Abstract
Assays to measure DNA methylation, which are important in epigenetic research and clinical diagnostics, typically rely on conversion of unmethylated cytosine to uracil by sodium bisulfite. However, no study has comprehensively evaluated the precision and performance characteristics of sodium bisulfite conversion and subsequent quantitative methylation assay. We developed quantitative real-time polymerase chain reaction (MethyLight) to measure percentage of methylated reference (PMR, ie, degree of methylation) for the MGMT, MLH1, and CDKN2A (p16) promoters. To measure the precision of bisulfite conversion, we bisulfite-treated seven different aliquots of DNA from each of four paraffin-embedded colon cancer samples. To assess run-to-run variation, we repeated MethyLight five times. Bisulfite-to-bisulfite coefficient of variation (CV) of PMR ranged from 0.10 to 0.38 (mean, 0.21), and run-to-run CV of PMR ranged from 0.046 to 0.60 (mean, 0.31). Interclass correlation coefficients were 0.74 to 0.84 for the three loci, indicating good reproducibility. DNA mixing study with methylated and unmethylated DNA showed good linearity of the assay. Of 272 colorectal cancers evaluated, most showed PMR either <1 or >10, and promoter methylation (PMR >4) was tightly associated with loss of respective protein expression (P < 10−16). In conclusion, sodium bisulfite conversion and quantitative MethyLight assays have good precision and linearity and can be effectively used for high-throughput DNA methylation analysis on paraffin-embedded tissue.
Transcriptional inactivation by promoter CpG island methylation in tumor suppressor genes is an important mechanism in human carcinogenesis, and methylated cytosine has been called “the fifth base.”1,2,3 Aberrant CpG island methylation has been shown to occur in a nonrandom manner, and the pattern of methylation is tumor-specific.4 Given the importance of DNA methylation in cancer and various other diseases, assays to measure DNA methylation may be very useful in both research and clinical practice. DNA methylation of some tumor suppressor genes in breast cancer has been shown to be predictive of responsiveness to tamoxifen therapy.5 Recently, methylation and silencing of the MGMT gene in glioblastoma has been associated with an increased benefit from temozolomide treatment.6 DNA methylation may be a useful marker for cancer diagnosis, screening, surveillance in high-risk individuals, monitoring of minimal residual disease, and determining optimal therapeutic options.2,7,8,9,10,11,12 Therefore, there are increasing demands for reliable assays to measure DNA methylation, particularly for formalin-fixed paraffin-embedded tissue.
A number of methods to determine DNA methylation status in tumor tissues have been developed. For formalin-fixed, paraffin-embedded tissue, methylation-specific polymerase chain reaction (MSP) after sodium bisulfite conversion13 is widely used. However, because of the qualitative nature of the assay, MSP cannot reliably distinguish low levels of methylation from high levels of methylation. Furthermore, MSP does not appear to be highly reproducible for some samples that may have low levels of methylation, and it is difficult to assess performance characteristics. Thus, clinical use of qualitative MSP is a major concern in terms of quality control and assurance. Quantitative measurement of methylation is important because low levels of methylation (below the threshold of transcriptional silencing) may not be biologically important. Hence, a variety of quantitative assays to measure DNA methylation have been developed, including combined bisulfite restriction analysis,14 restriction ligation-mediated polymerase chain reaction (PCR),15 methylation-sensitive single nucleotide primer extension (Ms-SNuPE),16 ion-pair reverse-phase high performance liquid chromatography,17 denaturing high performance liquid chromatography,18 pyrosequencing,19,20,21 MALDI-TOF,22 and real-time PCR.5,23,24,25,26,27,28,29,30,31 Most of these methods use DNA treated with sodium bisulfite. Therefore, sodium bisulfite treatment is a critical step in the measurement of DNA methylation. However, reproducibility of the sodium bisulfite conversion step has not been well investigated. Sodium bisulfite treatment presents a harsh environment for DNA molecules, and previous data suggest that ∼84 to 96% of DNA is degraded during 4 hours of bisulfite treatment at 55°C.32 Assurance of reproducibility in this critical bisulfite treatment step is essential in applying quantitative methylation assays to clinical practice.
In this study, we evaluated the precision and performance characteristics of sodium bisulfite treatment and MethyLight, a real-time PCR assay.5,26,28,33 Our data indicate that both sodium bisulfite conversion and MethyLight assays have good reproducibility and precision and can be effectively used to quantify DNA methylation in paraffin-embedded tumor tissue.
Materials and Methods
Tissue Specimens
Tissue collection and analysis in this study were approved by the Dana-Farber Harvard Cancer Center and Brigham and Women’s Hospital Institutional Review Boards. Formalin-fixed, paraffin-embedded tissue samples of four cases (designated as case 1 through case 4) of colon cancer were anonymized after collection from the archival file of Department of Pathology, Brigham and Women’s Hospital. For the correlation study of methylation and immunohistochemistry, formalin-fixed, paraffin-embedded tissue samples of colorectal cancer were collected from participants of the Nurses’ Health Study and Health Professional Follow-up Study cohorts.34,35 Informed consents from all participants were obtained. Tumors were selected from these cohorts, based on availability of adequate tumor tissue samples and assay results at the time of this study, resulting in the inclusion of a total of 274 cases of colorectal cancer.
Sodium Bisulfite Treatment and DNA Extraction
Hematoxylin and eosin (H&E)-stained slides of the tumors were reviewed, and areas of tumor were marked, to exclude pure normal tissue and enrich tumor DNA. H&E-stained tumor sections (10 μm × 14 sections) were scraped off slides from each of case 1 through case 4 and suspended in 140 μl of tissue lysate solution (100 mmol/L Tris, pH 8, 10 mmol/L ethylenediaminetetraacetic acid, pH 8, 1 mg/ml proteinase K, and 0.05 mg/ml tRNA). The lysate solutions were incubated overnight at 50°C. The lysate was aliquoted into seven tubes, each containing 18 μl of tissue lysate, and stored at −20°C until sodium bisulfite modification was performed. For methylation and immunohistochemistry study (using cohort colorectal cancer cases), H&E-stained tissue sections (depending on tissue and tumor size, in average, large tumor tissue 10 μm × 1 section) from each case were scraped off slides, suspended in 20 μl of the tissue lysate, and incubated at 50°C overnight. The tissue lysate was then stored at −20°C until sodium bisulfite modification was performed.
Sodium metabisulfite (1.9 g) was dissolved in mixture of 3.2 ml of 0.44 mol/L NaOH at 50°C. Then, 0.5 ml of 1 mol/L hydroquinone was added to the dissolved sodium bisulfite mixture. An 18-μl aliquot of the tissue lysate was denatured at 100°C for 10 minutes and chilled on ice. Then, 2 μl of 3 mol/L NaOH was added and incubated at 42°C for 20 minutes. The bisulfite solution (120 μl) was added (total volume of 140 μl) and incubated at 50°C for 15 hours in the dark. The bisulfite-converted DNA was recovered using a Qiagen QIAamp viral RNA mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions with some modifications. Buffer AVL/carrier RNA (560 μl) was added to the 140 μl of bisulfite-converted DNA sample and incubated at room temperature for 10 minutes. Ethanol (560 μl) was then added, and after extensive mixing, the mixture was loaded onto the provided spin columns in consecutive 630-μl aliquots. After each loading, the columns were centrifuged at full speed (21,000 × g) for 1 minute. Both the filtrate and spin column were saved, and both filtrates were passed through the column a second time in the same manner to increase the yield of recovery. The spin column was then washed with 500 μl of buffer AW1, followed by centrifugation at 21,000 × g for 1 minute. Buffer AW2 (500 μl) was then added to the column, and the column was centrifuged at 21,000 × g for 4 minutes to wash the column and eliminate possible buffer AW2 carry over. DNA in the spin column was eluted by the addition of 40 μl of buffer AVE, followed by a 1-minute incubation at room temperature and centrifugation at 7600 × g for 1 minute. This elution step was repeated with a second 40-μl volume of buffer AVE. Fifty μl of 0.2 mol/L NaOH was added to the 80-μl pooled eluate for 15 minutes at room temperature to desulphonate the sample, and then 10 μl of 1 mol/L HCl was added to for neutralization. Buffer AVL/carrier RNA (560 μl) was then added to the 140-μl sample mixture, and the recovery procedure was repeated with a new spin column. The eluted DNA (80-μl volume) was then used for MethyLight analysis.
Quantitative Real-Time PCR (MethyLight)
Real-time PCR assays to measure DNA methylation (MethyLight) have been described.5,26,28 Briefly, three sets of primers and probes designed specifically for bisulfite-converted DNA were used: a set for the gene of interest and two sets for ACTB and COL2A1 to normalize for the amount of input DNA. We used an ABI 7300 real-time PCR instrument (Applied Biosystems, Foster City, CA). Primers and probes for CDKN2A, MGMT, ACTB, and COL2A1 were previously described.5 The MLH1 forward primer is 5′-AGG AAG AGC GGA TAG CGA TTT-3′, the MLH1 reverse primer is 5′-TCT TCG TCC CTC CCT AAA ACG-3′, and the MLH1 probe is 6FAM-5′-CCC GCT ACC TAA AAA AAT ATA CGC TTA CGC G-3′-BHQ1. The amount of methylated DNA (PMR, percentage of methylated reference36) at a specific locus was calculated by dividing the GENE:ACTB (or COL2A1) ratio of a sample by the GENE:ACTB (or COL2A1) ratio of SssI-treated human genomic DNA (presumably fully methylated) and multiplying by 100. Reactions using SssI-treated DNA were used to normalize for any difference in amplification efficiencies between GENE and ACTB (or COL2A1).
Measuring the Precision of Sodium Bisulfite Conversion and MethyLight Assay
Figure 1 shows our strategy to measure precision and reproducibility of bisulfite conversion of DNA and quantitative MethyLight assay. We performed bisulfite conversion on seven different aliquots from each of the four cell lysate samples (case 1 through case 4), starting on 7 different days. Thus, from each cell lysate, we have seven separate bisulfite-converted DNA samples (designated as B1 through B7), which ideally should give at least similar values for methylation at a specific locus. We measured PMR for each locus by the MethyLight assay on each of these seven separate bisulfite-DNA samples. For each case, we measured the coefficient of variation (CV) of these seven PMR values on B1 through B7, which would primarily depend on variations in bisulfite conversion. We repeated the MethyLight assays five times (designated as M1 through M5) and determined PMR values on the exact same set of bisulfite-converted DNA samples (B1 through B7) on 5 different days. We calculated the CV of these five PMR values (M1 through M5) on each one of the seven bisulfite-converted DNA samples (B1 through B7). Thus, these seven CVs would primarily depend on day-to-day variations of each MethyLight assay.
Figure 1.
Precision study for sodium bisulfite conversion and MethyLight. Bisulfite conversion was performed on seven different aliquots from each of the four cell lysate samples (cases 1 through 4), starting on 7 different days. Each sample was assessed for PMR at each locus by the MethyLight assay. MethyLight assays were repeated for each bisulfite-treated DNA sample on 5 different days, and PMR values were again determined.
We also evaluated reproducibility of standard curves, which affects run-to-run precision. We examined five standard curves in five MethyLight repeats (M1 to M5 in Figure 1). To obtain PMR variations solely due to variations of standard curves, we also performed a separate MethyLight run with five standard curves in a single plate and calculated five different PMR values for each sample-marker combination, using each one of the five different standard curves.
DNA Mixing Study to Assess Assay Linearity
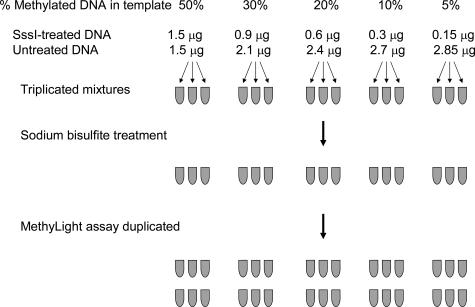
We performed a mixing study to assess linearity of sodium bisulfite treatment and quantitative MethyLight assays (Figure 2). We first treated human peripheral blood leukocyte DNA (Promega, Madison, WI) with SssI to methylate all CpG sites. Assuming near complete methylation and no loss of DNA during SssI treatment, we prepared DNA mixtures in triplicates (each containing 1 μg of template DNA) for SssI-treated DNA:untreated DNA as follows: 50:50, 30:70, 20:80, 10:90, and 5:95 (Figure 2). We used 100% SssI-treated DNA as control reference DNA to calculate PMR as described above. We treated DNA mixtures in triplicates with sodium bisulfite and duplicated MethyLight assays, resulting in six data points of PMR values for each dilution. Linear regression analysis was performed to assess assay linearity.
Figure 2.
DNA mixing study to assess linearity of MethyLight. Human peripheral blood leukocyte DNA (Promega) was first treated with M.SssI to methylate all CpG sites (near complete methylation and no loss of DNA was assumed). Mixtures of SssI-treated DNA:untreated DNA were prepared in triplicate (each containing 1 μg of template DNA) as shown. One hundred percent SssI-treated DNA served as methylated reference DNA. Each DNA mixture was bisulfite-treated and assayed in duplicate by MethyLight. Linear regression analysis was performed to assess assay linearity.
Immunohistochemistry
Immunohistochemistry for MLH1, p16 (CDKN2A), and MGMT was previously described.37 Immunohistochemistry was evaluated while blinded from methylation data.
Statistical Analysis
For statistical analysis, paired t-test to compare means, F-test to assess the equality of variances38 and linear regression analysis to assess assay linearity were performed using the Microsoft Excel 2000 Analysis ToolPak. For F-test, raw PMR values were log-transformed to make distributions close to normal. Interclass correlation coefficients were calculated to assess assay reproducibility using the SAS program (version 9.1; SAS Institute, Cary, NC). To assess the association between promoter methylation and loss of protein expression, Fisher’s exact test was performed using the SAS program. All P values were two-sided.
Results
Precision of Sodium Bisulfite Conversion and Quantitative Real-Time PCR (MethyLight)
We measured the threshold cycle (Ct) in our MethyLight assay for each of the bisulfite-converted samples (cases 1 through 4, samples B1 through B7) in each of the five MethyLight runs (M1 through M5) (Figure 1). Case 1 showed positive reaction (Ct <40) in MLH1, cases 2 and 3 showed positive reaction in CDKN2A and MGMT, and case 4 showed positive reaction in CDKN2A and MLH1. Table 1 shows representative Ct values (case 2, samples B1 through B7) for each of the five different MethyLight analyses (M1 through M5) for CDKN2A. We designated SD(B) as bisulfite-to-bisulfite SD (SD) (within-run variation in seven bisulfite-treated samples B1 to B7) in a particular MethyLight run, and SD(M) as run-to-run SD (within-sample variation by five runs M1 to M5) on a particular sample (Figure 1 and Table 1). Table 2 shows SD(B) and SD(M) for all positive markers and control genes ACTB and COL2A1 in cases 1 through 4, which appeared comparable with SDs of Ct values that have been described in various real-time PCR assays.39,40,41 SD(B) and SD(M) of Ct values for COL2A1 on the four cases were significantly smaller than those for ACTB (P = 0.0004 by paired t-test).
Table 1.
Ct Values of Case 2 in MethyLight for CDKN2A (p16)
| Different MethyLight runs
|
Mean of Ct values | SD(M) | ||||||
|---|---|---|---|---|---|---|---|---|
| M1 | M2 | M3 | M4 | M5 | ||||
| Different bisulfite treatments | B1 | 31.35 | 31.15 | 31.02 | 31.17 | 31.20 | 31.18 | 0.12 |
| B2 | 30.86 | 30.79 | 30.49 | 30.82 | 30.75 | 30.74 | 0.15 | |
| B3 | 31.41 | 31.19 | 31.42 | 31.33 | 31.22 | 31.31 | 0.11 | |
| B4 | 31.05 | 31.04 | 30.95 | 31.07 | 30.67 | 30.96 | 0.17 | |
| B5 | 31.07 | 30.94 | 31.07 | 31.15 | 31.18 | 31.08 | 0.09 | |
| B6 | 30.46 | 30.49 | 30.54 | 30.75 | 30.53 | 30.55 | 0.11 | |
| B7 | 30.41 | 30.22 | 30.09 | 30.18 | 30.33 | 30.25 | 0.13 | |
| Mean of Ct values | 30.94 | 30.83 | 30.80 | 30.92 | 30.84 | |||
| SD(B) | 0.39 | 0.36 | 0.45 | 0.39 | 0.36 | |||
Each of B1 through B7 represents each bisulfite-converted sample, and each of M1 through M5 represents each of MethyLight assay results on 5 different days. SD(M) refers to MethyLight-to-MethyLight SD within each sample, and SD(B) refers to bisulfite-to-bisulfite SD within each MethyLight run.
We obtained the PMR based on standard curves in each run, Ct of a gene of interest (GENE), Ct of ACTB (or COL2A1) as a control reaction to normalize for the amount of input DNA, and reactions for both GENE and ACTB (or COL2A1) of SssI-treated DNA to normalize for any difference in PCR efficiencies between GENE and ACTB (or COL2A1). Representative PMR values are listed in Table 3(case 3, samples B1 to B7) in the five different MethyLight runs (M1 to M5) for MGMT. We designated CV(B) as bisulfite-to-bisulfite CV, ie, within-run variation in seven bisulfite-treated samples B1 to B7, and CV(M) as MethyLight-to-MethyLight run-to-run CV, ie, within-sample variation by five runs M1 to M5.
Table 3.
PMR Values of Case 3 in MethyLight for MGMT Using COL2A1 as a Reference
| Different MethyLight runs
|
Mean of PMR | SD | CV(M) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| M1 | M2 | M3 | M4 | M5 | |||||
| Different bisulfite treatments | B1 | 36.75 | 28.71 | 33.57 | 43.92 | 22.69 | 33.13 | 8.04 | 0.24 |
| B2 | 39.52 | 35.63 | 40.11 | 42.61 | 31.90 | 37.95 | 4.21 | 0.11 | |
| B3 | 35.53 | 36.22 | 30.21 | 44.78 | 29.74 | 35.30 | 6.07 | 0.17 | |
| B4 | 36.16 | 40.01 | 33.42 | 42.43 | 41.46 | 38.70 | 3.80 | 0.10 | |
| B5 | 39.08 | 37.83 | 33.56 | 61.04 | 37.44 | 41.79 | 10.96 | 0.26 | |
| B6 | 31.62 | 36.00 | 34.49 | 46.19 | 32.56 | 36.17 | 5.85 | 0.16 | |
| B7 | 30.26 | 34.54 | 40.80 | 51.03 | 36.73 | 38.67 | 7.89 | 0.20 | |
| Mean of PMR | 35.56 | 35.56 | 35.16 | 47.43 | 33.22 | ||||
| SD | 3.50 | 3.50 | 3.86 | 6.67 | 6.10 | ||||
| CV(B) | 0.10 | 0.10 | 0.11 | 0.14 | 0.18 | ||||
Abbreviations: CV(B), bisulfite-to-bisulfite coefficient of variation (CV) within each run; CV(M), run-to-run CV within each bisulfite-treated sample.
As described previously, PMR values of ≤4 indicated methylation-negative.33 In our analysis, negative markers typically showed PMR ≤1 and were most frequently undetected (Ct >45 and PMR = 0). There were a total of five case-negative marker combinations (case 1, CDKN2A; case 1, MGMT; case 2, MLH1; case 3, MLH1; and case 4, MGMT). Within a total of 175 PMR results (five case-marker combinations times seven bisulfite-treated samples times five MethyLight runs) for these negative reactions, when using COL2A1 as a control, 160 runs (91%) showed PMR values of 0, 14 runs (8%) showed a PMR <1, and only one run (0.6%) showed PMR >1 (PMR 1.01). However, when using ACTB as a control, 4 of 175 negative reactions (2.3%) showed PMR >1 (PMR ranging from 1.10 to 3.59). Notably with either reference, none of these 175 presumed negative reactions showed PMR >4.
Mean and variance of PMR values in MethyLight using ACTB as a control were compared to those using COL2A1 as a control (Table 4). For each case-marker combination, we obtained PMR values 35 times (seven different bisulfite-treated samples times five MethyLight runs) using each one of ACTB and COL2A1 as a control. Distributions of PMR values became closer to normal distributions after log transformation of raw PMR values. Thereafter, we assessed the equality of variances in the 35 repeated runs. PMR variances using ACTB were significantly larger than those using COL2A1 in four of seven case-marker combinations (P < 0.003) (Table 4), indicating COL2A1 as a superior control. CVs of PMR values in MethyLight using COL2A1 as a control are shown in Table 5. Both bisulfite-to-bisulfite variation [CV(B)] and run-to-run variation [CV(M)] were similar, ranging from 0.05 to 0.6. Therefore, MethyLight assay showed fair reproducibility in terms of both bisulfite-to-bisulfite variations [CV(B)] and run-to-run variations [CV(M)]. To assess within-sample variation of PMR values, interclass correlation coefficients were calculated as 0.84, 0.74, and 0.79 for CDKN2A, MLH1, and MGMT, respectively, indicating fair reproducibility of MethyLight assays.
Table 4.
Mean and CV of PMR Values in MethyLight
|
CDKN2A
|
MLH1
|
MGMT
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Reference gene | Mean of PMR* | CV of PMR* | P value, F-test for the equality of variances | Mean of PMR* | CV of PMR* | P value, F-test for the equality of variances | Mean of PMR* | CV of PMR* | P value, F-test for the equality of variances | |
| Case 1 | ACTB | 122.05 | 0.48 | P = 0.08 | ||||||
| COL2A1 | 48.27 | 0.47 | ||||||||
| Case 2 | ACTB | 941.16 | 0.71 | P = 0.0004 | 140.69 | 0.64 | P = 0.2 | |||
| COL2A1 | 345.7 | 0.35 | 56.07 | 0.42 | ||||||
| Case 3 | ACTB | 8.95 | 0.60 | P = 0.002 | 64.92 | 0.61 | P = 8 × 10−8 | |||
| COL2A1 | 5.25 | 0.30 | 37.39 | 0.19 | ||||||
| Case 4 | ACTB | 369.79 | 0.52 | P = 4 × 10−6 | 32.87 | 0.33 | P = 0.9 | |||
| COL2A1 | 115.11 | 0.21 | 11.22 | 0.31 | ||||||
Blanks indicate negativity for methylation in a particular case-marker combination.
The mean and CV of PMR values are based on 35 different results (seven different bisulfite-treated samples times five different MethyLight runs) for each case-marker combination.
Table 5.
CV of PMR Values in MethyLight Using COL2A1 as a Reference
|
CDKN2A
|
MLH1
|
MGMT
|
|||||
|---|---|---|---|---|---|---|---|
| Mean of CV | Range | Mean of CV | Range | Mean of CV | Range | ||
| Case 1 | CV(B) | 0.23 | 0.11–0.32 | ||||
| CV(M) | 0.51 | 0.47–0.57 | |||||
| Case 2 | CV(B) | 0.18 | 0.12–0.23 | 0.30 | 0.21–0.35 | ||
| CV(M) | 0.36 | 0.29–0.48 | 0.44 | 0.30–0.60 | |||
| Case 3 | CV(B) | 0.29 | 0.23–0.38 | 0.13 | 0.10–0.18 | ||
| CV(M) | 0.22 | 0.046–0.48 | 0.18 | 0.10–0.26 | |||
| Case 4 | CV(B) | 0.17 | 0.15–0.20 | 0.21 | 0.062–0.31 | ||
| CV(M) | 0.17 | 0.085–0.26 | 0.31 | 0.16–0.52 |
Abbreviations: CV(B), bisulfite-to-bisulfite CV within each run; CV(M), run-to-run CV within each bisulfite-treated sample.
To examine variations of standard curves, which contribute to run-to-run variations, we made five standard curves in a single plate. For each sample-marker combination, we calculated five different PMR values, using each one of the five different standard curves. Slopes of the five standard curves ranged −3.23 to −3.57 (mean, −3.41; SD, 0.14), intercepts ranged 31.63 to 31.88 (mean, 31.79; SD, 0.10), and correlation coefficients (r) ranged 0.9928 to 0.9971 (mean, 0.9953; SD, 0.0025). CVs of the five different PMR values for each sample-positive marker combination ranged 0.00368 to 0.136 (mean, 0.0629; median, 0.0515). These data indicate that run-to-run PMR variations were partly due to variations in standard curves, which were small. In MethyLight runs M1 through M5, slopes of standard curves ranged −3.19 to −3.78 (mean, −3.52; SD, 0.20), intercepts ranged 31.56 to 32.96 (mean, 32.25; SD, 0.44), and correlation coefficients (r) ranged 0.9857 to 0.9990 (mean, 0.9934; SD, 0.0041). These data indicate good reproducibility in standard curves.
Assay Linearity
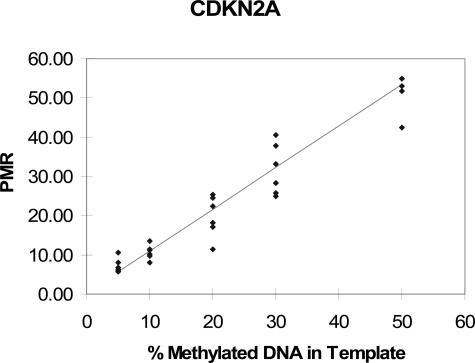
We performed repeated measurements of PMR values on DNA mixtures containing 50%, 30%, 20%, 10%, and 5% of methylated (M-SssI-treated) DNA (Figures 2and 3). Correlation coefficients and P values for the test of a linear association were as follows: r = 0.962, P = 3 × 10−17 for CDKN2A; r = 0.913, P = 2 × 10−12 for MLH1; and r = 0.904, P = 10−9 for MGMT. Thus, our MethyLight assay showed fair to good linearity for all three genes.
Figure 3.
DNA mixing study and linear regression analysis to assess linearity of MethyLight. DNA mixing assays were performed as described in Materials and Methods and Figure 2. The 100% SssI-treated DNA sample served as methylated reference DNA to calculate PMR. Linear regression analysis to assess assay linearity was performed.
Distribution of PMR Values and Correlation with Loss of Protein Expression
We performed MethyLight assays for the CDKN2A, MLH1, and MGMT promoters and immunohistochemistry to assess protein loss in 274 colorectal cancer cases. Among these, 272 cases (99.2%) demonstrated successful amplification in MethyLight assays, and only two cases (0.8%) failed to amplify. More than half of the tumors showed PMR = 0 (no methylation) for each marker, and most methylation-positive cases showed relatively high PMR values (Table 6). There were only rare cases with PMR values between 3 and 5. As such, the MethyLight assay for each gene appears to distinguish two distinct subsets of colorectal cancer, reflecting methylation-negative and -positive tumors. Our previous MethyLight analyses have defined methylated promoters as those with PMR >4 and unmethylated promoters as PMR ≤4.33 The bimodal distributions of PMR values in Table 6 appear to support a cutoff of 4.
Table 6.
Distributions of PMR Values of CDKN2A (p16), MLH1, and MGMT in Colorectal Cancer and Loss of Protein Expression
| PMR (methylation) | CDKN2A expression
|
MLH1 expression
|
MGMT expression
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Loss | Present | Total | Loss | Present | Total | Loss | Present | Total | |
| 0 | 5 | 139 | 144 | 3 | 215 | 218 | 17 | 108 | 125 |
| 0–1 | 1 | 12 | 13 | 0 | 6 | 6 | 1 | 6 | 7 |
| 1–3 | 1 | 5 | 6 | 0 | 1 | 1 | 1 | 3 | 4 |
| 3–4
|
0
|
0
|
0
|
0
|
2
|
2
|
0
|
3
|
3
|
| 4–5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 5–10 | 0 | 3 | 3 | 3 | 2 | 5 | 3 | 0 | 3 |
| 10–50 | 4 | 7 | 11 | 27 | 0 | 27 | 10 | 7 | 17 |
| >50 | 41 | 18 | 59 | 13 | 0 | 13 | 56 | 27 | 83 |
| Total | 52 | 184 | 236 | 46 | 226 | 272 | 88 | 158 | 243 |
Notes: The distributions of PMR values support PMR cutoff of 4. P values for statistical significance in association of PMR >4 with loss of expression are: P = 1 × 10−21 for CDKN2A, P = 6 × 10−44 for MLH1, and P = 1 × 10−17 for MGMT.
We compared promoter methylation status defined by PMR values for CDKN2A, MLH1, and MGMT to loss of protein expression assessed by immunohistochemistry (Table 6). Notably, intact protein expression was observed in 96% (156 of 163), 99% (224 of 227), and 86% (120 of 139) of samples with PMR ≤4 for the promoters of CDKN2A, MLH1, and MGMT, respectively. Loss of protein expression was observed in 62% (45 of 73), 96% (43 of 45), and 66% (69 of 104) of samples with PMR >4 for CDKN2A, MLH1, and MGMT, respectively. Using a PMR cutoff of 4, promoter methylation was significantly associated with loss of respective protein expression (P = 1 × 10−21 for CDKN2A, P = 6 × 10−44 for MLH1, and P = 1 × 10−17 for MGMT).
Discussion
Promoter CpG island methylation and transcriptional inactivation of tumor suppressor genes are thought to be important carcinogenic mechanisms. Assays to measure DNA methylation may be potentially very useful in clinical practice, because many tumor suppressor genes have been shown to be methylated and functionally silenced in a variety of human neoplasias.2,3 DNA methylation may be a useful marker for predicting prognosis and monitoring efficacy of adjuvant therapy in cancer patients10,11 and for evaluating risks for tumor recurrence in surveillance of high- or low-risk individuals.8,9,12 A variety of assays to measure DNA methylation have been developed for paraffin-embedded tissue, and many of these methods rely on sodium bisulfite treatment of genomic DNA from tumor tissue.7 However, precision and performance characteristics of sodium bisulfite treatment and measurement of methylation have not been comprehensively evaluated.
In this study, we evaluated precision of sodium bisulfite conversion as well as subsequent real-time PCR (MethyLight) assays for the CDKN2A (p16), MLH1, and MGMT promoters. Advantages of MethyLight technology include its quantitative and high-throughput nature and relatively simple assay procedures, which do not require opening of tubes containing PCR products. All of these features are extremely beneficial in clinical molecular diagnostics. Any of these properties is not a feature of qualitative MSP, which has been widely used for methylation detection. As for disadvantages of MethyLight, it cannot quantify methylation at the individual nucleotide level; rather, it can assess methylation levels at the primer sites and/or a probe site as a whole. On the other hand, bisulfite-sequencing, such as bisulfite-pyrosequencing,19,20,21 can achieve resolution at the individual nucleotide level. However, in contrast to MethyLight, pyrosequencing involves end-point analysis of PCR products and requires procedures that need opening of PCR tubes.
To measure the relative amount of methylated DNA, we divided the measured amount of a methylated gene of interest by the amount of COL2A1 (or ACTB) for input DNA. One should also be aware of PCR bias in quantitative molecular assays,42 so we further normalized for a potential difference in PCR efficiencies between the gene of interest and COL2A1 by dividing the gene:COL2A1 ratio in a sample of interest by that ratio in SssI-treated DNA (presumably fully methylated). For all three genes tested, we showed good precision in threshold cycle (Ct) values in terms of both bisulfite-to-bisulfite variation (among seven bisulfite-treated aliquots B1 to B7) in the same run and MethyLight-to-MethyLight run-to-run variation (among five repeated MethyLight runs M1 to M5) in the same bisulfite-treated DNA sample. COL2A1 reactions, as well as PMR values based on COL2A1 reactions, showed much smaller variances than ACTB. Thus, COL2A1 is a superior control gene to ACTB in terms of precision of MethyLight assays.
We also demonstrated that run-to-run CVs of PMR values were larger than bisulfite-to-bisulfite CVs of PMR values. This likely reflects the introduction of other sources of variations when we measured run-to-run CVs of PMR, including different standard curves. Thus, we also examined variations of standard curves in a single plate as well as between different MethyLight runs. We demonstrated that variations of standard curves were small and acceptable. Other sources of run-to-run variations are independent control reactions (COL2A1 or ACTB) in different MethyLight runs. A gene that showed small variation in repeated amplification reactions, such as COL2A1, would be suitable for a control. Normalizing methylation measurement for input DNA by a control reaction is necessary to compare quantitative methylation data across samples. Quality and quantity of DNA and the degree of bisulfite conversion can vary from sample to sample.
To assess acceptability of the precision of sodium bisulfite conversion and subsequent MethyLight assays, we measured PMR values (degrees of methylation) for the CDKN2A, MLH1, and MGMT promoters in 272 cases of colon cancer. We demonstrated that overall distributions of PMR values for all three loci were bimodal (either PMR ≤1 or >10) with only rare cases showing PMR values between 3 and 5. For most cases, CVs of PMR were sufficiently low to definitively differentiate methylation-positive tumor from methylation-negative tumor, with a PMR cutoff of 4. Thus, our data indicate that precision of sodium bisulfite conversion and quantitative Methy-Light assays is acceptable, and we can reliably determine the status of methylation for most cases by a single MethyLight run.
In addition, for each of the three genes tested, promoter methylation was strongly associated with loss of respective protein expression. Low PMR values between 0 and 4 were principally unrelated to loss of expression and, therefore, seemed to represent biologically insignificant low levels of methylation. There were infrequent cases with loss of expression without promoter methylation and cases with intact protein expression despite PMR values >10. The former may reflect different mechanisms of gene silencing, including gene deletion or mutation, and the latter may reflect partial methylation or mono-allelic methylation. Difficulty in interpreting immunohistochemistry, particularly for MGMT and CDKN2A (p16), may also contribute to some of the discrepant results.
In conclusion, sodium bisulfite conversion is reproducible, and subsequent quantitative real-time PCR methylation assays have acceptable precision. Quantitative promoter methylation data are highly correlated with loss of protein expression. Carefully validated quantitative MethyLight assays will be useful in both research and clinical molecular diagnostics.
Note Added in Proof
We have shown that MethyLight assays are useful for molecular classification of colorectal cancer, in particular for the determination of CpG island methylator phenotype, using a large number of samples.43
Table 2.
Standard Deviations (SD) of Ct Values in MethyLight
|
CDKN2A
|
MLH1
|
MGMT
|
ACTB
|
COL2A1
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean of SD | Range | Mean of SD | Range | Mean of SD | Range | Mean of SD* | Range | Mean of SD* | Range | ||
| Case 1 | SD(B) | 0.41 | 0.21–0.67 | 0.39 | 0.24–0.82 | 0.26 | 0.20–0.37 | ||||
| SD(M) | 0.96 | 0.58–1.1 | 0.45 | 0.14–0.93 | 0.19 | 0.095–0.44 | |||||
| Case 2 | SD(B) | 0.39 | 0.36–0.45 | 0.48 | 0.31–0.75 | 0.40 | 0.20–0.54 | 0.33 | 0.19–0.46 | ||
| SD(M) | 0.12 | 0.093–0.17 | 0.56 | 0.37–0.86 | 0.61 | 0.44–0.78 | 0.27 | 0.15–0.48 | |||
| Case 3 | SD(B) | 0.51 | 0.40–0.65 | 0.21 | 0.10–0.31 | 0.53 | 0.39–0.76 | 0.19 | 0.14–0.32 | ||
| SD(M) | 0.51 | 0.36–0.67 | 0.17 | 0.076–0.29 | 0.50 | 0.27–0.74 | 0.19 | 0.13–0.21 | |||
| Case 4 | SD(B) | 0.35 | 0.34–0.36 | 0.38 | 0.23–0.62 | 0.33 | 0.14–0.55 | 0.18 | 0.15–0.22 | ||
| SD(M) | 0.17 | 0.089–0.31 | 0.48 | 0.23–0.97 | 0.46 | 0.28–0.67 | 0.16 | 0.052–0.26 |
Paired t-test, P = 0.0004.
Blanks indicate negativity for methylation in a particular case-marker combination.
Abbreviations: SD(B), bisulfite-to-bisulfite SD within each run; SD(M), run-to-run SD within each bisulfite-treated sample.
Acknowledgments
We thank Neal Lindeman for critical reading of the manuscript and helpful discussion.
Footnotes
Supported by the National Institutes of Health (grants P01 CA87969-03 and P01 CA55075-13).
P.W.L. is shareholder and Scientific Advisory Board Member of Epigenomics, AG, which has a commercial interest in the development of DNA methylation markers for disease detection and diagnosis. None of the work described in this manuscript was supported by Epigenomics, AG.
References
- Costello JF, Vertino PM. Methylation matters: a new spin on maspin. Nat Genet. 2002;31:123–124. doi: 10.1038/ng0602-123. [DOI] [PubMed] [Google Scholar]
- Laird PW. Cancer epigenetics. Hum Mol Genet. 2005;14(Spec no 1):R65–R76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC, Schuller DE, Yu L, Bloomfield CD, Caligiuri MA, Yates A, Nishikawa R, Su Huang H, Petrelli NJ, Zhang X, O’Dorisio MS, Held WA, Cavenee WK, Plass C. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Siegmund KD, Muller HM, Fiegl H, Marth C, Muller-Holzner E, Jones PA, Laird PW. Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer Res. 2004;64:3807–3813. doi: 10.1158/0008-5472.CAN-03-3852. [DOI] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- Shen L, Kondo Y, Rosner GL, Xiao L, Hernandez NS, Vilaythong J, Houlihan S, Krouse RS, Prasad AR, Einspahr J, Buckmeier J, Alberts DS, Hamilton SR, Issa JP. MGMT promoter methylation and the field defect in sporadic colorectal cancer. J Natl Cancer Inst. 2005;97:1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- Giovannucci E, Ogino S. DNA methylation, field effects, and colorectal cancer. J Natl Cancer Inst. 2005;97:1317–1319. doi: 10.1093/jnci/dji305. [DOI] [PubMed] [Google Scholar]
- Fiegl H, Millinger S, Mueller-Holzner E, Marth C, Ensinger C, Berger A, Klocker H, Goebel G, Widschwendter M. Circulating tumor-specific DNA: a marker for monitoring efficacy of adjuvant therapy in cancer patients. Cancer Res. 2005;65:1141–1145. doi: 10.1158/0008-5472.CAN-04-2438. [DOI] [PubMed] [Google Scholar]
- Muller HM, Millinger S, Fiegl H, Goebel G, Ivarsson L, Widschwendter A, Muller-Holzner E, Marth C, Widschwendter M. Analysis of methylated genes in peritoneal fluids of ovarian cancer patients: a new prognostic tool. Clin Chem. 2004;50:2171–2173. doi: 10.1373/clinchem.2004.034090. [DOI] [PubMed] [Google Scholar]
- Woodson K, Weisenberger DJ, Campan M, Laird PW, Tangrea J, Johnson LL, Schatzkin A, Lanza E. Gene-specific methylation and subsequent risk of colorectal adenomas among participants of the polyp prevention trial. Cancer Epidemiol Biomarkers Prev. 2005;14:1219–1223. doi: 10.1158/1055-9965.EPI-04-0726. [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steigerwald SD, Pfeifer GP, Riggs AD. Ligation-mediated PCR improves the sensitivity of methylation analysis by restriction enzymes and detection of specific DNA strand breaks. Nucleic Acids Res. 1990;18:1435–1439. doi: 10.1093/nar/18.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalgo ML, Jones PA. Quantitative methylation analysis using methylation-sensitive single-nucleotide primer extension (Ms-SNuPE). Methods. 2002;27:128–133. doi: 10.1016/s1046-2023(02)00064-6. [DOI] [PubMed] [Google Scholar]
- Matin MM, Baumer A, Hornby DP. An analytical method for the detection of methylation differences at specific chromosomal loci using primer extension and ion pair reverse phase HPLC. Hum Mutat. 2002;20:305–311. doi: 10.1002/humu.10118. [DOI] [PubMed] [Google Scholar]
- Deng D, Deng G, Smith MF, Zhou J, Xin H, Powell SM, Lu Y. Simultaneous detection of CpG methylation and single nucleotide polymorphism by denaturing high performance liquid chromatography. Nucleic Acids Res. 2002;30:E13. doi: 10.1093/nar/30.3.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann K, Brinckmann A, Toliat MR, Ritter H, Nurnberg P. Evaluation of a potential epigenetic biomarker by quantitative methyl-single nucleotide polymorphism analysis. Electrophoresis. 2002;23:4072–4079. doi: 10.1002/elps.200290023. [DOI] [PubMed] [Google Scholar]
- Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by pyrosequencing. Biotechniques. 2003;35:152–156. doi: 10.2144/03351md02. [DOI] [PubMed] [Google Scholar]
- Schatz P, Dietrich D, Schuster M. Rapid analysis of CpG methylation patterns using RNase T1 cleavage and MALDI-TOF. Nucleic Acids Res. 2004;32:e167. doi: 10.1093/nar/gnh165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelker DL, Yamashita K, Tokumaru Y, Mimori K, Howard DL, Tanaka Y, Carvalho AL, Jiang W-W, Park HL, Kim MS, Osada M, Mori M, Sidransky D. PGP9.5 promoter methylation is an independent prognostic factor for esophageal squamous cell carcinoma. Cancer Res. 2005;65:4963–4968. doi: 10.1158/0008-5472.CAN-04-3923. [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Henrique R, Hoque MO, Mambo E, Ribeiro FR, Varzim G, Oliveira J, Teixeira MR, Lopes C, Sidransky D. A quantitative promoter methylation profile of prostate cancer. Clin Cancer Res. 2004;10:8472–8478. doi: 10.1158/1078-0432.CCR-04-0894. [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Usadel H, Henrique R, Oliveira J, Lopes C, Nelson WG, Sidransky D. Quantitation of GSTP1 methylation in non-neoplastic prostatic tissue and organ-confined prostate adenocarcinoma. J Natl Cancer Inst. 2001;93:1747–1752. doi: 10.1093/jnci/93.22.1747. [DOI] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Danenberg PV, Laird PW. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res. 1999;59:2302–2306. [PubMed] [Google Scholar]
- Zeschnigk M, Bohringer S, Price EA, Onadim Z, Masshofer L, Loh-mann DR. A novel real-time PCR assay for quantitative analysis of methylated alleles (QAMA): analysis of the retinoblastoma locus. Nucleic Acids Res. 2004;32:e125. doi: 10.1093/nar/gnh122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomassin H, Kress C, Grange T. MethylQuant: a sensitive method for quantifying methylation of specific cytosines within the genome. Nucleic Acids Res. 2004;32:e168. doi: 10.1093/nar/gnh166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell SE, Distler J, Goodman NS, Mooney SH, Kluth A, Olek A, Schwope I, Tetzner R, Ziebarth H, Berlin K. A real-time PCR assay for DNA-methylation using methylation-specific blockers. Nucleic Acids Res. 2004;32:e10. doi: 10.1093/nar/gnh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand K, Qu W, Ho T, Clark SJ, Molloy P. Conversion-specific detection of DNA methylation using real-time polymerase chain reaction (ConLight-MSP) to avoid false positives. Methods. 2002;27:114–120. doi: 10.1016/s1046-2023(02)00062-2. [DOI] [PubMed] [Google Scholar]
- Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:E65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads CA, Lord RV, Kurumboor SK, Wickramasinghe K, Skinner ML, Long TI, Peters JH, DeMeester TR, Danenberg KD, Danenberg PV, Laird PW, Skinner KA. Fields of aberrant CpG island hypermethylation in Barrett’s esophagus and associated adenocarcinoma. Cancer Res. 2000;60:5021–5026. [PubMed] [Google Scholar]
- Fuchs CS, Willett WC, Colditz GA, Hunter DJ, Stampfer MJ, Speizer FE, Giovannucci EL. The influence of folate and multivitamin use on the familial risk of colon cancer in women. Cancer Epidemiol Biomarkers Prev. 2002;11:227–234. [PubMed] [Google Scholar]
- Colditz GA, Hankinson SE. The Nurses’ Health Study: lifestyle and health among women. Nat Rev Cancer. 2005;5:388–396. doi: 10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- Eads CA, Lord RV, Wickramasinghe K, Long TI, Kurumboor SK, Bernstein L, Peters JH, DeMeester SR, DeMeester TR, Skinner KA, Laird PW. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61:3410–3418. [PubMed] [Google Scholar]
- Ogino S, Brahmandam M, Cantor M, Namgyal C, Kawasaki T, Kirkner G, Meyerhardt J, Loda M, Fuchs CS. Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucious component. Mod Pathol. 2006;19:59–68. doi: 10.1038/modpathol.3800482. [DOI] [PubMed] [Google Scholar]
- Rosner B. Fundamentals of Biostatistics. Boston: Duxbury Press,; 2006 [Google Scholar]
- Grace MB, McLeland CB, Gagliardi SJ, Smith JM, Jackson WE, III, Blakely WF. Development and assessment of a quantitative reverse transcription-PCR assay for simultaneous measurement of four amplicons. Clin Chem. 2003;49:1467–1475. doi: 10.1373/49.9.1467. [DOI] [PubMed] [Google Scholar]
- Gruber F, Falkner FG, Dorner F, Hammerle T. Quantitation of viral DNA by real-time PCR applying duplex amplification, internal standardization, and two-color fluorescence detection. Appl Environ Microbiol. 2001;67:2837–2839. doi: 10.1128/AEM.67.6.2837-2839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmann S, Taube T, Paal K, Graf VEH, Geilen W, Seifert G, Eckert C, Henze G, Seeger K. Specific reverse transcription-PCR quantification of vascular endothelial growth factor (VEGF) splice variants by LightCycler technology. Clin Chem. 2001;47:654–660. [PubMed] [Google Scholar]
- Ogino S, Wilson RB. Quantification of PCR bias caused by a single nucleotide polymorphism in SMN gene dosage analysis. J Mol Diagn. 2002;4:185–190. doi: 10.1016/S1525-1578(10)60702-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Cantor M, Kawasaki T, Brahmandam M, Kirkner G, Weisenberger DJ, Campan M, Laird PW, Loda M, Fuchs CS: CpG island methylator phenotype (CIMP) of colorectal cancer is best characterized by quantitative DNA methylation analysis and prospective cohort studies. Gut, In Press [DOI] [PMC free article] [PubMed] [Google Scholar]