Abstract
Alveolar rhabdomyosarcoma (ARMS) is a soft tissue cancer in which chromosomal translocations generate PAX3-FKHR and PAX7-FKHR gene fusions. To improve the approach for fusion detection in archival samples, we developed a real-time reverse transcriptase-polymerase chain reaction assay for these fusion transcripts. By incorporating consensus primers and gene-specific probes, both presence and subtype of the fusion were determined in one assay. We applied this approach to a convenience sample of 78 formalin-fixed, paraffin-embedded ARMS tumors from the Intergroup Rhabdomyosarcoma Study (IRS)-III clinical trial and obtained satisfactory results in 59 (76%) cases. The distribution of fusion types was 35 (59%) PAX3-FKHR, 11 (19%) PAX7-FKHR, and 13 fusion-negative (22%). In a subsequent clinical analysis, we found that IRS-III ARMS cases analyzed for fusion status had a significantly improved outcome compared to IRS-III ARMS cases that were not available for fusion analysis. The basis of this outcome could not be explained by known prognostic clinical factors, and multivariate analysis confirmed that our convenience sample was not representative of the whole IRS-III cohort. In conclusion, although these robust assays provide new opportunities for correlative studies of archival material, our first application illustrates an important limitation of using a convenience sample for molecular-clinical correlative studies.
In bone and soft tissue sarcomas, the identification of recurrent chromosomal translocations that are associated with specific sarcoma categories has provided opportunities for advancing both basic biology research and clinical practice.1 These translocations break within or adjacent to cellular genes and juxtapose portions of these genes to generate gene fusions. At the expression level, these fusion genes are transcribed into fusion transcripts and are ultimately translated into fusion proteins to produce a novel oncogenic activity within the cell.
In the myogenic soft tissue tumor alveolar rhabdomyosarcoma (ARMS), the characteristic 2;13 or variant 1;13 chromosomal translocation fuses the PAX3 or PAX7 gene with the FKHR gene to generate the PAX3-FKHR or PAX7-FKHR gene fusion, respectively.2 At the molecular pathology level, reverse transcriptase (RT)-polymerase chain reaction (PCR) assays have been developed to efficiently detect these (as well as other) fusion products within tumor specimens.3 Initial studies comparing specimens of defined histological categories have demonstrated the diagnostic utility of these assays. Beyond these diagnostic studies, a second level of analysis is trying to analyze the clinical difference between PAX3-FKHR, PAX7-FKHR, and fusion-negative ARMS tumors. In a retrospective analysis of frozen tissue specimens from the Intergroup Rhabdomyosarcoma Study (IRS)-IV, there were several differences identified between PAX3-FKHR and PAX7-FKHR tumors.4 The PAX7-FKHR tumors tended to occur in younger patients, were locally less invasive, and when metastatic at diagnosis, were associated with a better outcome than PAX3-FKHR tumors.
To analyze further these differences between PAX3-FKHR and PAX7-FKHR tumors as well as to explore further the clinical characteristics of the fusion-negative subset of ARMS patients, more cases need to be studied at the genotypic and phenotypic level. Although prospective studies certainly provide optimal sample and data collection, time is required to accrue a sufficient number of cases and to determine clinical outcome. As an alternative, we performed a retrospective analysis of fusion status in an earlier clinical study, IRS-III (1984 to 1991),5 from which only formalin-fixed, paraffin-embedded (FFPE) archival specimens were available. Although we had previously developed a RT-PCR protocol for working with FFPE material, the protocol relied on agarose gel electrophoresis for initial analysis, Southern blot hybridization for higher sensitivity analysis, and slot blot analysis for subtyping and required up to 8 working days to completely work up a case.6 In this report, we describe a highly efficient and rapid real-time RT-PCR approach for analysis of FFPE samples and describe our experience with the analysis of FFPE samples from the IRS-III clinical trial.
Materials and Methods
Acquisition of ARMS Specimens
For patients entered on the IRS-III study, the review diagnoses from the IRS Pathology Committee were retrieved, and cases of ARMS were identified. The criteria used for the histopathological diagnosis of ARMS in the IRS-III study were based on the characteristics described by Horn and Enterline7 with the additional stated requirement for alveolar pattern in at least 50% of the lesion. The paraffin blocks or derivative materials from these ARMS cases were retrospectively requested from the participating institutions by the Children’s Oncology Group Biopathology Center and/or a study coordinator, and 78 samples of the paraffin-embedded tumors were received. This set of specimens constitutes a convenience sample, which is defined as a “group of individuals believed to be representative of the population from which it was selected, but chosen because it is close at hand rather than randomly selected.”8
Extraction of RNA
Starting with FFPE blocks, tissue sections measuring 10 μm in thickness were cut on a microtome and deparaffinized in Citrisolve (Fisher Scientific, Pittsburgh, PA) followed by extraction with ethanol. A sufficient number of tissue sections to give ∼1 mm3 (1 cm2 × 10 μm) of tissue was incubated overnight at 50°C in a 250-μl volume of reaction mixture containing 1 μg/μl proteinase K, 20 mmol/L Tris pH 8.0, 20 mmol/L ethylenediaminetetraacetic acid, and 1% sodium dodecyl sulfate. RNA was then extracted with TRIzol LS (Invitrogen, Carlsbad, CA), precipitated in isopropanol using linear acrylamide (Ambion, Austin, TX) as carrier, and then redissolved in 10 μl of water.
Standard Real-Time RT-PCR Assay for PAX3/PAX7-FKHR
A 0.5-μl aliquot of extracted RNA was pretreated for 60 minutes at 37°C with 3 U DNase I (Roche, Indianapolis, IN) and 10 U RNase Inhibitor (Applied Biosystems, Foster City, CA) in 3.5-μl final volume of a 2 mmol/L MgCl2 solution. After inactivating the DNase for 5 minutes at 90°C, RT and PCR reagents (Applied Biosystems) were added to give final concentrations of 1× TaqMan buffer A, 5.5 mmol/L MgCl2, 1.5 mmol/L GeneAmp dNTP blend (with dUTP), 0.4 μmol/L PAX3/7–3 forward primer, 0.4 μmol/L FKHR reverse primer, 0.1 μmol/L 3F3 probe, 0.1 μmol/L 7F probe, 0.4 U/μl RNase inhibitor, 0.25U/μl Multiscribe RT, and 0.025 U/μl AmpliTaq Gold DNA polymerase in a 50-μl final volume (Table 1). These RT-PCR mixtures were assayed in an ABI Prism 7700 sequence detection system (Applied Biosystems) with cycling conditions of one cycle of 30 minutes at 48°C followed by one cycle of 10 minutes at 95°C followed by 40 cycles of 15 seconds at 95°C alternating with 1 minute at 65°C.
Table 1.
Probes and Primers
| Name | Type | Sequence |
|---|---|---|
| PAX3/7–3 | Forward Primer | 5′-CCTCCAACCMCATGAACCC-3′ |
| FKHR | Reverse Primer | 5′-CCTTCATTCTGCACACGAATGA-3′ |
| 3F3 | Probe | VIC-5′-TGGCAATGGCCTCTCACCTCAGAATT-3′-TAMRA |
| 7F | Probe | 6FAM-5′-AGCAACGGCCTGTCTCCTCAGAATTCA-3′-TAMRA |
| GAPDHsh | Forward Primer | 5′-ATTCCACCCATGGCAAATTC-3′ |
| GAPDHsh | Reverse Primer | 5′-TGGGATTTCCATTGATGACAAG-3′ |
| GAPDHsh | Probe | 6FAM-5′-CAAGCTTCCCGTTCTCAGCC-3′-TAMRA |
| FKHR-RT | RT Primer | 5′-CTGGATTGAGCATC-3′ |
| 13CF3 | Forward Primer | 5′-GCAGATCTACGAGTGGATGG-3′ |
| 13CR1 | Reverse Primer | 5′-AACTGTGATCCAGGGCTGTC-3′ |
| PAX3/7–1 | Forward Primer | 5′-CCGACAGCAGCTCTGCCTAC-3′ |
Real-Time RT-PCR Assay for GAPDH
Using the DNase I treatment protocol above, 4.5 μg of RNA from a normal lymphoblast cell line were treated in a final volume of 10.5 μl, and 0.5 μl of RNA from tissue sections was treated as described above. After inactivating the DNase for 5 minutes at 90°C, the control lymphoblast RNA was serially diluted to generate a standard curve consisting of 1, 0.3, 0.1, 0.03, 0.01, 0.003, and 0.001 μg; RT and PCR reagents were added to give final concentrations of 1× TaqMan buffer A, 5.5 mmol/L MgCl2, 1.5 mmol/L GeneAmp dNTP blend (with dUTP), 0.2 μmol/L GAPDHsh forward primer, 0.2 μmol/L GAPDHsh reverse primer, 0.05 μmol/L GAPDHsh probe, 0.4 U/μl RNase inhibitor, 0.25 U/μl Multiscribe RT, and 0.025 U/μl AmpliTaq Gold DNA polymerase in a 50-μl final volume (Table 1). These RT-PCR mixtures were assayed with cycling conditions of one cycle of 30 minutes at 48°C followed by one cycle of 10 minutes at 95°C followed by 40 cycles of 15 seconds at 95°C alternating with 1 minute at 60°C.
Sensitive Real-Time RT-PCR Assay for PAX3/PAX7-FKHR
A 1.0-μl aliquot of extracted RNA was pretreated with DNase I as described above in a final volume of 7 μl. For RT with an FKHR-specific primer, reagents were added to give final concentrations of 1× PCR buffer II, 5 mmol/L MgCl2, 0.5 mmol/L dNTP mix, 5 μmol/L FKHR-RT primer, and 2.5 U/μl MuLV RT in a 25-μl final volume (Table 1). This reaction mixture was incubated for 45 minutes at 42°C on a GeneAmp 2400 thermal cycler (Applied Biosystems), and the reaction was inactivated by incubating for 5 minutes at 99°C followed by cooling to 5°C.
For the first PCR, half of the cDNA was assayed for wild-type FKHR with 13CF3 and 13CR1 primers, and the other half was assayed for PAX3/PAX7-FKHR with PAX3/7-1 and FKHR primers (Table 1). The first PCR was performed by adding reagents to give final concentrations of 1× PCR buffer II, 1.5 mmol/L MgCl2, 1 μmol/L of each PCR primer, and 0.025 U/μl AmpliTaq DNA polymerase. These PCR mixtures were assayed on a GeneAmp 2400 thermal cycler with cycling conditions of 40 cycles of 1 minute at 94°C, 1 minute at 65°C, and 2 minutes at 72°C followed by 1 cycle of 10 minutes at 72°C and then cooling at 4°C. For the seminested PCR reaction, a 3.5-μl aliquot of a 1:25 dilution of the PAX3/7-FKHR PCR reaction was mixed with PCR reagents to give final conditions as described above for the standard real-time RT-PCR assay, with the exception that the RT and RNase inhibitor were omitted from this PCR reaction mixture. These PCR mixtures were assayed on an ABI Prism 7700 sequence detection system with cycling conditions of one cycle of 10 minutes at 95°C followed by 40 cycles of 15 seconds at 95°C alternating with 1 minute at 65°C.
Statistical Analysis
Clinical characteristics and outcome were retrieved from the Children’s Oncology Group Statistics and Data Center. Failure-free survival was measured from the time of diagnosis until progression, relapse after response, or death (if occurring before relapse or progression). Patients who did not fail were censored at their date of last contact. Survival was measured from the time of diagnosis until death, or until the date of last contact, if the patient was alive at last report. All deaths were counted as failures, whether or not they were disease-related. Estimates of the time-to-event distributions were calculated using the Kaplan-Meier method. Comparisons of outcome among patient subsets were made using the log-rank test, and the Cox proportional hazards regression model was used for multivariate analysis. Comparisons of demographic and disease characteristics among patient subsets were made using the χ2 test for association or, where appropriate due to sparseness, Fisher’s exact test.
Results
Development and Application of Real-Time RT-PCR Assay for ARMS Gene Fusions
To analyze the gene fusion status of FFPE specimens from the IRS-III clinical trial in a more rapid manner than possible with our previously described agarose gel/Southern and slot blot hybridization-based assay,6 we turned to the methodology of real-time RT-PCR. In our assay design, the RT and PCR steps are performed in one reaction system without intermediate reagent addition. The FKHR primer serves as both the RT primer as well as a PCR primer. In addition, two differentially labeled detection probes from the PAX3/PAX7-FKHR region internal to the primers are added to the reaction system, one PAX3-specific and the other PAX7-specific. As the PCR with the forward consensus PAX3/PAX7 and FKHR reverse primers proceeds, the two probes not only permit detection of accumulating PCR products but also permit the products to be subtyped as PAX3-FKHR or PAX7-FKHR. To determine which negative results are meaningful, a separate real-time RT-PCR quantitative analysis of GAPDH expression is performed. Based on our data in fusion-positive samples, we set a GAPDH expression level equivalent to that found in 1 ng of the control lymphoblast line as the minimum cutoff for a satisfactory result. In this analysis, the general working scheme is extraction of the FFPE sample on the first day and proteinase K digestion overnight. RNA extraction is performed on the second day, and the RT-PCR reaction is set up later that day. Finally, results are retrieved and analyzed on the third day. Therefore, results are available within 3 working days.
We applied the real-time RT-PCR assay to 78 separate FFPE samples. In this cohort, we obtained satisfactory results in 59 cases (76%). In three of these cases with borderline or inconsistent GAPDH levels, we verified our findings using a higher sensitivity assay consisting of a two-step seminested protocol. In the first step, we performed a separate RT step in which an FKHR-specific short RT primer optimized for 42°C was used. In the next step, the RT product was split to perform two simultaneous standard PCR steps—one to assay for wild-type FKHR expression as an internal control for RNA quality and a second to generate the first stage of the PAX3/PAX7-FKHR product by use of a consensus PAX3/PAX7 forward primer and a reverse FKHR primer. If evidence of a wild-type FKHR product was detected on agarose gel electrophoresis, then a second seminested step was performed by assaying an aliquot of a dilution of the first stage product by the real-time assay described above.
Analysis of Patient and Tumor Characteristics
In the 59 ARMS cases with satisfactory PCR data, the frequency of the fusion subtypes was 35 cases (59%) with a PAX3-FKHR fusion, 11 cases (19%) with a PAX7-FKHR fusion, and 13 fusion-negative cases (22%). This distribution of fusion subtypes is quite similar to the distribution that we previously determined in an analysis of frozen samples from ARMS cases in the IRS-IV clinical trial4 and supports the validity of our real-time RT-PCR methodology applied to FFPE specimens.
In our previous study of fusion subtypes in IRS-IV,4 we identified several associations between the specific fusion subtypes and clinical parameters (Table 2). In this study, the subset of PAX7-FKHR-positive ARMS tumors was smaller, and we thus had less power than that of our previous study in the examination of these statistical associations. However, even if we did not reach statistical significance, we were interested in examining whether fusion status was associated with patient or tumor characteristics in this cohort of ARMS tumors. An examination of the relationship of age and fusion status demonstrated a trend that was consistent with the previously observed association of PAX7-FKHR-positive tumors in younger patients. The data also indicated a possible trend for lower invasiveness in PAX7-FKHR-positive tumors compared to PAX3-FKHR-positive tumors, which was seen in the IRS-IV clinical study. However, it should be noted that there is incomplete data available for invasiveness, and the level of statistical confidence suffered accordingly. Finally, in a comparison of the three fusion subtypes, there is an association of fusion status with site and nodal status. The association with site is characterized by a low incidence of fusion-negative tumors in the extremities and high incidence in nonbladder/prostate genitourinary sites compared to the two fusion-positive categories. The association with nodal status is characterized by a very low frequency of nodal involvement in the fusion-negative tumors compared to the two fusion-positive categories. However, it should be noted that these latter two associations were not seen in our previous study of IRS-IV cases. Otherwise, we again found no association of fusion status with gender, race, tumor size, stage, or clinical group (data not shown).
Table 2.
Patient Characteristics Compared by Fusion Status
| Category | PAX3 (n = 35) | PAX7 (n = 11) | Negative (n = 13) | Overall P value | PAX3 versus PAX7 P value | |
|---|---|---|---|---|---|---|
| Age | <9 | 20 (57%) | 10 (91%) | 7 (54%) | 0.098 | 0.069 |
| 10+ | 15 (43%) | 1 (9%) | 6 (46%) | |||
| Gender | Male | 20 (57%) | 7 (64%) | 8 (62%) | 1.0 | 1.0 |
| Female | 15 (43%) | 4 (36%) | 5 (38%) | |||
| Race | White | 27 (77%) | 9 (82%) | 12 (92%) | 0.58 | 1.00 |
| Nonwhite | 8 (23%) | 2 (18%) | 1 (8%) | |||
| Stage | 1 | 3 (9%) | 3 (27%) | 5 (38%) | 0.15 | 0.29 |
| 2 | 9 (26%) | 4 (36%) | 4 (31%) | |||
| 3 | 10 (29%) | 1 (18%) | 3 (23%) | |||
| 4 | 13 (37%) | 2 (18%) | 1 (8%) | |||
| No data | 0 | 1 | 0 | |||
| Group | I | 8 (23%) | 2 (27%) | 4 (31%) | 0.24 | 0.31 |
| II | 7 (20%) | 5 (45%) | 4 (31%) | |||
| III | 6 (17%) | 1 (9%) | 4 (31%) | |||
| IV | 14 (40%) | 2 (18%) | 1 (8%) | |||
| No data | 0 | 1 | 0 | |||
| Size | < = 5 cm | 17 (55%) | 7 (70%) | 9 (69%) | 0.59 | 0.48 |
| > 5 cm | 14 (45%) | 3 (30%) | 4 (31%) | |||
| No data | 4 | 1 | 0 | |||
| Nodal status | NB0 | 16 (53%) | 7 (70%) | 12 (92%) | 0.046 | 0.47 |
| NB1 | 14 (47%) | 3 (30%) | 1 (8%) | |||
| No data | 5 | 1 | 0 | |||
| Tumor invasiveness | T-1 | 10 (40%) | 6 (75%) | 9 (69%) | 0.12 | 0.12 |
| T-2 | 15 (60%) | 2 (25%) | 4 (31%) | |||
| No data | 10 | 3 | 0 | |||
| Site group | Head/Neck | 3 (9%) | 3 (27%) | 1 (8%) | 0.004 | 0.24 |
| PM* | 5 (14%) | 0 (0%) | 3 (23%) | |||
| GU nonBP | 0 (0%) | 0 (0%) | 4 (31%) | |||
| Extremity | 19 (54%) | 7 (64%) | 2 (15%) | |||
| Other | 8 (23%) | 1 (9%) | 3 (23%) |
PM, parameningeal; GU, genitourinary; BP, bladder-prostate
Analysis of Patient Outcome
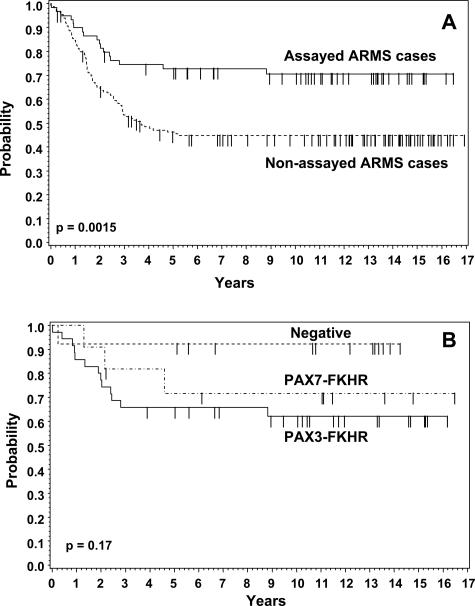
Before analyzing patient outcome among the fusion subtypes and other subsets in this patient cohort, we examined whether the ARMS cases with available fusion data were representative of the full set of ARMS cases from IRS-III. Our comparison of both overall survival and failure-free survival between the 59 ARMS cases with gene fusion data and the remaining 151 eligible and centrally reviewed IRS-III ARMS cases without fusion data revealed statistically significant differences (Figure 1A). The estimated 5-year overall survival rates in the groups with and without gene fusion data are 79% (95% CI: 68 to 89%) and 52% (95% CI: 44 to 60%), respectively, and the estimated 5-year failure-free survival rates are 74% (95% CI: 62 to 85%) and 46% (95% CI: 38 to 54%), respectively. To account for this difference between the two groups, we compared them in regard to known prognostic clinical variables. However, the two groups did not differ significantly in regard to distribution of these prognostic variables (data not shown).
Figure 1.
Kaplan-Meier plots of failure-free survival for ARMS patients registered on IRS-III. A: Comparison of outcome between cases with available fusion data (n = 59) and cases without available fusion data (n = 151). B: Comparison of outcome among cases with available fusion data separated according to fusion status: PAX3-FKHR (n = 35), PAX7-FKHR (n = 11), and fusion-negative (n = 13).
To explore whether this association between the availability of fusion data and outcome could be explained by known prognostic variables, Cox regression was used for multivariate analysis of overall and failure-free survival. Backwards selection was used to obtain the best model. Variables were retained in the model if P values were less than 0.05. The specific variables included in the model selection were age (<9 or 10+), gender, race (white or nonwhite), stage (1 or 2 or 3 or 4), group (I or II or III or IV), tumor size (less than or equal to 5 cm or greater than 5 cm), nodal status (N0 or N1), tumor invasiveness (T1 or T2), and tumor site category (favorable or unfavorable). There was complete information on 200 cases for inclusion in this analysis. In analyzing this data set, we found that, in addition to the availability of fusion data, race and stage are significant predictors of both overall and failure-free survival. Furthermore, after adjusting for race and stage, we determined that the availability of fusion data are also a significant independent predictor of overall and failure-free survival (Table 3).
Table 3.
Multivariate Analysis, IRS-III ARMS Patients, after Adding Fusion Data to Model
| Category | HR | 95% CI | P value | |
|---|---|---|---|---|
| Overall Survival | ||||
| Race | White | 1 | 0.013 | |
| Nonwhite | 1.9 | 1.1 to 3.1 | ||
| Stage | 1 | 1 | <0.0001 | |
| 2 | 0.9 | 0.3 to 2.7 | ||
| 3 | 2.8 | 1.2 to 6.8 | ||
| 4 | 9.8 | 4.1 to 23.2 | ||
| Fusion data | Yes | 1 | 0.00027 | |
| No | 2.4 | 1.3 to 4.1 | ||
| Failure-free survival | ||||
| Race | White | 1 | 0.051 | |
| Nonwhite | 1.6 | 1.0 to 2.6 | ||
| Stage | 1 | 1 | <0.0001 | |
| 2 | 1.1 | 0.4 to 2.9 | ||
| 3 | 2.6 | 1.2 to 5.9 | ||
| 4 | 7.9 | 3.5 to 17.6 | ||
| Fusion data | Yes | 1 | 0.0075 | |
| No | 2.1 | 1.2 to 3.5 | ||
To further investigate this relationship, we hypothesized that the clinical case volume of the submitting institution might influence outcome because for many clinical conditions, outcome is associated with case volume. In our analysis, institutions participating in IRS-III were divided into three groups based on the number of IRS-III cases that were entered and treated by those institutions: low (less than or equal to 5 IRS III accruals), intermediate (6 to 14 IRS-III accruals), and high (greater than or equal to 15 IRS-III accruals). We first compared accrual with fusion status using the Mantel-Haenszel test for trend and found that submission for fusion status tends to be more likely to occur in institutions with higher accrual (P = 0.067). However, when accrual category was added to the overall and failure-free survival models in the above-described Cox regression analysis, accrual category was not found to be an independent predictor of overall or failure-free survival after adjusting for other significant predictors and fusion status (data not shown). Therefore, the case volume of the submitting institutions does not explain the association between fusion status data and clinical outcome.
We next investigated the relationship of outcome to fusion subtype in the IRS-III ARMS cases with available fusion data, recognizing the fact that we are starting with a selected group with an overall good outcome. For both failure-free and overall survival, there was not a statistically significant difference in outcome among the three subtypes (Figure 1B). In our analysis, the fusion-negative subset appeared to have the best outcome among the three subsets, although in pair-wise analysis, this outcome difference did not reach statistical significance (eg, PAX3-FKHR versus fusion-negative in failure-free survival, P = 0.067). Furthermore, it should be noted that in our previous study of IRS-IV ARMS cases, the fusion-negative subset appeared to have an intermediate outcome between the PAX3-FKHR and PAX7-FKHR subsets. Although our previous study of IRS-IV cases demonstrated interesting findings when outcome was compared in locoregional and metastatic subsets, such subset analysis is not feasible in this study because of the low number of group IV cases in the PAX7-FKHR (n = 2) and fusion-negative (n = 1) categories.
Finally we addressed the issue of whether this association of submitted/assayed ARMS cases with improved outcome also extended to our previous study of IRS-IV ARMS cases. In IRS-IV, frozen material was prospectively collected and centrally banked.4 We found no statistically significant difference for overall or failure-free survival between 69 ARMS cases from IRS-IV with gene fusion data and the remaining 177 eligible IRS-IV ARMS cases without fusion data (data not shown). In addition, in a multivariate analysis, the availability of fusion data in these IRS-IV cases was not a significant predictor of overall or failure-free survival after adjusting for the other significant predictors.
Discussion
In this study of FFPE specimens from IRS-III, we showed that the application of a real-time RT-PCR approach is a rapid and efficient means for determining PAX3-FKHR and PAX7-FKHR fusion status in archival samples. In particular, the ability to generate satisfactory RT-PCR results in 76% of ARMS cases that were from 13- to 20-year-olds highlights the robust quality of this assay. In ARMS, the utility of these assays is further facilitated by the invariant size and composition of the PAX3-FKHR and PAX7-FKHR fusion transcripts. This situation contrasts with the combinatorial diversity seen in the case of Ewing’s sarcoma family of tumors in which the EWS-FLI1 and EWS-ERG fusion transcripts vary in size and composition,9 thereby complicating the design of a definitive real-time RT-PCR assay for these fusion products.
The currently applied histopathological criteria for diagnosing ARMS differ from the criteria used at the time of the IRS-III trial, which required alveolar pattern in at least 50% of the lesion for a diagnosis of ARMS. The International Classification of Rhabdomyosarcoma system, published in 1995, proposed that any focus with alveolar pattern was sufficient for a histopathological diagnosis of ARMS,10 and thus some cases previously diagnosed as embryonal (or sometimes called mixed embryonal/alveolar rhabdomyosarcoma) are now reclassified as ARMS. This new classification was first instituted by the IRS Pathology Committee in 1995 toward the end of the IRS-IV study (1991 to 1997). In our previously published study of gene fusions in IRS-IV cases, the majority of samples were obtained from the period before the new diagnostic criteria were instituted and thus we anticipate that relatively few cases with small foci of alveolar histology were analyzed in that IRS-IV molecular diagnostic study. The gene fusion status and corresponding outcome of the cases with relatively small foci of alveolar histology therefore have not yet been adequately investigated but are currently being analyzed in ongoing studies of the Children’s Oncology Group Soft Tissue Sarcoma Committee.
An unexpected feature of this study that complicated interpretation of outcome was the finding that the group of FFPE cases available for analysis was not representative of the overall group of IRS-III ARMS cases. In particular, the IRS-III cases available for RT-PCR analysis were associated with a better outcome than the remaining unavailable cases. A further surprising aspect of this selection bias was that the basis of it could not be explained by a difference between the two groups in regard to a known prognostic parameter. This issue was demonstrated by the finding that availability of material for RT-PCR analysis was determined on multivariate analysis to be a significant independent predictor of outcome. Although we hypothesized that a difference in some parameter among the submitting institutions could be considered, the obvious possibility of volume of IRS-III ARMS cases was excluded. There are other possible factors, such as socioeconomic status, distance from treating center, and insurance status, which could have influenced the outcome, but these factors cannot be assessed with the available data. In addition, it is unlikely that the selection bias is due to chance because of the magnitude of the effect and level of statistical significance. Finally, we note that there was no evidence of such a selection bias in our molecular diagnostic study of IRS-IV cases, but the basis for this difference between the IRS-III and IRS-IV studies is unknown.
The salient issues to be considered are the opportunities that these molecular diagnostic assays present and the necessary cautions that should be exercised before pursuing those opportunities. With robust assays that permit high-efficiency use of archival specimens such as available in pathology departments and cooperative group banks, there are certainly opportunities to perform retrospective studies of a reasonably large number of cases. However, as this study exemplifies, there are as limitations to this approach when one is interested in molecular and clinical outcome associations. Convenience samples may interject bias, and, thus, the available samples and the entire clinical cohort must be analyzed for significant differences in known prognostic variables and for differences in outcome. These analyses should be performed at the first stage of investigation for best determination of which questions can be reliably answered with the available material.
Footnotes
Supported in part by the National Institutes of Health (grants CA89461, CA98543, CA98413, and CA24507). A complete listing of grant support for research conducted by C.C.G. and P.O.G. before initiation of the COG grant in 2003 is available online at: http://www.childrensoncologygroup.org/admin/grantinfo.htm.
References
- Barr FG, Ladanyi M. Sarcomas. Leonard DGB, editor. Philadelphia: Saunders,; Diagnostic Molecular Pathology. 2003:53–76. [Google Scholar]
- Barr FG. Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma. Oncogene. 2001;20:5736–5746. doi: 10.1038/sj.onc.1204599. [DOI] [PubMed] [Google Scholar]
- Barr FG, Chatten J, D’Cruz CM, Wilson AE, Nauta LE, Nycum LM, Biegel JA, Womer RB. Molecular assays for chromosomal translocations in the diagnosis of pediatric soft tissue sarcomas. JAMA. 1995;273:553–557. [PubMed] [Google Scholar]
- Sorensen PH, Lynch JC, Qualman SJ, Tirabosco R, Lim JF, Maurer HM, Bridge JA, Crist WM, Triche TJ, Barr FG. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children’s oncology group. J Clin Oncol. 2002;20:2672–2679. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- Crist W, Gehan EA, Ragab AH, Dickman PS, Donaldson SS, Fryer C, Hammond D, Hays DM, Herrmann J, Heyn R, Jones PM, Lawrence W, Newton W, Ortega J, Raney RB, Ruymann FB, Tefft M, Webber B, Wiener E, Wharam M, Vietti TJ, Maurer HM. The Third Intergroup Rhabdomyosarcoma Study. J Clin Oncol. 1995;13:610–630. doi: 10.1200/JCO.1995.13.3.610. [DOI] [PubMed] [Google Scholar]
- Edwards RH, Chatten J, Xiong QB, Barr FG. Detection of gene fusions in rhabdomyosarcoma by reverse transcriptase-polymerase chain reaction assay of archival samples. Diagn Mol Pathol. 1997;6:91–97. doi: 10.1097/00019606-199704000-00004. [DOI] [PubMed] [Google Scholar]
- Horn RC, Jr, Enterline HT. Rhabdomyosarcoma: a clinicopathological study and classification of 39 cases. Cancer. 1958;11:181–199. doi: 10.1002/1097-0142(195801/02)11:1<181::aid-cncr2820110130>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Petrie A, Sabin C. Oxford: Blackwell Science Ltd; Medical Statistics at a Glance. 2000 [Google Scholar]
- Zucman J, Melot T, Desmaze C, Ghysdael J, Plougastel B, Peter M, Zucker JM, Triche TJ, Sheer D, Turc-Carel C, Ambros P, Combaret V, Lenoir G, Aurias A, Thomas G, Delattre O. Combinatorial generation of variable fusion proteins in the Ewing family of tumours. EMBO J. 1993;12:4481–4487. doi: 10.1002/j.1460-2075.1993.tb06137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton WA, Jr, Gehan EA, Webber BL, Marsden HB, van Unnik AJ, Hamoudi AB, Tsokos MG, Shimada H, Harms D, Schmidt D, Ninfo V, Cavazzana AO, Gonzalez-Crussi F, Parham DM, Reiman HM, Asmar L, Beltangady MS, Sachs NE, Triche TJ, Maurer HM. Classification of rhabdomyosarcomas and related sarcomas. Pathologic aspects and proposal for a new classification—an Intergroup Rhabdomyosarcoma Study. Cancer. 1995;76:1073–1085. doi: 10.1002/1097-0142(19950915)76:6<1073::aid-cncr2820760624>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]