Abstract
Recent studies have shown the hedgehog and Wnt families of signaling proteins to be associated with tumor initiation, growth, and survival. However, these pathways remain unexplored in ovarian endometrioid adenocarcinoma (OEA). Here, we describe a novel TaqMan low-density array to examine the expression of 26 and 20 genes in the hedgehog and Wnt pathways, respectively, in six matched snap-frozen and formalin-fixed, paraffin-embedded (FPE) OEA specimens. Expression values were normalized to uninvolved ovarian epithelium. Gene expression in matched frozen and FPE tissues demonstrated significant concordance (r = 0.92, P < 0.0001). However, comparison of amplified and unamplified RNA from frozen OEA tissues revealed an altered molecular profile in amplified RNA. Amplification of RNA from FPE tissues was not successful. The expression of Desert hedgehog (DHH), Indian hedgehog (IHH), Hedge-hog interacting protein (HHIP), Wnt10B, Wnt9B, and Wnt inhibitory factor (WIF1) were tumor-specific with no detectable expression in normal ovarian epithelium. In addition, several genes were significantly (P < 0.025) down-regulated in OEA, including cyclin E2, Porcupine, c-Myc, and Axin 2 (4.8-, 3.6-, 2.9-, and 1.9-fold, respectively). TaqMan low-density array provides an effective multivariate technique for examining gene expression in RNA isolated from either snap-frozen or archival FPE tissues and can identify tumor-specific genes, possibly leading to novel treatments.
Ovarian cancer is distinguished by particularly aggressive local invasiveness potential and in the United States, remains the fourth highest cause of cancer death in women.1,2 Epithelial ovarian cancer is the major ovarian malignancy consisting of four histological subtypes including serous, mucinous, endometrioid, and clear cell with ovarian endometrioid adenocarcinoma (OEA) being the second most common.3 Unfortunately, ∼80% of patients diagnosed with advanced OEA die within 5 years.4 In addition, the limited knowledge of the molecular mechanisms involved in the development and clinical progression of OEA have hampered attempts to develop novel rationally designed treatment paradigms.
Activation of the hedgehog signaling pathway, normally involved in embryogenesis, can lead to tumor formation and is necessary for tumor survival in several types of cancer (including medulloblastoma, basal cell carcinoma, small-cell lung cancer, and breast cancer).5,6,7,8,9,10 Recent studies have also implicated hedgehog signaling as an early mediator of tumorigenesis in cancers of the digestive tract, particularly pancreatic adenocarcinoma.11,12 Collectively, these studies suggest that abrogation of the hedgehog pathway may provide a novel, targeted therapeutic approach. Interestingly, the hedgehog pathway has not been examined in OEA. The Wnt pathway, also involved in embryogenesis, has been found to possess similarities to the hedgehog pathway with respect to posttranslational modification, secretion, signaling mechanisms, and tumorigenesis.13,14 Recent studies have shown that increased expression of components in the Wnt pathway have been implicated in ovarian tumorigenesis, although the exact molecular mechanisms remain to be elucidated.15,16
Despite recent advances in gene quantitation technology, there is limited analysis of large clusters of genes, such as the hedgehog and Wnt families, in ovarian carcinomas in part because of the limited availability of fresh frozen tissue. Conversely, formalin-fixed, paraffin-embedded (FPE) tissues, derived from institutional archives, offer a more readily available alternative to frozen tissue in most cancer treatment/research facilities. Several studies have shown that real-time quantitative (RTQ) polymerase chain reaction (PCR) can be used to quantify gene expression from RNA isolated from FPE tissue.17,18,19,20,21,22 However, these studies have been limited by RTQ’s ability to quantify only one gene at a time in a single RNA sample. In addition, validation of gene expression profiles obtained from frozen versus FPE tissue has been problematic due to the difficulty in obtaining matched frozen and paraffin-embedded samples. The capabilities of RTQ have recently been expanded with the development of TaqMan low-density arrays (TLDAs) (Applied Biosystems, Foster City, CA), which are able to determine the expression of multiple, user-defined gene clusters simultaneously.
In the current study, we examine whether TLDA could be used for the analysis of archival, paraffin-embedded tissues by correlating the expression of 26 and 20 genes in the hedgehog and Wnt pathways, respectively, in six matched snap-frozen and FPE OEA specimens. In addition, expression profiles of amplified versus unamplified RNA were compared to determine whether the small amounts of RNA available from needle biopsies or laser capture-microdissected samples could be increased and quantified while conserving the expression profile. These analyses represent the first multivariate examination of the hedgehog and Wnt pathways in OEA.
Materials and Methods
Tissue Collection and Processing
Six ovarian endometrioid tumors were obtained from the Department of Pathology at the University of Alabama at Birmingham using an institutional review board-approved protocol. The samples were collected in the operating room and sectioned into three pieces. The central piece was immediately snap-frozen in liquid nitrogen and stored at −80°C before RNA isolation. The other two end pieces were immediately fixed in neutral-buffered formalin for 6 to 18 hours before paraffin embedding. Paraffin-embedded tissues were cut into 20-μm sections and stored at room temperature before RNA isolation. Normal ovarian surface epithelium (OSE) that harbored no neoplasms was scraped from the ovaries of two unrelated patients and immediately snap-frozen in liquid nitrogen and stored at −80°C before RNA isolation.
RNA Extraction
Total RNA was isolated from frozen tissues using Trizol reagent (Invitrogen, Carlsbad, CA) as per the manufacturer’s instructions. RNA was then DNase treated and purified using the RNeasy mini kit (Qiagen, Hilden, Germany) as per the manufacturer’s instructions. RNA was eluted in 30 μl of RNase-free water and stored at −80°C.
Paraffin tissue sections were deparaffinized by incubation with 800 μl of xylene and 400 μl of 100% ethanol. The samples were then centrifuged and the supernatant was removed. Tissue pellets were washed with 1 ml of 100% ethanol and dried for 10 minutes at 55°C. The RNA isolation that followed was performed using the Roche High Pure RNA paraffin kit (Roche Diagnostics, Mannheim, Germany) as per the manufacturer’s instructions. RNA was eluted in 30 μl of RNase-free water and stored at −80°C.
Houskeeping Gene Variability
Assessment of housekeeping gene variability between normal and neoplastic ovarian tissues was performed as previously described.23 Briefly, the RNA concentrations of both the OEA and OSE samples were determined spectrophotometrically by A260 measurement and adjusted to 20 ng/μl to ensure that differences in housekeeping gene expression were not because of variability in RNA concentrations. Because of the low amounts of tissue obtained from the two scraped OSE specimens, their RNA was combined to obtain a sufficient concentration for further analysis. The concentration of each sample was confirmed by densitometry and RNA integrity (degradation) was verified by electrophoresis and ethidium bromide staining on a 1% agarose gel. Primers and probe for the ribosomal protein, large, P0 (RPLP0) (NM_001002) gene were obtained from Applied Biosystems (Foster City, CA) and used according to the manufacturer’s instructions. The concentration of all RNA samples (OSE and OEA) was then determined using RPLP0 and linear regression analysis of a standard curve derived from known concentrations of normal ovary total RNA (Ambion, Austin, TX) as previously described by our laboratory.23,24
RNA Amplification
A fixed amount of 20 ng of total RNA isolated from either frozen or paraffin-embedded OEA was amplified using the Ovation Nanosample RNA amplification system (NuGEN Technologies, Inc., San Carlos, CA), Full Spectrum Global amplification kit (System Biosciences, Mountain View, CA), MessageAmp aRNA kit (Ambion) and RiboAmp RNA amplification kit (Arcturus, Mountain View, CA) as per the manufacturers’ instructions. The yield obtained from each amplification procedure was assessed by the RPLP0 housekeeping gene.
Reverse Transcription
Before cDNA synthesis, all RNA samples, amplified and unamplified, were diluted to 4 ng/μl using RNase-free water containing 12.5 ng/μl of total yeast RNA (Ambion) as a carrier. cDNA was prepared using the High Capacity cDNA archive kit (Applied Biosystems) as per the manufacturer’s instructions. The resulting cDNA samples were used immediately for TLDA analysis.
PCR Amplification Efficiency
For RNA isolated from frozen and FPE tissues, a standard curve was prepared using 20, 10, 5, 2.5, and 1.25 ng RNA. RTQ was performed for the RPLP0 housekeeping gene. The slope to each standard curve was then calculated and the efficiency of PCR amplification was determined using the equation e = 10(−1/slope). In a PCR reaction that is 100% efficient, the amount of amplicon doubles at each cycle such that an e of 2 represents 100% PCR amplification efficiency.
TLDA
TLDA Design
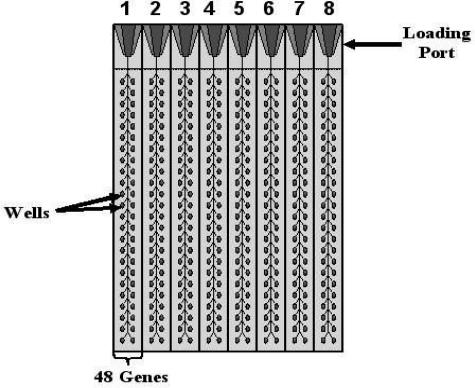
For each card of the low-density array, there are eight separate loading ports that feed into 48 separate wells for a total of 384 wells per card (Figure 1). Each 2-μl well contains specific, user-defined primers and probes, capable of detecting a single gene. In this study, the TLDA card was configured into eight identical 48-gene sets (Figure 1). Genes were chosen based on literature reviews of the hedgehog, Wnt, and cell cycle molecular pathways and their involvement in tumorigenesis.5,6,13,14,25 Each set of 48 genes (Table 1) also contains two housekeeping genes, RPLP0 and 18S (a mandatory control designed into each card by the manufacturer). In this study, however, RPLP0 was used exclusively as the housekeeping gene.
Figure 1.
TLDA design using a 48-gene template.
Table 1.
Hedgehog and Wnt Gene Expression in Ovarian Endometrioid Adenocarcinoma
| Gene | Average frozen* | P value† | Ratio‡ | Average paraffin* | P value† | Ratio‡ |
|---|---|---|---|---|---|---|
| Desert hedgehog (DHH) | X | 6/6 | X | 3/6 | ||
| Hedgehog interacting protein (HHIP) | X | 2/6 | X | 2/6 | ||
| Indian hedgehog (IHH) | X | 4/6 | X | 3/6 | ||
| Wnt inhibitory factor 1 (WIF1) | X | 5/6 | X | 4/6 | ||
| Wingless-type MMTV integration site family, member 10B (WNT1) | X | 5/6 | X | 4/6 | ||
| Wingless-type MMTV integration site family, member 9B (WNT9B) | X | 1/6 | X | 1/6 | ||
| Porcupine (PPN) | 0.28 ± 0.11 | <0.0001 | 6/6 | 0.48 ± 0.39 | 0.021 | 6/6 |
| Cyclin E2 (CCNE2) | 0.21 ± 0.20 | <0.0001 | 6/6 | 0.38 ± 0.45 | 0.020 | 6/6 |
| c-Myc oncogene (MYC) | 0.35 ± 0.39 | 0.010 | 6/6 | 0.12 ± 0.08 | 0.000 | 6/6 |
| Axin 2 (AXIN2) | 0.54 ± 0.29 | 0.011 | 6/6 | 0.54 ± 0.23 | 0.005 | 6/6 |
| GLI pathogenesis-related 1 (GLIPR1) | 0.46 ± 0.29 | 0.006 | 6/6 | 2.16 ± 2.53 | 0.312 | 6/6 |
| HER-2/Neu (ERBB2) | 1.39 ± 2.03 | 0.654 | 6/6 | 0.36 ± 0.51 | 0.027 | 6/6 |
| Patched (PTCH) | 0.83 ± 0.63 | 0.536 | 6/6 | 1.64 ± 0.55 | 0.035 | 6/6 |
| Epidermal growth factor receptor (EGFR) | 1.09 ± 0.86 | 0.802 | 6/6 | 2.90 ± 2.07 | 0.074 | 6/6 |
| Wingless-type MMTV integration site family, member 7A (WNT7A) | 7.01 ± 11.00 | 0.238 | 4/6 | 16.49 ± 18.06 | 0.090 | 4/6 |
| Smoothened (SMO) | 1.61 ± 1.44 | 0.345 | 6/6 | 2.96 ± 2.29 | 0.091 | 6/6 |
| Frizzled homolog (FRZD1) | 5.56 ± 10.29 | 0.328 | 6/6 | 9.34 ± 10.46 | 0.108 | 6/6 |
| Patched 2 (PTCH2) | 0.66 ± 0.56 | 0.194 | 6/6 | 5.60 ± 5.80 | 0.110 | 6/6 |
| Glioma-associated oncogene (GLI) | 3.84 ± 6.20 | 0.312 | 6/6 | 12.99 ± 15.39 | 0.114 | 5/6 |
| Kringle-containing transmembrane protein 1 (KREMEN1) | 1.22 ± 1.29 | 0.692 | 6/6 | 0.68 ± 0.43 | 0.126 | 6/6 |
| Platelet-derived growth factor receptor-α (PDGFRA) | 10.49 ± 7.80 | 0.031 | 6/6 | 12.79 ± 15.84 | 0.128 | 6/6 |
| Glioma-associated oncogene 3 (GLI3) | 1.22 ± 1.09 | 0.645 | 6/6 | 6.16 ± 7.06 | 0.134 | 6/6 |
| Glioma-associated oncogene 2 (GLI2) | 3.17 ± 4.47 | 0.289 | 6/6 | 5.41 ± 6.47 | 0.156 | 6/6 |
| Low-density lipoprotein receptor-related protein 6 (LRP6) | 1.85 ± 1.57 | 0.241 | 6/6 | 4.35 ± 4.99 | 0.161 | 6/6 |
| Cyclin B1 (CCNB1) | 0.46 ± 0.42 | 0.026 | 6/6 | 0.61 ± 0.58 | 0.163 | 6/6 |
| p21, Cip1 (CDKNA1) | 1.08 ± 1.30 | 0.881 | 6/6 | 2.93 ± 2.97 | 0.172 | 6/6 |
| Frizzled related protein (FRZB) | 1.60 ± 2.37 | 0.563 | 6/6 | 2.38 ± 2.16 | 0.178 | 6/6 |
| K-ras oncogene 2 (KRAS2) | 1.00 ± 0.85 | 0.992 | 6/6 | 0.75 ± 0.41 | 0.185 | 6/6 |
| Receptor-like tyrosine kinase (RYK) | 1.05 ± 0.93 | 0.896 | 6/6 | 2.39 ± 2.37 | 0.209 | 6/6 |
| Cyclin D1 (CCND1) | 0.67 ± 0.53 | 0.182 | 6/6 | 1.71 ± 1.24 | 0.218 | 6/6 |
| Sonic hedgehog (SHH) | 0.88 ± 1.41 | 0.840 | 3/6 | 5.26 ± 7.76 | 0.237 | 3/6 |
| Cyclin D3 (CCND3) | 2.97 ± 3.33 | 0.208 | 6/6 | 4.23 ± 6.18 | 0.257 | 6/6 |
| Wingless-type MMTV integration site family, member 8A (WNT8A) | 0.83 ± 1.95 | 0.837 | 2/6 | 0.45 ± 1.10 | 0.278 | 1/6 |
| Suppressor of fused (SUFU) | 0.54 ± 0.45 | 0.052 | 6/6 | 1.56 ± 1.13 | 0.279 | 6/6 |
| Cyclin D2 (CCND2) | 1.10 ± 1.29 | 0.860 | 6/6 | 2.51 ± 3.21 | 0.302 | 6/6 |
| Low-density lipoprotein receptor-related protein 5 (LRP5) | 0.61 ± 0.32 | 0.029 | 6/6 | 1.34 ± 0.84 | 0.368 | 6/6 |
| E2F1 transcription factor (E2F1) | 1.01 ± 0.73 | 0.987 | 6/6 | 2.14 ± 3.16 | 0.416 | 6/6 |
| Retinoblastoma 1 (RB1) | 1.20 ± 0.68 | 0.511 | 6/6 | 1.24 ± 0.87 | 0.537 | 6/6 |
| Wingless-type MMTV integration site family, member 1 (WNT1) | 0.25 ± 0.60 | 0.028 | 1/6 | 2.66 ± 6.51 | 0.560 | 1/6 |
| Wingless-type MMTV integration site family, member 7B (WNT7B) | 0.60 ± 0.92 | 0.338 | 5/6 | 0.68 ± 1.44 | 0.610 | 4/6 |
| Axin 1 (AXIN1) | 0.73 ± 0.54 | 0.278 | 6/6 | 1.18 ± 0.82 | 0.621 | 6/6 |
| Cyclin E1 (CCNE1) | 0.92 ± 0.87 | 0.836 | 6/6 | 1.10 ± 1.56 | 0.881 | 6/6 |
| Insulin promotor factor 1 (IPF1) | NE | 0/6 | NE | 0/6 | ||
| Wingless-type MMTV integration site family, member 16 (WNT16) | NE | 0/6 | NE | 0/6 | ||
| Wingless-type MMTV integration site family, member 8B (WNT8B) | NE | 0/6 | NE | 0/6 | ||
| Wingless-type MMTV integration site family, member 9A (WNT9A) | NE | 0/6 | NE | 0/6 |
Values are expressed as the mean of six samples ± SD. Genes designated by X did not express in normal ovarian epithelium. Genes designated by NE (no expression) did not express in either normal or OEA.
P < 0.025 was considered statistically significant.
Ratio indicates OEA samples expressing gene/total samples examined.
TLDA Preparation
Each cDNA sample (100 μl) was added to an equal volume of 2× TaqMan Universal PCR Master Mix (Applied Biosystems). After gentle mixing and centrifugation, the mixture was then transferred into a loading port on a TLDA card (Applied Biosystems). Each of the six matched OEA samples were run in quadruplicate with one matching pair (frozen and FPE) on each card. Amplified RNA samples were also run in quadruplicate on separate cards. To distinguish tumor-specific activation of the hedgehog and Wnt pathways, RNA from two combined snap-frozen OSE samples were also analyzed. The array was centrifuged twice for 1 minute each at 1200 rpm (306 × g) to distribute the samples from the loading port into each well. The card was then sealed and PCR amplification was performed using an Applied Biosystems Prism 7900HT sequence detection system. Thermal cycler conditions were as follows: 2 minutes at 50°C, 10 minutes at 94.5°C, 30 seconds at 97°C, 1 minute at 59.7°C for 40 cycles.
TLDA Analysis
Expression values were calculated using the comparative CT method as previously described (User Bulletin No. 2, Applied Biosystems). Briefly, this technique uses the formula 2−ΔΔCT to calculate the expression of target genes normalized to a calibrator. The threshold cycle (CT) indicates the cycle number at which the amount of amplified target reaches a fixed threshold. CT values range from 0 to 40 (the latter representing the default upper limit PCR cycle number that defines failure to detect a signal). ΔCT values [ΔCT = CT (target gene) − CT (RPLP0)] were calculated for the combined frozen OSE sample and subsequently used as the calibrator, for which all gene expression values were assigned a relative value of 1.00, to determine comparative gene expression such that ΔΔCT = ΔCT (OEA sample) − ΔCT (OSE sample). A range for each expression value was calculated based on the SD (s) of the ΔΔCT value where 2−(ΔΔCT + s) is the lower limit and 2−(ΔΔCT − s) is the upper limit.
Validation of TLDA
Primers and probes for Desert Hedgehog (DHH) (NM_021044), Indian Hedgehog (IHH) (NM_002181), Sonic Hedgehog (SHH) (NM_000193), Patched (PTCH) (NM_000264), Patched 2 (PTCH2) (NM_003738), Smoothened (SMO) (NM_005631), Glioma-associated oncogene homolog 1 (GLI) (NM_005269), and Glioma-associated oncogene homolog 3 (GLI3) (NM_000168) were obtained from Applied Biosystems and used according to the manufacturer’s instructions. RTQ was then performed for these eight genes on the six matched frozen and FPE OEA samples as well as the combined OSE sample using an ABI Prism 7900HT sequence detection system. Gene expression was calculated using the comparative CT method.
Statistical Analysis
All statistical analyses were conducted with SAS Ver. 9.1 (SAS Institute, Cary, NC). Because of the small sample size, a more stringent P < 0.025 was used to establish statistically significant differences rather than 0.05, which is typically used. To check the reproducibility of TLDA, the coefficient of variance (CV) was calculated for each gene in each sample. The CT values of all 48 genes examined in both snap-frozen and FPE tissue were measured four times (four replicates) for each of the six matched OEA samples. The average and range of CVs for the four replicates of each gene were calculated for ΔCT values, which were used instead of CT values so that different amounts of RNA added to each of the four replicates would not be reflected in the average CV.
To examine the correlation between matched frozen and FPE tissue, average ΔCT values for the four replicates of each gene were calculated in each of the six matched OEA samples. A two-dimensional plot was then created depicting ΔCT values from the six frozen samples as the explanatory variable (x) and ΔCT values from the six paraffin-embedded samples as the dependent variable (y). Linear regression analysis and Pearson correlation was then performed to determine the agreement in gene expression between frozen and FPE tissue. A similar comparison of amplified and unamplifed RNA was performed, however CT values were used instead of ΔCT values to illustrate the significant number of genes that could not be detected after amplification.
To distinguish significant differences between expression levels in OEA relative to normal tissue, a one-sample, two-sided t-test was applied to compare the average expression level of the six samples to the normalized ovarian epithelium (which was assigned an expression level of 1.00 for each of the genes examined). The significance level for this test is 0.025.
Results
Quantitation of Housekeeping Gene Expression in Human Tissues Using RTQ
Comparative analysis demonstrated no significant (P < 0.01) difference in RPLP0 expression between normal (OSE) and neoplastic (OEA) tissues.
Reliability of TLDA
For either frozen or FPE samples, the average coefficient of variance (CV) for all four replicates, using ΔCT values, is ∼5%, with a SD of 5%, over the 48 genes examined. In calculating the average CV, genes with a CT value of 40, the default upper limit PCR cycle number that defines no signal, as well as RPLP0 and 18S, were excluded. Those genes that failed to express (CT = 40) are designated as NE in Table 1 and include insulin promoter factor 1 (IPF1), Wnt16, Wnt8B, and Wnt9A.
PCR Amplification Efficiency
Analysis of the standard curves for RPLP0 amplification in both frozen and FPE OEA RNA yielded slopes of −3.00 and −3.11, respectively. The PCR amplification efficiency was calculated as 108% for frozen tissue and 105% for FPE tissue.
Correlation of Gene Expression in Matched Frozen and FPE Tissue
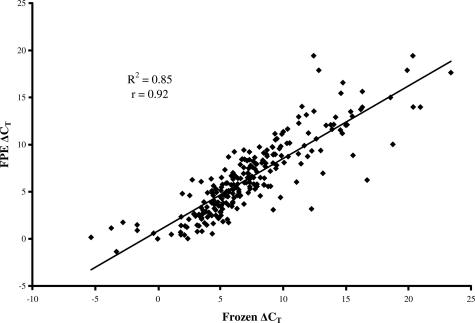
To determine whether archival FPE tissue is suitable for use with TLDA, we examined the correlation of 48 genes in frozen versus FPE OEA. As shown in Figure 2, a two-dimensional plot depicting average ΔCT values from the six frozen (x) and matching six FPE samples (y) demonstrates a significant (P < 0.0001) linear correlation (r2 = 0.85) with a Pearson correlation coefficient (r) of 0.92.
Figure 2.
Correlative plot of mean ΔCT values obtained by TLDA analysis of 48 genes in six frozen (x) compared to six matching paraffin-embedded OEA samples (y). Regression analysis demonstrated a coefficient of determination (r2) of 0.85 and a Pearson correlation coefficient (r) of 0.92 (P < 0.0001).
Correlation of Gene Expression in Amplified and Unamplified RNA
To examine whether commercially available kits could be used to amplify small amounts of RNA without altering the expression profile, we examined the correlation of 48 genes in matched amplified versus unamplified frozen and FPE OEA samples. Total RNA from frozen OEA successfully amplified using the Ovation, MessageAmp RiboAmp, and Full Spectrum amplification protocols and resulted in increases of 448-, 110-, 2757-, and 333-fold, respectively. Repeated attempts to amplify total RNA isolated from FPE tissue were unsuccessful using the four protocols examined.
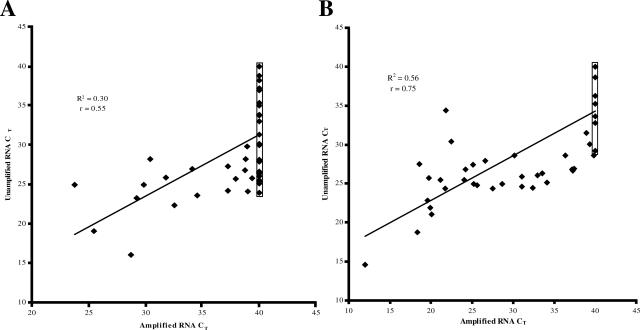
The conservation of gene expression in matched amplified versus unamplified (template) RNA from frozen OEA samples was examined using TLDA. As shown in Figure 3A, a two-dimensional plot depicting average CT values from the amplified (x) and unamplified (y) frozen OEA sample using the Ovation amplification system demonstrates a weak correlation with r = 0.55 (P < 0.0001). Twenty-four of a total forty-two genes (57%) that expressed in unamplified RNA were undetectable after amplification (as shown by the boxed data points). Similar results were obtained for RNA amplified using the MessageAmp and RiboAmp kits (data not shown). Figure 3B depicts a comparison of the same frozen OEA sample using the Full Spectrum amplification system and demonstrates a stronger correlation with r = 0.75 (P < 0.0001) and only 12 of 42 genes (29%) becoming undetectable after amplification. Although CT values were plotted instead of ΔCT values to emphasize undetectable genes in amplified RNA (boxed), identical r2 and r values were obtained when ΔCT values were used (data not shown).
Figure 3.
Correlative plot of CT values obtained by TLDA analysis of 48 genes in amplified (x) compared to unamplified (template) (y) using the Ovation Nanosample RNA amplification system with a coefficient of determination (r2) of 0.30 and a Pearson correlation coefficient (r) of 0.53 (P < 0.0001) (A); and the Full Spectrum Global RNA amplification kit with a coefficient of determination (r2) of 0.56 and a Pearson correlation coefficient (r) of 0.75 (P < 0.0001) (B).
Expression of Hedgehog and Wnt Genes in OEA
The expression of 46 independent genes in the hedgehog and Wnt pathways across six pairs of matched frozen and FPE OEA samples are summarized in Table 1. For both the frozen and FPE categories, each gene expression value represents an average of the six samples ± SD. Thus, the standard deviations in Table 1 reflect the range in expression over the six samples examined. Gene expression and statistical analysis show that 11 genes from frozen and 10 genes from FPE (Table 1, bolded) were differentially expressed in OEA compared to OSE (91% agreement).
Three members of the hedgehog pathway (DHH, IHH, HHIP) and three members of the Wnt pathway (Wnt9B, Wnt10B, WIF1) were found to be tumor-specific in both frozen and FPE tissues. The average expression, SD and P value for each of these genes could not be calculated because none of them express in the OSE calibrator. These six genes are designated as X in Table 1. Several other hedgehog- and Wnt-related genes were significantly lower in OEA (both frozen and FPE) compared to normal ovarian epithelium including cyclin E2 (CCNE2), Porcupine (PPN), c-Myc (MYC), and Axin 2 (AXIN2). The proapoptotic Gli-pathogenesis-related protein (GLIPR1) was significantly lower in frozen but not FPE OEA compared to normal ovarian epithelium. The difference in expression of the remaining 19 and 12 genes in the hedgehog and Wnt pathways, respectively, were not found to be significantly different due to their range in expression over the samples examined (Table 1). Insulin promoter factor 1 (IPF1), Wnt16, Wnt8B, and Wnt9A did not express in either OSE or OEA (designated as NE in Table 1).
Validation of TLDA
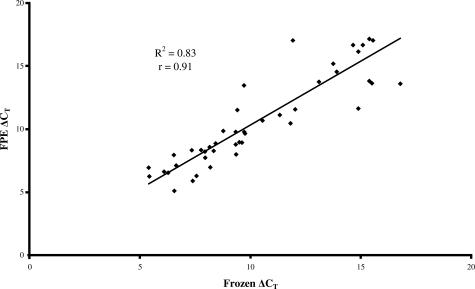
To validate the gene expression results obtained with TLDA, 8 genes (DHH, IHH, SHH, PTCH, PTCH2, SMO, GLI, GLI3) of the original 46 were individually analyzed in both frozen and FPE samples using TaqMan RTQ. A similar gene expression correlation (r = 0.91, P < 0.01) (Figure 4) between matched frozen and FPE tissues was obtained. When compared separately, gene expression values for these eight genes were not significantly different from the values obtained with TLDA in either frozen or FPE OEA (data not shown).
Figure 4.
Correlative plot of ΔCT values obtained by RTQ analysis of eight genes (DHH, IHH, SHH, PTCH, PTCH2, SMO, GLI, GLI3) in six frozen (x) compared to six matching paraffin-embedded OEA samples (y). Regression analysis demonstrated a coefficient of determination (r2) of 0.83 and a Pearson correlation coefficient (r) of 0.91 (P < 0.0001).
Discussion
Initial studies examined the reliability and amplification efficiency of the TLDA method and demonstrated an intra-assay CV of ∼5% with almost equal PCR amplification efficiency in RNA isolated from matched frozen and FPE tissues (108% and 105%, respectively). Pearson correlation (r) between the six matched fresh frozen and FPE OEA tissues for all tested genes was 92% (Figure 2). This agreement was further validated when similar gene expression correlation (r = 0.91, Figure 4) between the matched OEA samples was obtained by individually analyzing 8 genes of the original 46 using RTQ. This multivariate analysis is unique in that both snap-frozen and matched archival FPE specimens, routinely processed through the University of Alabama at Birmingham’s Department of Pathology, were examined simultaneously. Although encouraging, future studies involving the analysis of expression profiles in FPE tissues should consider the variability in the stability of any particular gene being examined and must be independently validated.
Based on these promising preliminary data, we examined whether RNA amplification could be used in combination with TLDA. RNA amplification is a method whereby nanogram amounts of total RNA (usually obtained from needle biopsies or laser-capture microdissected clinical samples) undergo a multistep process for linear amplification of the mRNA fraction. Pearson correlation (r) between matched amplified versus unamplified RNA isolated from frozen OEA varied among the four amplification protocols tested from a low of 53% to a high of 75% (Figure 3). The three amplification protocols (Ovation, MessageAmp, RiboAmp) using polydT oligomer priming (which requires the 3′ poly-A tail in template mRNA to bind with a modified oligo-dT primer) gave the poorest correlations of 53% (Figure 3A), 44%, and 55% (data not shown), respectively. The Full Spectrum amplification protocol, which utilizes a random hexamer (N6) priming method that does not require an intact 3′ poly-A tail, demonstrated the best correlation of 75% between amplified and unamplified RNA (Figure 3B). Unfortunately, RNA isolated from FPE samples failed to amplify using any of the four procedures and correlative studies could not be conducted. Collectively, these data suggest random hexamer priming may have an advantage over oligo-dT priming when comparing expression profiles in snap-frozen tissues but that sheared or degraded RNA (such as that obtained from FPE tissues) cannot be amplified using these protocols.
To identify potential tumor-specific therapeutic targets, we used TLDA to quantify the expression of 26 and 20 genes in the hedgehog and Wnt pathways, respectively, in six matched frozen and FPE OEA specimens (Table 1). These pathways, normally involved in embryogenesis, have both been implicated in cancer initiation and are similar in terms of posttranslational modification, secretion, and some signaling mechanisms and may have evolved from a common pathway.13 Recent studies have shown activation of the hedgehog signaling pathway in adult tissues can initiate and sustain tumor growth; however, this pathway has never been examined in OEA.5,6,7,8,9,10 Inhibition of the hedgehog pathway by small molecule inhibitors such as cyclopamine has been shown to be effective in decreasing tumor growth and is a promising new therapeutic strategy.26,27 Similarly, Wnt signaling is involved in normal follicular development and ovarian function.28 Because OSE is believed to be the origin of ovarian adenocarcinomas and Wnts have been implicated in oncogenic transformation of epithelial cells, it is thought that aberrant expression of this pathway could lead to ovarian carcinogenesis.16 Quantitation of hedgehog- and Wnt-associated genes in OEA revealed several tumor-specific genes including the two hedgehog ligands, DHH and IHH, and hedgehog pathway regulator, HHIP, as well as the two Wnt ligands, Wnt10B and Wnt9B, and Wnt pathway regulator, WIF1 (Table 1). Interestingly, these genes did not consistently express in all of the OEA samples. Other genes directly involved in the hedgehog and Wnt pathways including Smoothened (SMO), Glioma-associated oncogene (GLI), GLI2, GLI3, and Wnt7A, Frizzled homolog (FRZD1), low-density lipoprotein receptor-related protein 6 (LRP6), and Frizzled related protein (FRZB) were all overexpressed in both frozen and FPE OEA in comparison to normal OSE, but did not reach statistical significance. Similar gene expression results were obtained when eight differentially expressed genes (DHH, IHH, SHH, PTCH, PTCH2, GLI, GLI3, SMO) were examined individually using RTQ in both frozen and FPE samples (data not shown). In relation to statistically significant and tumor-specific genes, the differing genetic profiles among the OEA samples suggest variable activity of the hedgehog and Wnt pathways in this cancer. Thus, future studies involving individual analysis of a larger population of both normal and cancer patients to distinguish tumor-specific differences from interindividual variation are warranted. These studies could then offer the potential of identifying patients with advanced OEA who would benefit from anti-hedgehog or anti-Wnt therapy such as cyclopamine and nonsteroidal anti-inflammatory drugs, respectively.27,29,30
The TLDA methodology presented in this study represents a robust and reproducible technique for quantifying gene expression in tens to hundreds of independent genes concurrently in RNA samples isolated from either frozen or FPE tissues. This approach represents a significant advance in multivariate gene analysis that is less time and labor intensive than individually analyzing single genes by RTQ. The correlation of gene expression profiles between matched frozen and FPE tissues offers the exciting possibility that archival, paraffin-embedded tissues (a more abundant alternative to frozen tissue available from most cooperative groups) may be examined to identify specific therapeutic targets and/or prognostic indicators. The importance of these multivariate analyses have been emphasized by recent studies that have shown that examination of genes acting collectively in a specific pathway, such as the hedgehog or Wnt pathway, offers more information about clinical outcome than examination of individual genes.31,32,33,34 In the current study, TLDA analysis was used to quantify the expression of hedgehog and Wnt-related genes in OEA and determined that elements of both these pathways are expressed in a subset of the patient samples examined. These analyses could potentially be used to identify patients with advanced OEA who would benefit from specifically targeted anti-hedgehog and/or anti-Wnt therapy.
Footnotes
Supported by the National Institutes of Health (grants CA101955-01 and CA086359-06).
References
- Greenlee RT, Murray T, Bolden S, Wingo PA. Cancer statistics, 2000. CA Cancer J Clin. 2000;50:7–33. doi: 10.3322/canjclin.50.1.7. [DOI] [PubMed] [Google Scholar]
- Engel J, Eckel R, Schubert-Fritschle G, Kerr J, Kuhn W, Diebold J, Kimmig R, Rehbock J, Holzel D. Moderate progress for ovarian cancer in the last 20 years: prolongation of survival, but no improvement in the cure rate. Eur J Cancer. 2002;38:2435–2445. doi: 10.1016/s0959-8049(02)00495-1. [DOI] [PubMed] [Google Scholar]
- Goodman MT, Correa CN, Tung KH, Roffers SD, Cheng Wu X, Young JL, Jr, Wilkens LR, Carney ME, Howe HL. Stage at diagnosis of ovarian cancer in the United States, 1992–1997. Cancer. 2003;97:2648–2659. doi: 10.1002/cncr.11347. [DOI] [PubMed] [Google Scholar]
- McGuire WP, Hoskins WJ, Brady MF, Kucera PR, Partridge EE, Look KY, Clarke-Pearson DL, Davidson M. Cyclophosphamide and cisplatin versus paclitaxel and cisplatin: a phase III randomized trial in patients with suboptimal stage III/IV ovarian cancer (from the Gynecologic Oncology Group). Semin Oncol. 1996;23:40–47. [PubMed] [Google Scholar]
- Wicking C, Smyth I, Bale A. The hedgehog signalling pathway in tumorigenesis and development. Oncogene. 1999;18:7844–7851. doi: 10.1038/sj.onc.1203282. [DOI] [PubMed] [Google Scholar]
- Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- Berman DM, Karhadkar SS, Hallahan AR, Pritchard JI, Eberhart CG, Watkins DN, Chen JK, Cooper MK, Taipale J, Olson JM, Beachy PA. Medulloblastoma growth inhibition by hedgehog pathway blockade. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- Gailani MR, Stahle-Backdahl M, Leffell DJ, Glynn M, Zaphiropoulos PG, Pressman C, Unden AB, Dean M, Brash DE, Bale AE, Toftgard R. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nat Genet. 1996;14:78–81. doi: 10.1038/ng0996-78. [DOI] [PubMed] [Google Scholar]
- Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- Kubo M, Nakamura M, Tasaki A, Yamanaka N, Nakashima H, Nomura M, Kuroki S, Katano M. Hedgehog signaling pathway is a new therapeutic target for patients with breast cancer. Cancer Res. 2004;64:6071–6074. doi: 10.1158/0008-5472.CAN-04-0416. [DOI] [PubMed] [Google Scholar]
- Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, Qi YP, Gysin S, Fernandez-del Castillo C, Yajnik V, Antoniu B, McMahon M, Warshaw AL, Hebrok M. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman DM, Karhadkar SS, Maitra A, Montes De Oca R, Gerstenblith MR, Briggs K, Parker AR, Shimada Y, Eshleman JR, Watkins DN, Beachy PA. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- Nusse R. Wnts and Hedgehogs: lipid-modified proteins and similarities in signaling mechanisms at the cell surface. Development. 2003;130:5297–5305. doi: 10.1242/dev.00821. [DOI] [PubMed] [Google Scholar]
- Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- Wu R, Zhai Y, Fearon ER, Cho KR. Diverse mechanisms of beta-catenin deregulation in ovarian endometrioid adenocarcinomas. Cancer Res. 2001;61:8247–8255. [PubMed] [Google Scholar]
- Rask K, Nilsson A, Brannstrom M, Carlsson P, Hellberg P, Janson PO, Hedin L, Sundfeldt K. Wnt-signalling pathway in ovarian epithelial tumours: increased expression of beta-catenin and GSK3beta. Br J Cancer. 2003;89:1298–1304. doi: 10.1038/sj.bjc.6601265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DP, Quirke P, Lewis F, Boylston AW, Sloan JM, Robertson D, Taylor GR. Detection of measles virus RNA in paraffin-embedded tissue. Lancet. 1989;1:1391. doi: 10.1016/s0140-6736(89)92837-7. [DOI] [PubMed] [Google Scholar]
- Jackson DP, Lewis FA, Taylor GR, Boylston AW, Quirke P. Tissue extraction of DNA and RNA and analysis by the polymerase chain reaction. J Clin Pathol. 1990;43:499–504. doi: 10.1136/jcp.43.6.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanta G, Schneider C. RNA extracted from paraffin-embedded human tissues is amenable to analysis by PCR amplification. Biotechniques. 1991;11:304–308. [PubMed] [Google Scholar]
- Finke J, Fritzen R, Ternes P, Lange W, Dolken G. An improved strategy and a useful housekeeping gene for RNA analysis from formalin-fixed, paraffin-embedded tissues by PCR. Biotechniques. 1993;14:448–453. [PubMed] [Google Scholar]
- Stanta G, Bonin S, Perin R. RNA extraction from formalin-fixed and paraffin-embedded tissues. Methods Mol Biol. 1998;86:23–26. doi: 10.1385/0-89603-494-1:23. [DOI] [PubMed] [Google Scholar]
- Goldsworthy SM, Stockton PS, Trempus CS, Foley JF, Maronpot RR. Effects of fixation on RNA extraction and amplification from laser capture microdissected tissue. Mol Carcinog. 1999;25:86–91. [PubMed] [Google Scholar]
- Blanquicett C, Johnson MR, Heslin M, Diasio RB. Housekeeping gene variability in normal and carcinomatous colorectal and liver tissues: applications in pharmacogenomic gene expression studies. Anal Biochem. 2002;303:209–214. doi: 10.1006/abio.2001.5570. [DOI] [PubMed] [Google Scholar]
- Johnson MR, Wang K, Smith JB, Heslin MJ, Diasio RB. Quantitation of dihydropyrimidine dehydrogenase expression by real-time reverse transcription polymerase chain reaction. Anal Biochem. 2000;278:175–184. doi: 10.1006/abio.1999.4461. [DOI] [PubMed] [Google Scholar]
- Roy S, Ingham PW. Hedgehogs tryst with the cell cycle. J Cell Sci. 2002;115:4393–4397. doi: 10.1242/jcs.00158. [DOI] [PubMed] [Google Scholar]
- Incardona JP, Gaffield W, Kapur RP, Roelink H. The teratogenic Veratrum alkaloid cyclopamine inhibits sonic hedgehog signal transduction, Development. 1998;125:3553–3562. doi: 10.1242/dev.125.18.3553. [DOI] [PubMed] [Google Scholar]
- Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- Ricken A, Lochhead P, Kontogiannea M, Farookhi R. Wnt signaling in the ovary: identification and compartmentalized expression of wnt-2, wnt-2b, and frizzled-4 mRNAs. Endocrinology. 2002;143:2741–2749. doi: 10.1210/endo.143.7.8908. [DOI] [PubMed] [Google Scholar]
- Boon EM, Keller JJ, Wormhoudt TA, Giardiello FM, Offerhaus GJ, van der Neut R, Pals ST. Sulindac targets nuclear beta-catenin accumulation and Wnt signalling in adenomas of patients with familial adenomatous polyposis and in human colorectal cancer cell lines. Br J Cancer. 2004;90:224–229. doi: 10.1038/sj.bjc.6601505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbie Z, Muller PY, Heinimann K, Albrecht C, D’Orazio D, Bendik I, Muller H, Bauerfeind P. Expression of COX-2 and Wnt pathway genes in adenomas of familial adenomatous polyposis patients treated with meloxicam. Anticancer Res. 2002;22:2215–2220. [PubMed] [Google Scholar]
- Zhang Z, Bast RC, Jr, Yu Y, Li J, Sokoll LJ, Rai AJ, Rosenzweig JM, Cameron B, Wang YY, Meng XY, Berchuck A, Van Haaften-Day C, Hacker NF, de Bruijn HW, van der Zee AG, Jacobs IJ, Fung ET, Chan DW. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004;64:5882–5890. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zheng H, Yang X, Sun L, Xin Y. Effects of mutation and expression of PTEN gene mRNA on tumorigenesis and progression of epithelial ovarian cancer. Chin Med Sci J. 2004;19:25–30. [PubMed] [Google Scholar]
- Torng PL, Mao TL, Chan WY, Huang SC, Lin CT. Prognostic significance of stromal metalloproteinase-2 in ovarian adenocarcinoma and its relation to carcinoma progression. Gynecol Oncol. 2004;92:559–567. doi: 10.1016/j.ygyno.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Barbieri F, Lorenzi P, Ragni N, Schettini G, Bruzzo C, Pedulla F, Alama A. Overexpression of cyclin D1 is associated with poor survival in epithelial ovarian cancer. Oncology. 2004;66:310–315. doi: 10.1159/000078332. [DOI] [PubMed] [Google Scholar]