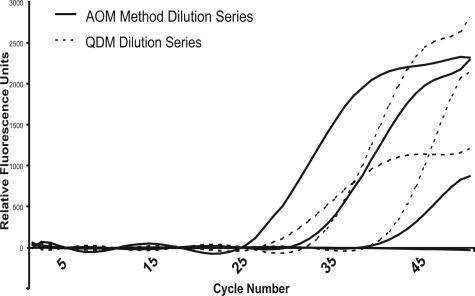
To assess the dynamic range for HSV PCR amplification in AOM tubes, a comparative serial dilution experiment was performed. Serial dilutions of HSV1-PC+ representing 100, 1000, 10,000, and 100,000 copies of HSV per ml of CSF were prepared. Each HSV1-PC+ dilution was extracted using the AOM method. A comparable dilution series of quantified HSV-1 DNA standard (ABI) was prepared in 50% AL buffer and filtered in AOM tubes. Master mixes for both dilution series were supplemented with 5 mmol/L sodium phosphate and real-time PCR was performed. The crossing thresholds were plotted against the viral DNA copies/ml. The data show that amplification of the HSV-1 DNA was linear from 100 to 100,000 copies/ml whereas amplification of the HSV CSF samples was linear only from 100 to 10,000 copies/ml (Figure 3).
Figure 3.
Dynamic range and extraction efficiency of AOM tubes. Quantified HSV-positive CSF (HSV1-PC+, 100 to 100,000 copies of HSV-1 per ml of CSF) dilutions were processed using the AOM method (solid line) and compared to quantified HSV-1 DNA (ABI) dilutions filtered into AOM tubes (dashed line, 100 to 100,000 copies of HSV-1 per ml). Error bars represent a single SD. All reactions run in triplicate.
Comparison of AOM and Qiagen Extraction
Experiments comparing the AOM method to the QIAamp DNA blood mini kit (QDM) were performed using HSV-positive CSF. Stock, 1:100, and 1:100,000 dilutions of HSV1-PC+ CSF were processed by both methods, amplified, and detected by real-time PCR. The crossing thresholds using the AOM method were 26.5, 33.4, and 40.8 for the undiluted, 1:100 dilution, and 1:100,000 dilution, respectively. The crossing thresholds using the QDM method were 28.6, 33.9, and 40.7 for the undiluted, 1:100 dilution, and 1:100,000 dilution, respectively (Figure 4).
AOM Clinical Comparison
Ten randomly chosen, deidentified, HSV-positive CSF clinical samples were processed using AOM and compared to clinical results. A pooled set of 10 HSV-negative samples was used as a negative control. The AOM method qualitatively reproduced the clinical results (10 of 10) using real-time PCR. The negative control did not amplify (data not shown).
Discussion
Previous reports have shown that nucleic acids from biological samples could be captured or localized to AOM and amplified by PCR.8,11 The goal of this study was to characterize the performance of a custom-designed, 0.2-ml PCR tube fitted with an AOM filter insert for single tube extraction, amplification, and detection of nucleic acids. A single tube nucleic acid extraction and amplification method based on embedded aluminum oxide and commercialized under the trade name Xtrana (Xtrana Inc., Broomfield, CO) has been previously described.12 The Xtrana method has been used for extraction, amplification, and detection of nucleic acids as well as sequencing and reverse transcription.13 The main limitation of the Xtrana method was suboptimal target sensitivity and detection, with observed crossing threshold lags of five to seven cycles (B. Linn, personal communication).13 Xtrana tubes were not currently commercially available for comparative studies with the AOM method.
Initial real-time PCR experiments with AOM tubes displayed CT shifts greater than five cycles compared to standard PCR tubes, suggesting reduced availability of the captured nucleic acids. In an effort to reduce the CT lag of AOM tubes compared to PCR tubes, a variety of manipulations were pursued. The denaturants dimethyl sulfoxide, dimethyl formamide, dithiothreitol, and formamide were supplemented into AOM master mix. The addition of 5% dimethyl sulfoxide to the AOM master mix displayed a CT gain of up to two cycles, but the resulting melting curves were adversely affected. The denaturants dimethyl formamide, dithiothreitol, and formamide had little or no effect on shifting the CT at concentrations of less than 2 to 5% w/v. Higher concentrations of the denaturants completely inhibited amplification presumably by DNA polymerase inhibition (data not shown). Base denaturation and mechanical disruption were also pursued with variable success. Eventually the addition of 5 mmol/L sodium phosphate buffer to the master mix was shown to be the simplest method to reproducibly increase PCR efficiency using AOM tubes (Figure 2). We hypothesize that the DNA is delocalized from the AOM by negatively charged phosphate ions competing against the nucleic acid phosphate backbone for positively charged sites on the membrane.
Experiments comparing the amplification efficiency of 1000 copies of HSV-1 DNA (ABI) in standard PCR tubes and AOM tubes displayed a 0.1 CT difference demonstrating that the AOM matrix does not significantly interfere with PCR or real-time acquisition of signal. A second set of experiments examined the availability of HSV-1 DNA for amplification when localized onto AOM by filtration and amplified in the presence of 5 mmol/L phosphate. Compared to control PCR tubes, a CT shift of 1.7 cycles was observed with HSV-1 DNA in the AOM tubes. This shift indicates a 75% reduction in amplifiable HSV 1 DNA with optimized phosphate conditions.
The current HSV-PCR clinical assay at ARUP has a lower detection limit of 150 to 300 copies of HSV virus per ml of CSF (J. Stevenson, personal communication). To determine the detection limits and dynamic range of the AOM technique, quantified HSV-positive CSF (HSV1-PC+) was analyzed using the AOM method (Figure 3). The AOM method reproducibly detected 100 copies of HSV per ml of CSF (equivalent to 10 copies of HSV-1 DNA per tube). The dynamic range of HSV-1 DNA (ABI) detection using AOM was linear from 100 to 100,000 copies/ml. In contrast, the dynamic range for detection of HSV1-PC+ was linear from 100 to 10,000 copies per ml of CSF. The theoretical nucleic acid binding capacity of AOM was calculated to determine whether nucleic acid saturation could be affecting the dynamic range of the AOM disposables. The theoretical maximum capacity of nucleic acid as a monolayer on the AOM disposable 2.5-mm2 filter is ∼4 ng).8 The genome size of HSV-1 is ∼152 kb, therefore 4 ng is equal to ∼40 amol, or 2.4 × 107 copies of target HSV-1 DNA. These calculations suggest that the disposable membrane has sufficient capacity to retain 100,000 copies of HSV. Clinical CSF samples typically contain human genomic DNA, and other potentially pathogenic bacterial or viral genomes. The loss of linearity at higher copy numbers of HSV in CSF samples might be attributed to other nucleic acids competing with HSV DNA, saturating the total nucleic acid binding sites on the AOM. Nucleic acids have been shown to filter through AOM when the maximum monolayer capacity has been exceeded.11 In contrast, other experiments using 10,000 copies (∼36 ng) of human genomic DNA have shown that mass quantities of nucleic acid exceeding the theoretical maximum capacity of the AOM can be filtered, amplified, and detected using AOM. In some cases, a stacking phenomena in which multiple layers of nucleic acid are present on the AOM has been observed.8,11 At present, the loss of linearity in the dynamic range at higher concentrations of HSV is not completely understood. AOM tubes may best be suited for qualitative assays, with linearity at the lower range being sensitive enough to make an estimate of viral load, but not accurate when high levels of viral target are present. A second-generation AOM filter with a larger diameter membrane would increase nucleic acid monolayer mass retention and improve flow rates.
To further characterize the AOM method, experiments were conducted to compare HSV extraction by the AOM method with that obtained using a silica-based method, the Qiagen QDM method. Because the AOM method relies on localization of nucleic acids on a membrane surface, and the QDM method is based on adsorption followed by elution, several assumptions were made. First, it was assumed that all HSV genomes were extracted from CSF by the QDM method and eluted in 80 μl, and all HSV genomes were extracted from CSF by the AOM method and delocalized during PCR. Based on these assumptions, it was further assumed that 10 μl of elutent from the QDM method contained the same amount of HSV DNA as that present in 25 μl of HSV-positive CSF extracted by the AOM method. In a comparative dilution series study, both methods displayed similar crossing thresholds for each dilution, supporting the concept that the AOM method of extraction and delocalization and the QDM method of extraction and elution present similar quantities of amplifiable HSV DNA for PCR (Figure 4).
In a pilot comparison of clinical samples, the AOM method showed complete concordance for the detection of HSV in 10 clinical CSF samples that had been previously reported as positive in the ARUP clinical method and a pooled negative CSF sample was negative using the AOM method. The clinical samples were sequenced and found to represent both HSV-1 and HSV-2, confirming that the technique can also detect HSV-2 (data not shown).
From the project’s inception, a major concern involved the ability to filter biological samples through the AOM without clogging the membrane. AOM is highly uniform and has a porosity of ∼50%,7,9,14 which yields flow rates up to 8 μl/second/mm2 for the AOM disposables. Multiple buffer formulas were investigated in an attempt to create a universal buffer that could efficiently extract various samples, and allow AOM membrane filtration. An α-casein lysis buffer, reported to counteract enzyme-processing inhibitors in CSF and urine,6 was modified for use with CSF. The nonproprietary, α-casein lysis buffer is well suited for the extraction of HSV DNA from CSF and yields comparable results to Qiagen AL buffer. Digestion of CSF samples in optimized buffers reduced clogging or abnormally slow filtration in the AOM tubes to ∼5%. A brief centrifugation to pellet any particulates was introduced after digestion that effectively eliminated any clogging, even in turbid CSF samples with high red blood cell counts. Alternatively, CSF samples can be lysed and processed at room temperature within a single AOM tube if the vacuum is greater than 760 mmHg at the source. Escherichia coli in LB media, CSF, urine, biological swabs, serum, plasma, and whole blood have all been filtered through AOM. Different buffers and lysis conditions are needed for each sample type, and filtration speeds ranged from 0.5 μl/second/mm2 for highly viscous plasma to the 8 μl/second/mm2 for CSF at 450 mmHg. Filtration efficiency can be increased by lowering input sample volume, which may be preferable under certain circumstances, or by increasing vacuum pressure. A second-generation AOM spin column with a larger surface area may minimize filtration issues with more complex samples.
The single tube extraction, amplification, and detection AOM method offers advantages to current techniques, providing fewer manipulations and less disposable waste compared to other extraction methods, characteristics desirable when analyzing infectious agents. The AOM tube does not interfere with real-time amplification or detection, and accurately detects HSV DNA in clinical samples. AOM can filter most biological samples tested under optimized conditions, but is better suited to low-viscosity samples such as CSF, or diluted high-viscosity samples such as high triglyceride plasma. The preferred embodiment of the AOM method would lend itself to accurately determining low copy numbers of target or a qualitative assay.
Acknowledgments
We thank Chris Link and Jacob Durtschi for comments regarding the manuscript, Jeff Stevenson and Bor-Chian Lin for personal communications, and Ray Dissonnette for fine adjustments to the AOM tube mold.
References
- Abravaya K, Huff J, Marshall R, Merchant B, Mullen C, Schneider G, Robinson J. Molecular beacons as diagnostic tools: technology and applications. Clin Chem Lab Med. 2003;41:468–474. doi: 10.1515/CCLM.2003.070. [DOI] [PubMed] [Google Scholar]
- Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Frisch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, Cold Springs Harbor Laboratory Press; 1989 [Google Scholar]
- Boom R, Sol C, Beld M, Weel J, Goudsmit J, Wertheim-van Dillen P. Improved silica-guanidiniumthiocyanate DNA isolation procedure based on selective binding of bovine alpha-casein to silica particles. J Clin Microbiol. 1999;37:615–619. doi: 10.1128/jcm.37.3.615-619.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford GP, Steele LM, Ondris-Crawford R, Iannacchione GS, Yeager CJ, Doane JW, Finotello D. Characterization of the cylindrical cavities of Anopore and Nuclepore membranes. J Chem Phys. 1992;96:7788–7796. [Google Scholar]
- Erali M, Durtschi JD, Voelkerding KV, Smith RE. Localization and imaging of nucleic acids on nanoporous aluminum oxide membranes. Clin Chem. 2004;50:1819–1821. doi: 10.1373/clinchem.2004.035170. [DOI] [PubMed] [Google Scholar]
- Elgort MG, Herrmann MG, Erali M, Durtschi JD, Voelkerding KV, Smith RE. Extraction and amplification of genomic DNA from human blood on nanoporous aluminum oxide membranes. Clin Chem. 2004;50:1817–1819. doi: 10.1373/clinchem.2004.035162. [DOI] [PubMed] [Google Scholar]
- Stevenson J, Hymas W, Hillyard D. The effect of sequence polymorphisms on the performance of two real time PCR assays for herpes simplex virus. J Clin Microbiol. 2005;43:2391–2398. doi: 10.1128/JCM.43.5.2391-2398.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RE, Voelkerding KV, Elgort MG, Durtshci J: Microporous materials and methods of making, using, and articles thereof. US patent application 20040253624. December 16, 2004. [Google Scholar]
- Kozwich D, Johansen KA, Landau K, Roehl CA, Woronoff S, Roehl PA. Development of a novel, rapid integrated Cryptosporidium parvum detection assay. Appl Environ Microbiol. 2000;66:2711–2717. doi: 10.1128/aem.66.7.2711-2717.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margraf RL, Page S, Erali M, Wittwer CT. Single-tube method for nucleic acid extraction, amplification, purification, and sequencing. Clin Chem. 2004;50:1755–1761. doi: 10.1373/clinchem.2004.035808. [DOI] [PubMed] [Google Scholar]
- Durtschi JD, Eralia M, Herrmann MG, Elgort MG, Voelkerding KV, Smith RE. Optically improved aluminum oxide membrane through electroless Ni modification. J Membr Sci. 2005;248:81–87. [Google Scholar]