Abstract
The recent discovery of a single point mutation in the JH2 pseudokinase domain of Janus kinase 2 (JAK2) in a considerable fraction of patients has shed light on the molecular pathomechanism in Philadelphia chromosome-negative chronic myeloproliferative disorders (Ph− CMPDs). We established a robust and reliable method for detection of the JAK2 mutation in bone marrow cells derived from archival bone marrow trephines based on polymerase chain reaction and subsequent restriction site analysis. In a series of proven Ph− CMPDs classified according to World Health Organization criteria (n = 79), we detected the JAK2 mutation in 90% of polycythemia vera, 22% of cellular prefibrotic chronic idiopathic myelofibrosis, 60% of advanced chronic idiopathic myelofibrosis, and 27% of essential thrombocythemia. JAK2 mutation was not detected in Ph+ chronic myeloid leukemia (n = 5), acute myeloid leukemia (n = 10), acute lymphoblastic leukemia (n = 10), secondary erythrocytosis (n = 10), or normal bone marrow (n = 10). Restriction site analysis was also suitable for unfixed cell populations derived from peripheral blood and bone marrow aspirates. Besides providing support in the differential diagnosis of reactive versus neoplastic myeloproliferations, this newly developed assay reveals considerable overlaps between histologically different disease entities, indicating that additional genetic alterations might be responsible for the established differences of CMPD subentities.
Chronic myeloproliferative disorders (CMPDs) comprise heterogenous stem cell malignancies that differ in their clinical course because of development of bone marrow fibrosis and evolution to blast crisis. According to the World Health Organization classification, CMPDs are referred to as polycythemia vera (PV), essential thrombocythemia (ET), chronic idiopathic myelofibrosis (cIMF) in the cellular, prefibrotic phase and with manifest myelofibrosis, and Philadelphia chromosome-positive chronic myeloid leukemia (Ph+ CML) along with more rare entities such as chronic neutrophilic leukemia and chronic eosinophilic leukemia.1 CMPDs arise from the clonal expansion of an as yet undefined hematopoietic progenitor cell, with subsequent overproduction of one or more of the formed elements of the blood.2 Except for Ph+ CML, the molecular mechanisms that induce sustained proliferation in Philadelphia chromosome-negative CMPDs (Ph− CMPDs) have been poorly understood3 until recently when a unique mutation in the Janus kinase 2 (JAK2) could be identified in the majority of patients with PV and a notable subset of patients with cIMF and ET.4,5,6,7
JAK2 is a cytoplasmic tyrosine kinase with a central role in initiating the signal transduction cascade after ligand binding to receptors of hematopoietic growth factors, cytokines, and interleukins. After autophosphorylation and building of docking sites, other kinases such as Src, signal transducer and activator of transcription (STATs), and related transcription factors become activated and efficiently regulate gene expression.8
In analogy to the uncontrolled tyrosine kinase activity of ABL that loses its autoinhibitory domain because of the reciprocal translocation t(9;22) and formation of the chimeric BCR-ABL protein in Ph+ CML, it has now been demonstrated that this single guanine-to-thymine point mutation in exon 12, encoding a valine-to-phenylalanine substitution at position 617 (Val617Phe), leads to a loss of function in the autoinhibitory JH2 pseudokinase domain of JAK2 and constitutive activity of its JH1 kinase domain.4,5,6,7,9 Subsequently, STATs and other transcription factors become up-regulated and force survival and proliferation of the affected clone even in the absence of an appropriate stimulus.10
Besides the major importance for subsequent investigations of JAK2-associated pathways and potential discovery of additional aberrations responsible for the formation of three rather distinct entities in Ph− CMPDs, the detection of JAK2 mutation will be of immense interest for the diagnostic setting and assessment of prognosis in hematopathology.
The aim of our study was 1) to establish a robust polymerase chain reaction (PCR) assay for amplification of the mutation-relevant site of the JH2 pseudokinase domain of JAK2 in bone marrow cells derived from archival bone marrow trephines, 2) to apply the JAK2 PCR on proven Ph− CMPD, secondary erythrocytosis, and normal hematopoiesis with subsequent restriction site analysis, 3) to reproduce results obtained by restriction site analysis through cloning/sequencing, and 4) to correlate the detection rate of JAK2 mutation in strictly World Health Organization-classified Ph− CMPD bone marrow trephines with published data obtained by JAK2 analysis of peripheral blood cells.
Materials and Methods
Bone Marrow Study Group
Formalin-fixed and paraffin-embedded (FFPE) bone marrow trephines with hematopathologically proven CMPD were retrieved from the bone marrow registry of the Institute of Pathology, Medizinische Hochschule Hannover. Bone marrow trephines were routinely fixed for 24 hours in a solution containing phosphate-buffered (pH 7.4) formalin. The decalcification step was performed in an EDTA-based solution (pH 7.5) for up to 48 hours. The study group (n = 127) comprised PV (n = 29), prefibrotic cellular cIMF (n = 18), advanced cIMF with manifest myelofibrosis (n = 20), ET (n = 15), Ph+ CML (n = 5), primary acute myeloid leukemia (n = 10), primary acute lymphoblastic leukemia (ALL, n = 10), secondary erythrocytosis clinically suspicious but without histological evidence for PV (n = 10), and 10 control cases showing normal hematopoiesis. According to the World Health Organization classification in close agreement with clinical data and presentation, patient bone marrow trephines were initially diagnosed in the years 2000 to 2004 to have CMPDs. As performed, cytogenetic analyses showed no chromosomal abnormalities within cIMF, ET, and PV cases. The prefibrotic stage of cIMF was distinguished from ET in the case of marked pleomorphism, atypias, clustering and dislocation of megakaryocytes, increase and hypolobulation of nuclei with decrease of chromatin density, marked increase of granulopoiesis, and slight focal increase of fiber density. ET was diagnosed in the case of occurrence of enlarged megakaryocytes showing hyperlobulation of nuclei and normal chromatin density but no significant pleomorphism, atypias and clustering, no significant increase of granulopoiesis, and a normal fiber content in bone marrow. cIMF cases were re-evaluated and subdivided into two groups depending on the degree of myelofibrosis as described previously.11 In particular, subtyping (myelofibrosis [MF]) of cIMF cases depends on the type and amount of fiber (reticulin versus collagen), the fiber spread, the appearance of sinusoidal sclerosis and intrasinusoidal hematopoiesis, and the existence of endophytic bone formation. Accordingly, cellular cIMF cases displayed no (MF0) or only a minor deposit of reticulin fibers (MF1), whereas manifest cIMF cases showed severe myelofibrosis with deposit of collagen, sclerotic sinusoids with sinusoidal hematopoiesis (MF2), and a remarkable osteosclerosis with bone apposition (MF3). Subsequently, two groups were assembled comprising cIMF in the cellular stage (MF0 and MF1) and cIMF with severe myelofibrosis and osteosclerosis (MF2 and MF3). In accordance with cIMF, other Ph− CMPD entities and control cases of the study group were re-evaluated by two expert hematopathologists before the JAK2 mutation study. Peripheral blood and bone marrow aspirates were available from two patients with proven diagnosis of PV and two patients with suspected CMPD for subsequent molecular analysis of JAK2 mutation status in fresh, unfixed cells.
RNA Extraction from FFPE Bone Marrow Trephines and Fresh, Unfixed Cell Populations
As we previously described,12,13 total RNA was extracted from total bone marrow cells after guanidinium isothiocyanate and proteinase K-based digestion and conventional organic extraction using phenol/chloroform. After precipitation with 2-propanol and glycogen as an RNA carrier, the RNA pellet was collected, washed with 70% ice-cold ethanol, and dissolved in 25 to 35 μl of diethylpyrocarbonate-treated H2O. Afterward, a second precipitation step with 1/10 volume of 3 mol/L sodium acetate (pH 5.2) and 2.5-fold volume of 100% ethanol was performed to increase purity. After overnight precipitation, samples were centrifuged, and the RNA pellet was washed and later dissolved in DEPC-treated H2O as described above. The RNA concentration was then measured spectrophotometrically. RNA from granulocytes and mononuclear cells was extracted with the RNeasy kit (Qiagen, Hilden, Germany) after isolation of cells using a Ficoll-Paque gradient according to standard procedures.
Synthesis of Complementary DNA
Total RNA (1 μg), pretreated with RNase-free (Rnase−) DNase (1 U/μg RNA; RQ1; Promega, Madison, WI), was transcribed into the complementary DNA using 500 ng of random hexamers (Amersham Pharmacia, Picattaway, CA) and 200 U of SuperScript II Rnase− Reverse Transcriptase (Invitrogen, Karlsruhe, Germany) in a volume of 20 μl following the manufacturer’s protocol. Negative controls were performed using water instead of Reverse Transcriptase.
Multiplex Reverse Transcriptase-PCR (RT-PCR) for Detection of Common BCR-ABL Fusion Transcripts in FFPE Bone Marrow Trephines
As previously described,14 a multiplex RT-PCR assay was applied to detect common BCR-ABL fusion transcripts in FFPE bone marrow cells. Primers for ABL control RT-PCR were as follows: ABL forward, 5′-AAAATGGCCAAGGCTGGG-3′, and ABL reverse, 5′-ACCAGG-AGTGTTTCTCCAGACTG-3′, generating a 69-bp product. Multiplex forward primers were located in Major-BCR b2 (5′-ATCCGTGGAGCTGCAGATG-3′), b3 (5′-GAGTCTC-CGGGGCTCTATGG-3′), and minor-BCR e1 (5′-AGATCTGGCCCAACGATGGCGA-3′) along with a mutual reverse primer located in ABL exon 2 (5′-TCAGATGCTACTGGCCGCTGAA-3′), generating products of 111 bp (b2a2), 98 bp (b3a2), and 73 bp (e1a2). By using a mutual reverse primer located in ABL exon 3 (5′-TGTGATTATAGCCTAAGACC-3′) in combination with forward primer b2, b3, and e1, more rare fusion transcripts such as b2a3 (118 bp), b3a3 (103 bp), and e1a3 (79 bp) could be detected. Cell lines BV173 (b2a2), K562 (b2a2), and SD-1 (e1a2) served as positive controls. ABL control and multiplex BCR-ABL detection assays were performed in a GeneAmp PCR System 2700 (Applied Biosystems, Weiterstadt, Germany). RT-PCR amplification was performed with a final reaction mixture of 25 μl containing 250 nmol/L of each primer, 0.5 U of Platinum Taq Polymerase (Invitrogen), 200 μmol/L each of dATP, dCTP, dTTP, and dGTP in 1× Platinum Taq reaction buffer with 1.5 mmol/L magnesium chloride, and 3.0 μl of cDNA. The reaction mixture was preheated at 95°C for 5 minutes, followed by 40 cycles at 95°C, 62°C, and 72°C, 30 seconds each. Final extension was performed for 5 minutes at 72°C. RT-PCR reactions were performed in duplicate with a single reverse transcriptase-negative sample. Single positive control reactions were performed for b2a2(a3), b3a2(a3), and e1a2(a3). Polyacrylamide gel-electrophoresis (8% PAGE) was performed on patient samples for separation and documentation of potential BCR-ABL products.
Amplification of JAK2
PCR amplification of JAK2 was performed in a GeneAmp PCR System 2700 (Applied Biosystems) with a final reaction mixture of 25 μl containing 250 nmol/L of each primer, 0.5 U of Platinum Taq Polymerase (Invitrogen), 200 μmol/L each of dATP, dCTP, dTTP, and dGTP in 1× Platinum Taq reaction buffer with 2.5 mmol/L magnesium chloride, and 3.0 μl of cDNA (corresponding to 150 ng of total RNA input). The reaction mixture was preheated at 95°C for 5 minutes, followed by 40 cycles at 95°C, 60°C, and 72°C, 30 seconds each. Final extension was performed for 5 minutes at 72°C.
Primers for JAK2 amplification are located within exons 12 and 13 of the JAK2 JH2 pseudokinase domain and cover the hotspot mutation site corresponding to nucleotide 1849 (see GenBank sequence AF001362). Primers for JAK2 were JAK2 forward, 5′-TATGATGAGCAAGCTTTCTCACAAG-3′, and JAK2 reverse, 5′-TCCAAATTTACAAACTCCTGAACC-3′, generating a 100-bp product. For location of JAK2 primers, see Figure 1. β-Glucuronidase (β-GUS) amplification was chosen as the control for sufficient cDNA synthesis, and primers were β-GUS forward, 5′-CTCATTTGGAATTTTGCCGATT-3′, and β-GUS reverse, 5′-CCGAGTGAGATCCCCTTTTTA-3′, generating an 81-bp product (GenBank sequence NM000181).
Figure 1.
Hotspot nucleotide at position 1849 of GenBank sequence AF001362 represents a guanine (enlarged, in bold and underlined). Forward and reverse primers for JAK2 PCR are italicized and underlined. Sequence illustration kept in Fast Alignment format to allow reproduction. The JAK2 PCR generates a 100-bp product.
Restriction Site Analysis
After visualization of the JAK2 PCR product on 6% PAGE, 10 μl of a given sample (approximately 1 μg DNA) was incubated with 4 U of the restriction enzyme BsaXI (2 U/μl; New England Biolabs, Beverly, MA) for 16 hours at 37°C in a PCR cycler with heated lid to avoid any sample evaporation. The BsaXI enzyme recognized a site (5′…. 9(N) A C (N)5 C T C C (N)10…. 3′) that included the hotspot nucleotide at position 1849 in GenBank sequence AF001362 (for demonstration of the relevant recognition site in the JH2 pseudokinase domain, see Figure 2A). For determination of BsaXI cuts in the respective JAK2 sequence, the NEBcutter 2.0 software tool was used (http://tools.neb.com/NEBcutter2/index.php). Samples analyzed by restriction site analysis were strictly accompanied by a positive control (PV patient with a proven JAK2 mutation) and a negative control (normal hematopoiesis).
Figure 2.
Identical sequence segment as shown in Figure 1 marked the recognition site for BsaXI. Relevant nucleotides are in bold. A: Triangles mark the two cut sites of BsaXI leading to digestion of the amplified JAK2 (entire amplicon size italicized) in fragments of 36, 34, and 30 bp. B: Hotspot guanine-to-thymine point mutation (mutated thymine in bold) destroys a relevant recognition site for BsaXI and prevents digestion of the JAK2 amplicon (italicized).
Cloning and Sequencing
After visualization on 6% PAGE, 4 μl of the JAK2 PCR product was inserted into a plasmid vector using the TOPO cloning kit (Invitrogen), followed by transformation into DH5-α-T1 cells. Recombinant plasmid DNA was isolated from overnight cultures of randomly selected bacterial colonies by means of QIAprep purification columns (Qiagen). Sequencing was conducted using the CycleReader AutoDNA Sequencing kit (Fermentas, St. Leon-Rot, Germany) and an automated nucleic acid sequence analyzer (LI-COR Bioscience, Lincoln, NE). The sequence covering the hotspot nucleotide in the JH2 pseudokinase domain of JAK2 from 5 (control cases) and 10 (PV and cIMF patients) randomly selected plasmids was aligned and compared with the GenBank sequence AF001362 of the respective part of the JH2 pseudokinase domain.
Restriction Site Analysis of JAK2 Clones
To determine the rate of clones carrying mutated JAK2 in a given sample, we first performed PCR using the M13 forward and reverse primer set pertinent to the TOPO cloning kit according to the manufacturer’s instructions. Briefly, the M13 primer set amplifies 90- and 76-bp sequences upstream and downstream of the inserted JAK2 sequence spanning 100 bp. PCR products (10 μl) were analyzed by 6% PAGE to allow visualization of the expected 266-bp PCR products. Afterward, 10 μl of PCR product of each clone was digested with BsaXI for 16 hours at 37°C, and restriction fragments were visualized using another 6% PAGE.
Results
Multiplex RT-PCR for Potential BCR-ABL Rearrangement
All CMPD cases were analyzed for detection of common BCR-ABL fusion transcripts to exclude a potential chromosomal translocation t(9;22). A multiplex RT-PCR approach was performed as described in Materials and Methods. Except for Ph+ CML cases and two cases with Ph+ ALL, no BCR-ABL fusion transcript could be detected within the study group.
JAK2 Could Be Sufficiently Amplified by RT-PCR in Bone Marrow Cells Derived from Archival Bone Marrow Trephines
The primer system covering the hotspot nucleotide in the JH2 pseudokinase domain was located within exons 12 and 13 and generated a 100-bp product, taking into account the inevitable fragmentation of RNA derived from FFPE samples. All cases analyzed showed positive control RT-PCR (β-GUS) and sufficient JAK2 amplification with no relevant side products (Figure 3).
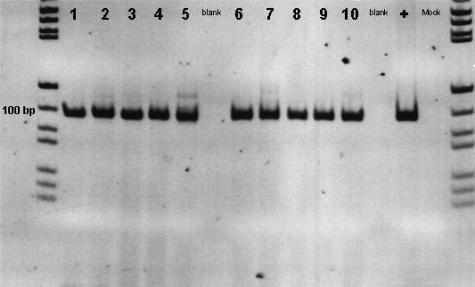
Figure 3.
JAK2 derived from bone marrow cells was amplified by RT-PCR and visualized by PAGE. Representative gel documentation showed the 100-bp JAK2 products without relevant side products. Lanes 1 to 4 represent JAK2 in PV patients; lane 5 represents JAK2 in a patient with cIMF; lanes 6 to 10 represent JAK2 in normal hematopoiesis; lane + represents JAK2 in cell line HL-60; and lane Mock represents PCR without template input. Note that the figure was digitally inverted.
Restriction Site Analysis of the JAK2 JH2 Pseudokinase Domain Region Harboring the Hotspot Nucleotide Discriminates Mutated from Unmutated Ph− CMPD and Non-Neoplastic Hematopoiesis
Extensive tests were performed to determine the appropriate concentration of BsaXI and time of incubation required for reliable digestion. In a series of control samples (n = 5), incubation with BsaXI was stopped at different time points (2, 4, 6, and 16 hours) to determine digestion status of unmutated JAK2 products. Complete digestion could regularly be demonstrated after 2 hours of incubation (Figure 4, A and B) in all control samples under study. Mutated cases were digested with an excess of BsaXI to ensure that neither a lower concentration of enzyme nor inadequate incubation time could impair completeness of digestion. Finally, JAK2 PCR products were digested with a twofold excess of BsaXI at 37°C for 16 hours. According to the manufacturer’s instructions, enzyme activity of BsaXI is ensured up to 16 hours.
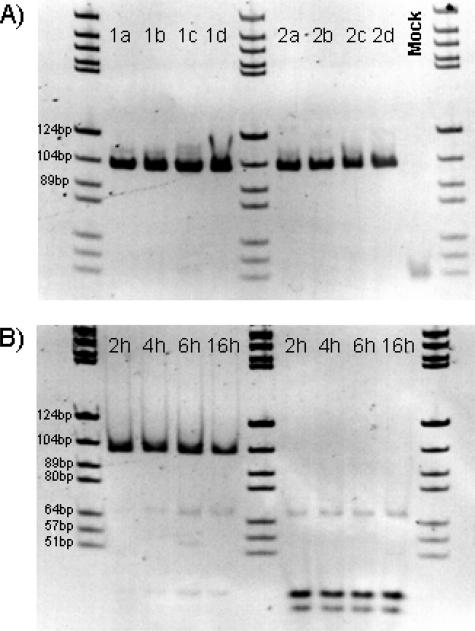
Figure 4.
A: JAK2 PCR products derived from a PV patient carrying the JAK2 mutation (1a–1d) and a control sample (2a–2d) were digested with 4 U of BsaXI for 2, 4, 6, and 16 hours. B: In the control sample, virtually complete digestion of the initial JAK2 product could be demonstrated after 2 hours of incubation, whereas in the PV patient, the JAK2 product was almost undigested even after 16 hours of incubation. Six-percent PAGE, DNA marker pBR322 BsuRI, and marker size fragments are partially labeled. Note that the figure is digitally inverted.
BsaXI cuts at position 36 (+3 nucleotide extension at 3′) and position 66 of the JAK2 PCR product. The expected JAK2 fragments after complete digestion were 36, 34, and 30 bp (Figure 2A, schematic illustration). The single guanine-to-thymine point mutation destroys the BsaXI recognition site and disables digestion (Figure 2B, schematic illustration).
After BsaXI digestion, the integrity of the JAK2 band varied in the group of patients harboring the mutation. Because clonal and non-clonal cells contributed to amplified JAK2, a predominant band of undigested JAK2, aside from digested fragments, could be demonstrated. In contrast, unmutated JAK2 in Ph− CMPD cases, Ph+ CML, acute myeloid leukemia, ALL, secondary erythrocytosis, and control hematopoiesis showed complete digestion. Representative PAGE discriminated mutated and unmutated cases (Figure 5). For a comprehensive compilation of results obtained by restriction site analysis throughout the study group, see Table 1.
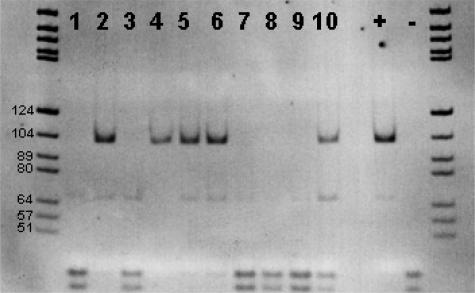
Figure 5.
A representative 6% PAGE loaded with JAK2 products after digestion with BsaXI according to Materials and Methods. Lanes 1 to 10 show mutated (2, 4, 5, 6, and 10) as well as unmutated (1, 3, 7, 8, and 9) cases with advanced cIMF together with a positive control (+) and a negative control (−). DNA marker pBR322 BsuRI, marker partially labeled. Note that the figure is digitally inverted.
Table 1.
Compilation of Results Determined by Restriction Site Analysis for JAK2 Mutation in Bone Marrow Cells Derived from CMPD and Non-Neoplastic Hematopoiesis
| Entity | Number of cases (n = 127) |
BsaXI restriction
|
JAK2 mutated (%) | |
|---|---|---|---|---|
| JAK2 mutated | JAK2 unmutated | |||
| PV | 29 | 26 | 3 | 90 |
| Cellular cIMF | 18 | 4 | 14 | 22 |
| Advanced cIMF | 20 | 12 | 8 | 60 |
| ET | 15 | 4 | 11 | 27 |
| Ph+ CML | 5 | 0 | 5 | 0 |
| AML | 10 | 0 | 10 | 0 |
| ALL | 10 | 0 | 10 | 0 |
| Secondary erythrocytosis | 10 | 0 | 10 | 0 |
| Normal hematopoiesis | 10 | 0 | 10 | 0 |
Restriction Site Analysis of the JAK2 JH2 Pseudokinase Domain Region Is Also Suitable on Unfixed Cell Populations Derived from Peripheral Blood and Bone Marrow Aspirates
JAK2 PCR and subsequent BsaXI restriction site analysis were also performed on fresh, unfixed granulocytes and mononuclear cells to confirm that this approach is suitable for both fixed and unfixed samples. Granulocyte and mononuclear cell fractions derived from peripheral blood and/or bone marrow aspirates from five patients (three patients with a known history of PV and two patients with suspected CMPDs) were analyzed according to Materials and Methods. Restriction site analysis clearly discriminated mutated cell fractions from unmutated fractions (Figure 6).
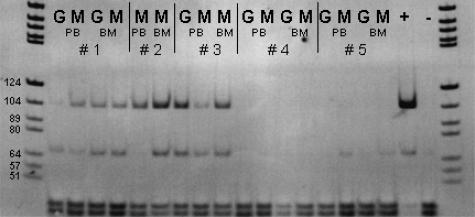
Figure 6.
Six-percent PAGE was loaded with BsaXI-digested JAK2 products derived from the granulocyte fraction and/or the mononuclear cell fraction of peripheral blood and/or bone marrow aspirates of four patients (1 to 3 with a history of PV; 4 and 5 with suspected CMPDs) along with a positive control (+), and a negative control (−). Patients 1 to 3 show residual undigested and therefore mutated JAK2 in all cell populations under investigation, whereas in patients 4 and 5, no mutated JAK2 was demonstrable (G, granulocytes; M, mononuclear cells; PB, peripheral blood; BM, bone marrow). Six-percent PAGE, DNA marker pBR322 BsuRI, and marker size fragments are partially labeled. Note that the figure is digitally inverted.
Sequencing of Selected Clones Derived from Patients with Ph− CMPD and Non-Neoplastic Hematopoiesis Confirmed the JAK2 Mutation Status as Determined by Restriction Site Analysis
To confirm the results as determined by restriction site analysis, we selected four patients with PV, two patients with cIMF (also shown in Figure 5, corresponding to lanes 2 and 10, respectively) and two samples showing normal hematopoiesis, and JAK2 from cell line HL-60 for cloning and sequencing purposes.
Ten clones derived from each patient with PV and cIMF and five clones from the control and HL-60 samples, respectively, were randomly selected and sequenced. Patients with PV and cIMF showed mutated clones ranging from 30 up to 100%. In contrast, none of the clones selected from the control samples or the cell line showed mutated JAK2 (Table 2).
Table 2.
Percentage of Mutated JAK2 Clones as Evidenced by Sequencing
| Number of clones | Sequencing
|
Clones showing JAK2 mutation (%) | ||
|---|---|---|---|---|
| JAK2 mutated | JAK2 unmutated | |||
| PV patient 1 | 10 | 4 | 6 | 40 |
| PV patient 2 | 10 | 10 | 0 | 100 |
| PV patient 3 | 10 | 6 | 4 | 60 |
| PV patient 4 | 10 | 10 | 0 | 100 |
| cIMF patient 1 | 10 | 3 | 7 | 30 |
| cIMF patient 2 | 10 | 10 | 0 | 100 |
| Normal bone marrow (control 1) | 5 | 0 | 5 | 0 |
| Normal bone marrow (control 2) | 5 | 0 | 5 | 0 |
| HL-60 | 5 | 0 | 5 | 0 |
Restriction Site Analysis of Randomly Selected Clones Enables Estimation of the Rate of Mutated JAK2 in a Given Sample
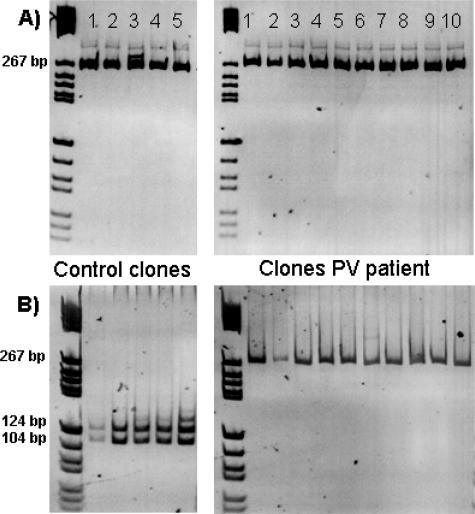
Digestion of PCR products derived from JAK2 inserted into the TOPO plasmid vector confirmed the results as determined by sequencing. Accordingly, patient samples showed up to 10 undigested clones, whereas control samples always showed complete digestion of the JAK2 clones. For example in a PV patient, none of 10 clones showed digested JAK2 in contrast to a control sample in which 5 selected clones showed complete digestion (Figure 7).
Figure 7.
M13 PCR products (10 μl) of JAK2 clones were loaded onto a 6% PAGE to allow visualization of the expected 266-bp amplicon according to Materials and Methods. Afterward, 10 μl of PCR product of each clone was digested with BsaXI for 16 hours at 37°C, and restriction fragments were visualized using another 6% PAGE. As shown in this example, 5 control clones showed complete digestion, whereas in 10 of 10 clones derived from a PV patient, no digestion could be demonstrated (A and B).
Discussion
Recent studies on the discovery of a single point mutation in the JH2 pseudokinase domain of JAK2 intriguingly uncovered a relevant underlying cause for autonomous proliferation of hematopoietic lineages in Ph− CMPDs. The detection rate of the JAK2 mutation was 65 to 97% in PV, 23 to 57% in ET, and 34 to 57% in cIMF.4,5,6,7 Most of the samples analyzed by the investigators represented sorted peripheral blood cells, ie, granulocytes, erythroblasts derived from CD34+ cells, and T cells. It could be demonstrated that the JAK2 mutation was acquired because T lymphocytes under investigation showed no JAK2 mutation even though other lineages in a given patient showed the mutation.4,5 The JAK2 mutation could also be detected in a small subset of patients with chronic myelomonocytic leukemia and myelodysplastic syndrome.15 Mutational analyses were performed on DNA extracted from the different lineages. Notably, the JAK2 mutation destroys a recognition site for the restriction enzyme BsaXI, leaving mutated PCR products undigested, whereas unmutated products showed complete digestion.4
Based on the latter finding, we conducted a study on the feasibility of JAK2 restriction site analysis in archival bone marrow trephines. We first established a RT-PCR based assay for amplification of a 100-bp sequence covering the hotspot nucleotide in the JH2 pseudokinase domain of JAK2 (Figures 1and 3). Essentially, a DNA-based assay is feasible as well because the hotspot is located in exon 12 of JAK2 and therefore analyzable at both the genomic and expression level. However, cDNA as the starting material offers the plasticity to analyze different targets apart from JAK2 on the expression level in the same sample, eg, exclusion of aberrant fusion transcripts such as BCR-ABL (see Materials and Methods and Results). We could demonstrate the sufficient and reliable JAK2 amplification independent of the mutation status in all samples analyzed. We then tested the enzyme BsaXI for efficiency in restriction site analysis in the JAK2 amplicon derived from bone marrow trephines showing normal hematopoiesis. As expected, the PCR product was completely digested after 2 to 4 hours of incubation using a concentration of 2 U of BsaXI. The next step was to investigate the BsaXI restriction site in the JAK2 amplicons derived from patients with Ph− CMPD. The single guanine-to-thymine point mutation would destroy a recognition site for BsaXI, therefore disabling digestion of the JAK2 amplicon (Figure 2, A and B). Using a twofold excess of enzyme and an incubation time of 16 hours, the majority of PV patients and a subset of ET and cIMF patients showed no or incomplete digestion of the JAK2 amplicon (Figure 5), indicating an underlying clone carrying the JAK2 mutation. An approximately 70-bp fragment could be regularly demonstrated after BsaXI digestion in both mutated and unmutated cases (Figures 4Band 5). In fact, BsaXI often cuts both recognition sites with different efficiencies, resulting in a subset of 70-bp fragments together with the expected fragments of 36, 34, and 30 bp (New England Biolabs, personal communication). However, different restriction site efficiencies solely created an additional fragment but did not lead to misinterpretation of the JAK2 mutation status because an existent clone inhibits any recognition of the mutated site.
We then aimed to prove the results obtained by restriction site analysis by use of a cloning/sequencing approach. We selected four PV patients, two cIMF patients, two control samples, and JAK2 from the cell line HL-60. Amplified JAK2 derived from two cIMF patients are also displayed in Figure 5 and showed either incomplete (Figure 5, lane 10) or virtually no digestion (Figure 5, lane 2). Alignment of the respective JAK2 sequence derived from five different clones in both control sample and HL-60 cells showed no mutation, whereas clones in cIMF showed the mutation to a varying extent. In the patient showing virtually no JAK2 digestion, 10 of 10 clones randomly selected showed the mutation (Table 2, cIMF patient 2). By contrast, in a cIMF patient showing both considerable integrity of the JAK2 amplicon and notable digestion fragments, only 3 of 10 clones showed the mutation (Table 2, cIMF patient 1). It has been demonstrated that one-third of PV patients were homozygous for the JAK2 mutation, whereas the majority harbored both unmutated and mutated alleles.5 Accordingly, one could expect a clear predominance of the mutated JAK2 amplicon in a patient homozygous for the JAK2 mutation in a defined cell population. Even though the neoplastic clone overgrowing the bone marrow might be predominant, total bone marrow cells still represent a mixture of cells including those carrying the unmutated JAK2, such as non-neoplastic fibroblasts16 or T lymphocytes.4
The assay described herein was also suitable for fresh unfixed cell populations. In three of five samples representing granulocytes and mononuclear cells from either peripheral blood and/or bone marrow aspirate, JAK2 amplification and subsequent restriction site analysis revealed the JAK2 mutation. Thus, the diagnostic value of this assay is not restricted to fixed tissues. We further aimed to prove whether the rate of JAK2 mutation as detected in peripheral blood cells4,5,6 could be reproduced in bone marrow samples classified on histological grounds according to the World Health Organization recommendations. Of the four studies on JAK2 mutation in Ph− CMPDs, only one used in part the World Health Organization criteria for classification. Therefore, it cannot be excluded that overlaps between World Health Organization, Polycythemia Vera Study Group, and the Italian Consensus Conference criteria used by these studies4,5,6,7 led to a misguided classification, particularly in cases showing cellular prefibrotic cIMF versus ET.17 As summarized in Table 1, the rate of mutated JAK2 as detected in bone marrow trephines by restriction site analysis correlated well with the frequency determined in peripheral blood cells.4,5,6 Hence, PV is the Ph− CMPD with the highest frequency of JAK2 mutation. Moreover, the diagnostic relevance for detection of the mutation status of JAK2 in reactive erythrocytosis with suspected PV could be clearly demonstrated because none of the reactive cases were mutated in contrast to the majority of PV cases.
It has been suggested that the existence of a neoplastic clone with or without mutated JAK2 in Ph− CMPDs might lead to revised classification5 because up to date diagnosis was mainly based on clinical and histomorphological criteria.18 JAK2 represents a promising biological marker, and it seems indispensable to investigate CMPD patients for a potential JAK2 mutation because it has been demonstrated that patients carrying the mutation have significantly longer duration of diseases and a higher rate of complication.7 In CMPDs, the bone marrow houses the neoplastic clone and additionally represents the only material available for diagnosis in patients with manifest myelofibrosis. Accordingly, we conclude that JAK2 restriction site analysis in bone marrow trephines is of high diagnostic relevance for discrimination of clonal and non-clonal myeloproliferations. Besides the diagnostic approach, future studies using this clear and concise assay might unmask different Ph− CMPD subgroups, allowing further correlation with histomorphology and time course of disease.
Acknowledgments
We thank the pioneering studies on JAK2 mutation in Ph− CMPDs. We also thank Ms. Gillian Teicke for critical reading of the manuscript.
Footnotes
Supported by a research grant from Deutsche Krebshilfe, Dr. Mildred Scheel Stiftung 10–2191 (to O.B. and H.K.).
References
- Tefferi A. The pathogenesis of chronic myeloproliferative diseases. Int J Hematol. 2001;73:170–176. doi: 10.1007/BF02981934. [DOI] [PubMed] [Google Scholar]
- Li CY. The role of morphology, cytochemistry and immunohistochemistry in the diagnosis of chronic myeloproliferative diseases. Int J Hematol. 2002;76(Suppl 2):6–8. doi: 10.1007/BF03165077. [DOI] [PubMed] [Google Scholar]
- Kralovics R, Buser AS, Teo SS, Coers J, Tichelli A, Van Der Maas AP, Skoda RC. Comparison of molecular markers in a cohort of patients with chronic myeloproliferative disorders. Blood. 2003;102:1869–1871. doi: 10.1182/blood-2003-03-0744. [DOI] [PubMed] [Google Scholar]
- Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, Scott MA, Erber WN, Green AR. Cancer Genome Project: acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, Garcon L, Raslova H, Berger R, Bennaceur-Griscelli A, Villeval JL, Constantinescu SN, Casadevall N, Vainchenker W. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, Adelsperger J, Koo S, Lee JC, Gabriel S, Mercher T, D’Andrea A, Frohling S, Dohner K, Marynen P, Vandenberghe P, Mesa RA, Tefferi A, Griffin JD, Eck MJ, Sellers WR, Meyerson M, Golub TR, Lee SJ, Gilliland DG. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- Rane SG, Reddy EP. JAKs, STATs and Src kinases in hematopoiesis. Oncogene. 2002;21:3334–3358. doi: 10.1038/sj.onc.1205398. [DOI] [PubMed] [Google Scholar]
- Kaushansky K. On the molecular origins of the chronic myeloproliferative disorders: it all makes sense. Blood. 2005;105:4187–4190. doi: 10.1182/blood-2005-03-1287. [DOI] [PubMed] [Google Scholar]
- Saharinen P, Takaluoma K, Silvennoinen O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol Cell Biol. 2000;20:3387–3395. doi: 10.1128/mcb.20.10.3387-3395.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr T, Büsche G, Choritz H, Länger F, Kreipe H. Evolution of myelofibrosis in chronic idiopathic myelofibrosis as evidenced in sequential bone marrow biopsy specimens. Am J Clin Pathol. 2003;119:152–158. doi: 10.1309/PTVG-B3DX-B8A8-M7KD. [DOI] [PubMed] [Google Scholar]
- Bock O, Kreipe H, Lehmann U. One-step extraction of RNA from archival biopsies. Anal Biochem. 2001;295:116–117. doi: 10.1006/abio.2001.5188. [DOI] [PubMed] [Google Scholar]
- Bock O, Lehmann U, Kreipe H. Quantitative intra-individual monitoring of BCR-ABL transcript levels in archival bone marrow trephines of patients with chronic myeloid leukemia. J Mol Diagn. 2003;5:54–60. doi: 10.1016/S1525-1578(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock O, Reising D, Kreipe H. Multiplex RT-PCR for the detection of common BCR-ABL fusion transcripts in paraffin-embedded tissues from patients with chronic myeloid leukemia and acute lymphoblastic leukemia. Diagn Mol Pathol. 2003;12:119–123. doi: 10.1097/00019606-200309000-00001. [DOI] [PubMed] [Google Scholar]
- Steensma DP, Dewald GW, Lasho TL, Powell HL, McClure RF, Levine RL, Gilliland DG, Tefferi A. The JAK2 V617F activating tyrosine kinase mutation is an infrequent event in both “atypical” myeloproliferative disorders and the myelodysplastic syndrome. Blood. 2005;106:1207–1209. doi: 10.1182/blood-2005-03-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A. Myelofibrosis with myeloid metaplasia. N Engl J Med. 2000;342:1255–1265. doi: 10.1056/NEJM200004273421706. [DOI] [PubMed] [Google Scholar]
- Michiels JJ, Thiele J. Clinical and pathological criteria for the diagnosis of essential thrombocythemia, polycythemia vera, and idiopathic myelofibrosis (agnogenic myeloid metaplasia). Int J Hematol. 2002;76:133–145. doi: 10.1007/BF02982575. [DOI] [PubMed] [Google Scholar]
- Brunning RD, Flandrin G, Imbert M, Pierre R, Thiele J, Vardiman JW. Jaffe ES, Harris NL, Stein H, Vardiman JW, editors. Lyon, France: IARC Press; Chronic myeloproliferative diseases. WHO Classification of Tumours. Pathology & Genetics. Tumours of Haematopoietic and Lymphoid Tissues. 2001:15–42. [Google Scholar]