Abstract
Over the last decade, fluorescence in situ hybridization (FISH) has become a firmly established technique in the diagnosis and assessment of lymphoid malignancies. However, this technique is not wide-ly used in the routine diagnostic evaluation of paraffin-embedded biopsies, most likely because of a perception that it is technically more demanding. There are also uncertainties regarding diagnostic thresholds and the way in which results should be interpreted. In this Review, we describe practical strategies for using FISH analysis to detect lymphoma-associated chromosomal abnormalities in routine paraffin-embedded lymphoma biopsies. Furthermore, we provide proposals on how FISH results should be interpreted (including how to calculate cutoff levels for FISH probes), recorded, and reported. An online appendix (available at http://jmd.amjpathol.org) details various simple, yet robust procedures for paraffin FISH analysis; it also provides additional information on the production of FISH probes, evaluating and reporting FISH results, sources for reagents and equipment, and troubleshooting. We hope that these suggestions will make FISH technology for the study of lymphoma biopsies more accessible to routine diagnostic and research laboratories.
Cytogenetic analysis, based on banding techniques, has historically proved to be invaluable for the detection of chromosomal abnormalities in tumor samples and is still considered to be the “gold standard” technique in tumor cytogenetics because it is the only technique providing a complete overview of all chromosomal changes within a tumor cell. However, the lack in availability of fresh material, the low mitotic index and/or percentage of neoplastic cells, the cytogenetic complexity, and the time-consuming nature of analysis all impose restrictions on the use of this technology for routine diagnosis.1,2
This is particularly true in the context of surgical biopsies, which reach the laboratory in an unfixed state in a few specialized centers but only rarely in peripheral laboratories. Information from conventional cytogenetic analysis can also, in part, be obtained by the use of polymerase chain reaction (PCR), Southern blotting, or comparative genomic hybridization techniques.3,4,5,6,7,8 However, the suitability of PCR and Southern blotting analysis for the detection of lymphoma-associated translocations is limited when chromosomal breakpoints are spread over a large genomic region, eg, as is the case for t(11;14)(q13;q32) translocations involving the CCND1 gene.4 Furthermore, because anomalies such as t(14;18)(q32;q21) and t(2;5)(p23;q35) translocations can be found in healthy individuals, the high sensitivity of the PCR technique can result in a degree of false-positive findings.9 Because comparative genomic hybridization analysis does not detect balanced translocations, it is of limited use for routinely diagnosing lymphomas in which the chromosomal translocations represent the commonest anomalies of diagnostic value. Most importantly, these techniques do not work as reliably on routine biopsies as they do on fresh tissue, and in practice, none of them are widely used for studying lymphoma biopsies in diagnostic laboratories.
Interphase fluorescence in situ hybridization (FISH) has long been used for characterizing hematological malignancies in bone marrow and blood samples, and several reports of its use on paraffin-embedded lymphoma biopsy material have appeared in the past 6 years (Table 1). However, it is still not widely used in routine diagnosis, probably because it is perceived to be technically demanding and costly. There are few guidelines or practical reviews for laboratories that wish to introduce this technique into routine practice.10
Table 1.
Examples of Published Reports of FISH Labeling of Paraffin-Embedded Tissue Sections for the Detection of Lymphoma-Related Chromosomal Abnormalities
| Lymphoma category | Chromosomal aberration(s) | Genes | Probe type* |
|---|---|---|---|
| ALK positive | t(2;5)(p23;q35) and variants | ALK and NPM (or other partners) | Break-apart5,40,41 and multicolor36 |
| Burkitt/Burkitt-like | t(8;14)(q24;q32) and variants | MYC and IGH (or other partners) | Break-apart42,43,44 |
| Diffuse large B cell | t(8;14)(q24;q32) and variants, t(14;18)(q32;q21), t(3q27) | MYC and IGH (or other partners), IGH/BCL2, and BCL6 | Break-apart45 and dual-fusion46 |
| Follicular | t(14;18)(q32;q21) and +3 | IGH and BCL2 | Break-apart,42 dual-fusion,8,46,47,48 and centromeric49 |
| Mantle cell | t(11;14)(q13;q32) | IGH/CCND1 | Break-apart42 and dual-fusion50 |
| MALT | t(11;18)(q21;q21),+3,+7, +12, and +18 | API2 and MALT1 | Break-apart, dual-fusion,51 and centromeric25 |
See text for descriptions of probe types.
In this Review, we describe strategies that are effective for identifying lymphoma-associated chromosomal abnormalities in routine paraffin-embedded tissue biopsies. We suggest approaches to the calculation of diagnostic cutoffs based on false positive rates and to the observation, recording, and interpretation of FISH analysis of this type of material. An online appendix (see supplemental material at http://jmd.amjpathol.org) details several protocols for FISH analysis and an extensive discussion on other factors important to the application of FISH to paraffin-embedded tissue.
We hope that these suggestions will make FISH technology for the study of paraffin-embedded lymphoma biopsies more accessible to routine diagnostic and research laboratories. It should be noted that this Review is dedicated to FISH on paraffin sections prepared from tissue biopsies rather than to leukemic samples. In the latter, conventional cytogenetic analyses supplemented by FISH are still the gold standard and should be routinely used.11
As a background to this practical review, it is valuable to consider the type of cytogenetic abnormalities that arise in human lymphoma and also the principles underlying their detection by the FISH technique in routine biopsies.
Cytogenetic Abnormalities in Lymphomas
A variety of primary and secondary nonrandom clonal cytogenetic abnormalities are found in lymphoid neoplasms, comprising translocations, inversions, insertions, duplications, amplifications, deletions, and aneusomy (eg, monosomy and trisomy).12 Characterization of the consequences of these changes at the DNA level has often provided the first step in the identification of lymphomagenesis-associated genes.13,14,15,16 Furthermore, many of the proteins encoded by these genes play important roles in diverse cellular functions such as apoptosis, regulation of cell growth, cell cycle control, and cell differentiation.15,16
Primary karyotypic changes in lymphoid neoplasms commonly juxtapose oncogenes to the potent transcriptional enhancers associated with IG and TCR loci in B and T cells, respectively, often resulting in elevated levels of protein expression and loss of normal mechanisms of control.13,15,16,17,18 Less commonly, fusion genes are created that encode novel hybrid proteins (eg, NPM-ALK in anaplastic large-cell lymphoma or API2-MALT1 in MALT lymphoma).13,19 Primary karyotypic abnormalities are often closely associated with an individual lymphoma subtype, and they can hence be of diagnostic value (Table 2). It should be noted, however, that not all cases in a particular lymphoma category necessarily harbor the expected translocation, eg, the t(14;18)(q32;q21) translocation, which is observed in only about 85% of follicular lymphomas,20 so its absence does not exclude this diagnosis. Also, some genetic abnormalities are seen in more than one category of hematological malignancy, eg, the t(8;14)(q24;q32) translocation found in Burkitt’s lymphoma but also, less commonly, in diffuse large-B-cell lymphoma, follicular lymphoma, mantle cell lymphoma, and other lymphomas. It is thus important to interpret the FISH results obtained from a lymphoma biopsy in the context of the patient’s clinical features and the pathology and immunohistology reports.
Table 2.
| Lymphoid malignancy | Chromosomal aberration(s) | Gene(s) involved |
|---|---|---|
| B-cell neoplasms | ||
| Burkitt’s lymphoma/leukemia | t(8;14)(q24;q32) | MYC/IGH |
| t(2;8)(p12;q24) and t(8;22)(q24;q11) | IGK/MYC and MYC/IGL | |
| Diffuse large B-cell lymphoma | t(3q27) | BCL6 |
| t(14q32) | IGH | |
| t(14;18)(q32;q21) | IGH/BCL2 | |
| t(8;14)(q24;q32) and variants | MYC/IGH (IGL/IGK) | |
| Follicular lymphoma | t(14;18)(q32;q21) and variants | IGH/BCL2 (IGL/IGK) |
| t(3;14)(q27;q32) | BCL6/IGH | |
| MALT lymphoma | t(11;18)(q21;q21) | API2/MALT1 |
| t(14;18)(q32;q21) | IGH/MALT1 | |
| t(1;14)(p22;q32) | BCL10/IGH | |
| +3/+3q | ||
| Mantle cell lymphoma | t(11;14)(q13;q32) | CCND1/IGH |
| T-cell neoplasms | ||
| Anaplastic large cell lymphoma | Rearrangements of 2p23 | ALK/various |
| Most commonly t(2;5)(p23;q35) | NPM/ALK | |
| Precursor T-cell lymphoblastic leukemia and | Rearrangements in 14q11 | TRA6/TRD6 |
| T-cell lymphoblastic lymphoma | Rearrangements in 7q35 | TRB6 |
This table does not include lymphoid malignancies in which paraffin-embedded material is not routinely used (eg B-cell chronic lymphocytic leukemia or multiple myeloma) nor a variety of anomalies (usually secondary events) that are rare and/or not relevant in the context of routine biopsy samples.
Secondary chromosomal changes occur more commonly in some types of lymphoma than others. They are characterized by multiple aberrations and can often be of prognostic value. For example, the t(8;14)(q24;q32) translocation is a primary aberration in Burkitt’s lymphoma, but it can also arise as a secondary aberration in follicular lymphoma,13,21 in which case, it is associated with poor prognosis.
Principles Underlying Interphase FISH
FISH methodology involves the binding of fragments of single-stranded DNA to complementary genomic target sequences in a cell or tissue preparation. These DNA probes are labeled, either directly or indirectly, with a fluorochrome, yielding a sharply defined fluorescent signal at the site of the target sequence within the nucleus.1,2 Most probes used in diagnostic laboratories are commercially available and are directly labeled with fluorochromes, and the present Review (and the online appendix) is based on this type of reagent. However, the online appendix also gives brief details of how to prepare “homemade probes” using a fluorescent label.
Types of Probes for the Detection of Translocations
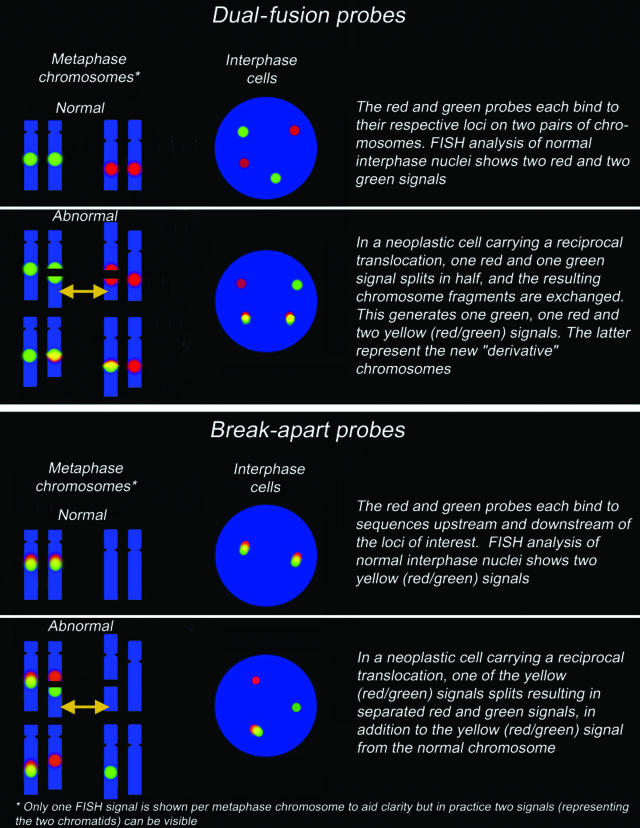
There are two fundamentally distinct categories of probes (Figure 1) for the detection of translocations. Such probes differ in terms of the type of information they yield, their sensitivity, and their ease of interpretation.
Figure 1.
Schematic representation of the characteristics of dual-fusion and break-apart probes. Depending on probe design (eg, the distance between the regions recognized) and the state of the genomic DNA at the time of fixation, a fused signal may appear either as a colocalized red and green signal or as a single yellow signal. When using break-apart probes, red/green signal pairs will occasionally appear to be slightly separated because of the secondary structure of the target DNA.
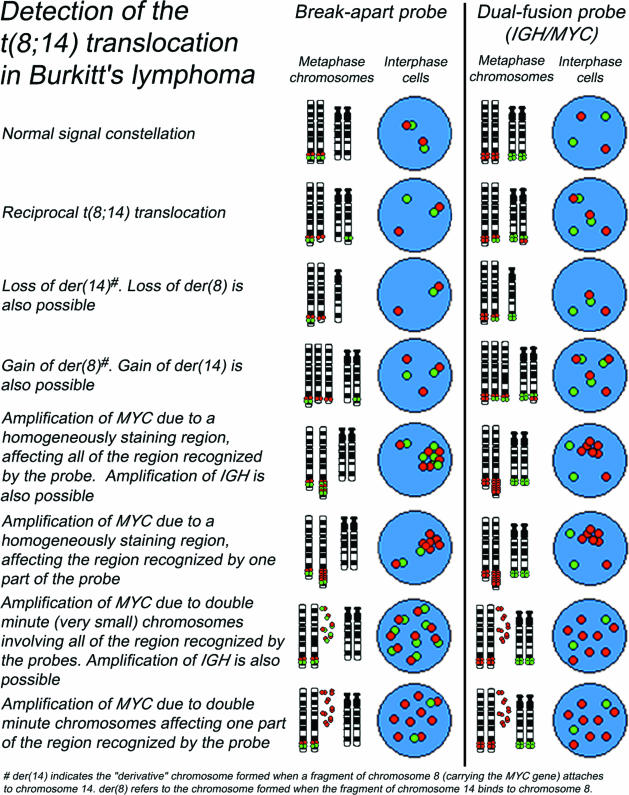
Dual-fusion probes consist of pairs of probes labeled in distinct colors, with each probe binding to a different chromosome. In contrast to single-fusion probes, which are located close to the translocation breakpoint and have a lower sensitivity, dual-fusion probes are designed to span the translocation breakpoint regions in the two different genes involved in a reciprocal translocation (Figure 1). For example, probes of different colors binding to the MYC (red) and IGH (green) genes are used to detect the t(8;14)(q24;q32) translocation. In a normal intact cell, two separate red and two separate green individual signals will be visible, whereas a reciprocal translocation t(8;14)(q24;q32) will generate two fused red/green signals (often appearing as single yellow signals), accompanied by one red and one green signal (representing the normal loci). It has to be added that this is the “classical” abnormal pattern: variant and complex patterns may also be seen (eg, because of gains, amplifications, or deletions). Any pattern differing from the signal patterns observed in normal cells should also be considered abnormal if it appears in a significant proportion of cells (Figures 2and 3). If the number, intensity, and location of signals in aberrant patterns is carefully evaluated and interpreted, it can provide valuable information on the underlying chromosomal change.
Figure 2.
Examples of normal and common variant signal constellations for t(8;14)(q24;q32) using break-apart and dual-fusion probes. Similar variant patterns may be seen in other translocations.
Figure 3.
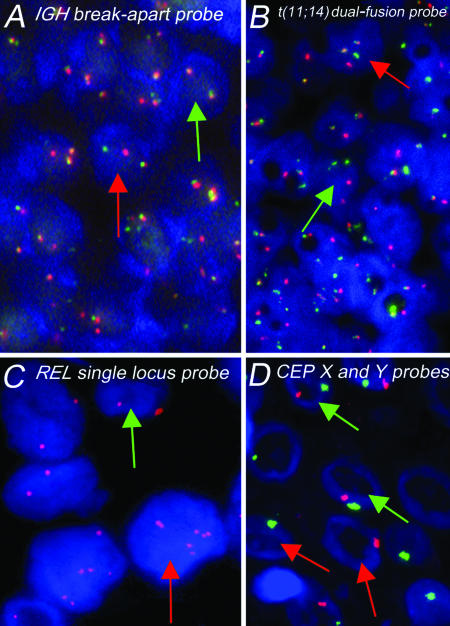
A and B: Photomicrographs of FISH patterns obtained in paraffin-embedded tissue sections with IGHbreak-apart and dual-fusion probes for t(11;14) in mantle cell lymphoma. A: A neoplastic cell with a split IGH locus (one fused, single red, and single green signals) is indicated by the red arrow. The green arrow indicates a normal cell (two fused signals). B: The red arrow indicates a neoplastic cell containing single red and green signals (corresponding to the normal CCND1 and IGH loci, respectively) together with two fused signals (corresponding to the genes fused by the translocation). A normal cell, with two red and two green signals, is indicated by the green arrow. C: Single locus probe for the detection of amplifications of the REL gene (2p16) in Hodgkin’s disease. Green arrow, normal diploid cell with two copies of the REL gene. Red arrow, larger cell showing gains (six copies) of the REL gene. D: Loss of nuclear material due to tissue sectioning demonstrated by FISH analysis of normal squamous epithelium using CEPs for chromosomes X and Y (red and green signals, respectively). The red arrows indicate cells containing only a single signal. However, two nuclei retain both signals (green arrows).
Break-apart probes also consist of pairs of two differently colored individual probes (ie, one red and one green). Each binds to sequences flanking the known breakpoint region in a locus or gene of interest (Figure 1), with the consequence that two sets of red/green-fused signals (representing the two alleles) will be visible in a normal diploid cell (provided the nucleus is intact). In an abnormal diploid cell, in which one allele has been split by a translocation, a separated red and green signal will be visible (hence the term “break-apart”) in addition to the normal fused signal. As for dual-fusion probes, any signal pattern deviating from the normal signal pattern described by the manufacturer or from those seen in normal cells should be considered abnormal (Figure 2) (see Principles of FISH Evaluation and online appendix for a discussion on cutoff values).
Which Type of Probe to Use: Break-Apart or Dual-Fusion?
Although break-apart probes can reveal breakage within a specific locus (and hence, by implication, that a translocation is present), they do not define the other gene involved. Moreover, because break-apart probes flank the locus of interest, small insertions leading to oncogenetic juxtapositions (eg, insertion of CCND1 into the IGH locus or insertion of IGH regulatory elements into CCND1) are likely to remain undetected. Although in this sense, they are less informative than dual-fusion probes, they have the advantage that, if the probe design is appropriate, they will give clearly abnormal results also for variant translocations in which different partners are involved, eg, t(8;14), t(2;8), t(8;22) for MYC. Furthermore, many laboratories prefer break-apart probes on the basis that the results are easier to interpret because the abnormality sought (separation of two signals) is readily recognized.10,11 However, normal signals may occasionally appear to be slightly separated when using break-apart probes, depending on the localization and secondary structure of the DNA, increasing the chances of a false-positive result. Thus, the “normal” pattern has to be carefully defined, eg, by evaluating the distance between signals in relation to the signal diameter. Depending on the probe design, the locus under study, and the material investigated, a signal pair might be scored positive only when a certain distance (eg, defined in relation to the signal diameters) is observed between signals (see Principles of FISH Evaluation).
A positive result using dual-fusion probes requires two fusion signals, an event that is very unlikely to occur by chance. Thus, dual-fusion probes are, strictly speaking, superior to break-apart probes regarding sensitivity because their false-positive rate on normal tissue is virtually zero.11,22,23 This issue can be especially relevant for detecting low levels of tumor cells in lymphomas with bone marrow or peripheral blood infiltration and for detecting minimal residual disease. However, in practice, this theoretic disadvantage is not usually significant when studying tissue sections because most lymphoma biopsies contain many neoplastic cells (with some exceptions, eg, Hodgkin’s lymphoma).
Types of Probes for the Detection of Copy Number Changes
Probes to detect copy number changes (eg, aneusomies, deletions, or amplifications) (Figure 3) are usually labeled in a single color and span the genomic region of interest. Such probes are either locus-specific (eg, REL and RB1) or detect repetitive sequences, like chromosome enumeration probes (CEPs), that bind to centromeric or pericentromeric satellites. CEPs can be used to detect aneusomies (ie, gains and losses of complete chromosomes) or for sex determination (for which the X and Y chromosomes are labeled in different colors) (Figure 3) after sex-mismatched bone marrow transplantation. Often CEPs will be combined with other types of probes. The commercially available IGH/MYC/CEP8 probe set (Vysis, Downers Grove, IL) contains a Spectrum Aqua (blue)-labeled probe for the centromere of chromosome 8 so that losses of the derivative 8 chromosome can be detected. Also, if the MYC gene is amplified, the inclusion of the CEP8 probe allows the level of amplification to be assessed as the ratio between MYC and CEP8 signals.
The major limitation of detecting copy number changes in tissue sections is that part of the cell can be lost during the sectioning process, leading to artificially induced deletions. This problem is irrelevant for the detection of amplifications, where extra copies or signal clusters will still be clearly visible. However, if the target chromosomal change is a deletion, one has to consider that the cutoff for such changes is high24 and has to be carefully determined using appropriate negative controls (see Principles of FISH Evaluation).
Preparation of Sections and FISH Labeling
Several publications in the past have recommended the use of sections thicker than those used for conventional histology, on the basis that this minimizes the number of nuclei that are truncated during sectioning.25,26,27 However, thick sections have their own disadvantages, principally the difficulty of interpreting signals in many different focal planes and distinguishing between individual nuclei. It must be considered that conventional sections will contain truncated nuclei, in which one or more FISH signals have been lost. As mentioned above, this is problematic for detecting deletions but not for amplifications and translocations. As a consequence, conventional sections (4 to 6 μm) are usually preferred by the authors.
All FISH techniques applied to routine tissue sections begin with dewaxing and dehydration steps, as for conventional (immuno)staining, and this is followed by a crucial “demasking” step in which the tissue is subjected to chemical and/or high-temperature treatment to make nuclear DNA sequences accessible to the probe. Three main methods are described in the online appendix, two using different types of pressure cooker and the third using chemical methods (see online appendix).
Most techniques include a subsequent proteolytic step, thus reducing background and enhancing signal visibility. Following these steps, FISH probes are added to the section and sealed under a coverslip. Samples are then briefly heated (70 to 90°C) to denature genomic DNA before overnight hybridization (37 to 45°C). These two incubation steps may be performed by moving slides from one oven/water bath to another or, more conveniently and reproducibly but also more expensively, by using dedicated hybridizing equipment into which slides are placed and not removed until hybridization is complete. Further details are given in the online appendix.
The demasking steps (whether by heating or chemical means) and subsequent proteolytic treatment are critical for obtaining readable FISH results. Insufficient pretreatment can result in weak or absent hybridization, whereas extended pretreatment can cause the tissue section to separate from the slide. Because tissue fixation and processing varies widely from sample to sample (and often within a single biopsy) and some specimens do not yield satisfactory results on first analysis, the technique may require repeating under different conditions (eg, longer demasking). Factors such as time from surgery to fixation, size of specimen, fixation parameters, and postfixation storage conditions may all potentially effect FISH quality (reviewed by Srinivason et al28). For instance, the use of buffered formalin is, in our experience, associated with the best hybridization signals. It is important to note that it may, therefore, be difficult or impossible to perform successful FISH analysis on some tissue biopsies, regardless of how many attempts are made.
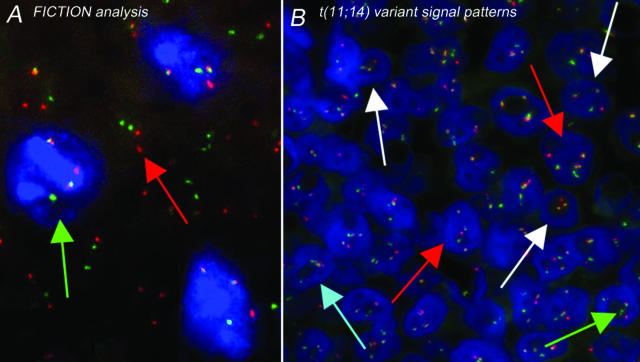
A discussion of the various techniques is included in the online appendix, including a protocol for “fluorescence immunophenotyping and interphase cytogenetics as a tool for investigation of neoplasms” (FICTION). This technique involves the combination of FISH and fluorescence immunophenotyping, thus allowing the simultaneous detection of genetic aberrations in immunologically defined cell populations (Figure 4).29,30
Figure 4.
Examples of FICTION and a variant FISH signal pattern. A: FICTION analysis of paraffin-embedded lymphoma tissue sections using an antibody against proliferating cells (MIB1, blue immunostaining) (see online appendix for protocol) and t(11;14) dual-fusion FISH probe without DAPI counterstain. Green arrow, MIB1-positive (proliferating) cell (immunostained blue) containing the IGH-CCND1 fusion, indicating the presence of a t(11;14) translocation (two fused, one red, and one green signal). The red arrow indicates a normal, ie, nonproliferating, cell. B: Mantle cell lymphoma showing a variant dual-fusion t(11;14) signal pattern. Red arrow, one red, one green, and three fused signals are present in contrast to the classic two fused, one red, and one green pattern. White arrows, variation of this pattern due to truncation artifacts and/or intraclonal heterogeneity. Green arrow, occasionally, two small probe signals corresponding to a single locus will appear slightly separated (dependent on the probe design, DNA secondary structure, and phase of the cell cycle). Care should be taken not to interpret these as separated signals as being indicative of an abnormality. Blue arrow, when the FISH signals in a neoplastic cell are grouped together, they might be scored as un-interpretable.
Principles of FISH Evaluation
When setting up a research or diagnostic FISH service for routine lymphoma biopsies, the person(s) responsible for interpreting the results should acquire experience of normal and abnormal signal patterns for each probe applied, using negative tissues (eg, tonsil from healthy individuals) and relevant positive samples (eg, lymphomas known to contain the abnormality under investigation). When evaluating the results of FISH labeling, several factors should be kept in mind: the architecture of the tissue, including local variations in neoplastic cell content, fixation, and cellularity within the section; the frequent presence of truncated nuclei; and the complex nature of genetic arrangements seen in some lymphoid neoplasms.
Choice of Area for Evaluation
FISH analysis is normally focused on areas richest in abnormal cells. However, the complete hybridized area should be screened for the presence of subclonal changes that might be of diagnostic and prognostic importance, eg, the presence of t(8;14) only in a subpopulation might indicate transformation into a more aggressive lymphoma. Thus, it may be useful to have a conventionally stained section at hand for reference. Variability will also be observed within the section in terms of tissue preservation and morphological detail. Thus, areas should be avoided where the borders of individual nuclei are not clearly identifiable and/or high cell density causes excessive nuclear overlap. At the same time that the quality of the tissue section is reviewed, the FISH signals should be assessed, looking for areas with bright, distinct signals and low background in which individual nuclei are clearly distinguishable.
Truncation Artifacts
When evaluating sections, it should be recognized that a substantial number of nuclei will have lost one or more of their target sequences recognized by conventional probes, a phenomenon that is more prominent in large-cell lymphomas. However, abnormal nuclei that have lost one or more signals due to truncation can also be recognized and interpreted. For example, when using break-apart probes, a nucleus with one fused (normal signal) and one single-color signal is suggestive of a translocation (on the assumption that the other translocated “broken-apart” sequence has been removed by sectioning). Similarly, using a dual-fusion probe, a nucleus with two single-color (normal) signals accompanied by only one fused signal would also suggest the presence of a translocation in a cell that has lost one fused sequence by tissue sectioning (this may also occur because of a chance colocalization of red and green signals in a normal cell).10 However, none of these patterns in a single cell are reliable indicators of classical translocations if they are not accompanied by a significant proportion of nuclei also showing an abnormal complement of signals (see below for discussion on cutoff values). When evaluating samples, it is also important to consider the possibility that signals are lost due to a chromosomal loss rather than a cell truncation.
Negative Controls and Establishment of Cutoff Values
One of the most critical factors affecting a proper interpretation of FISH signals in diagnostic samples is the establishment of cutoff values for the different probes used and for all signal patterns that might appear with a given assay. It is widely accepted that the diagnostic cutoff is calculated as the mean of false-positive findings in at least five healthy donors plus three times the standard deviation. For paraffin-embedded material, routine sections from normal lymphoid tissue (eg, tonsils) constitute appropriate negative controls. For a dual-fusion probe, the number of isolated and fused signals must be scored per cell. It also has to be considered that loss of one of the derivative chromosomes involved in the translocation is a nonrandom event in B-cell neoplasias, resulting in only one fusion instead of two. This pattern also commonly appears in negative controls as a result of random colocalizations of the loci under study, frequently leading to cutoff values of up to 15% for this constellation.
For break-apart probes, because the extension of the gap between red and green signals depends on the spreading of the breakpoints and on the probe design, a reproducible scoring system must be established. The authors’ strategy is to visually estimate the relative distance between the differentially colored probes of a signal pair within the nucleus and create several categories: 1) overlapping signals, 2) nonoverlapping signals, 3) distance between the signals more than one time the signal diameter, 4) distance between the signals more than two times the signal diameter, and 5) distance between the signals more than three times the signal diameter. In our view, a break-apart probe should ideally have a cutoff between 1 and 5% when the distance between flanking signals is twice or three times the estimated signal diameter.
For probes aimed at detecting copy number changes, the establishment of cutoffs for detecting deletions in paraffin-embedded material requires special consideration because sectioning induces truncation artifacts, and by definition, the cutoff will be higher than in samples containing intact nuclei. With regard to cutoff levels, it must always be taken into account that, for the vast majority of the assays, these are influenced by the condensation of the DNA (decondensed chromatin results in larger signals), the size of the nucleus (larger nuclei are more prone to sectioning artifacts), the assay design (the signal distance in break-apart assays depends partially on the genomic distance of the probes), the ploidy status (cells with supernumerary copies of the investigated loci have a higher likelihood of random colocalizations), the labeling scheme (combinatorial labeling can interfere with signal colocalization), and other variables. Several of these variables such as ploidy, nuclear size, and chromatin condensation can dramatically vary between normal control samples and tumor specimens. Thus, one should take into account that these factors cannot be properly modeled in negative controls. This is of particular importance in samples with low-tumor cell content or subclonal changes.
Complex Chromosomal Abnormalities
Lymphomas may contain more than one chromosomal abnormality, especially those that are acquired as secondary events. These are often “invisible” when cells are analyzed for specific abnormalities by the FISH technique, but on occasion, they can give rise to patterns that differ from the classical alterations (as described in the probe manufacturer’s literature). For example, cases of mantle cell lymphoma, follicular lymphoma, and Burkitt’s lymphoma will frequently show extra fusion signals and/or extra loci signals (Figures 2and 4). These “unusual” patterns should not be ignored but should be considered abnormal and interpreted in conjunction with pathology reports and relevant reports in the literature. It is therefore imperative that the specialist performing the analysis is aware of the possibility of variant signal patterns, and it is advisable to record accurately the FISH patterns in a series of 100 nuclei, and only then to draw conclusion from these patterns. If an observer views a slide looking only for an expected pattern, variant patterns may be ignored, and an incorrect diagnosis may be reached (a section on reporting FISH results has been included in the online appendix).
Future Directions and Conclusion
The establishment of, and adherence to, external quality control systems in laboratories using FISH analysis is essential; such initiatives have been previously described, and several are currently under way.31,32,33,34,35 It is also important to foster an interdisciplinary approach involving geneticists, pathologists, and clinicians when using FISH for diagnosis because the quantity and quality of data will be reflected in the accuracy of diagnosis and the efficacy of treatment.
The future will see increasing use of multicolor approaches36 and techniques that combine immunofluorescence and FISH.30,37 These techniques will enhance sensitivity and allow statements concerning both diagnosis and prognosis to be made on the basis of a single assay. FISH analysis can be performed on tissue microarrays,38 allowing larger numbers of cases to be simultaneously analyzed at lower cost. Furthermore, the development of improved automated FISH analysis systems, capable of counting signals by “tile sampling”,39 will make diagnosis less labor-intensive. In consequence, FISH techniques are likely to play an increasing role in the future of diagnosis and assessment of routine biopsies from hematological malignancies.
Supplementary Material
Acknowledgments
We thank Bridget Watson for her expert assistance with the preparation of this manuscript and the online appendix. We apologize to all colleagues whose work has not been quoted in the present manuscript.
Footnotes
Supported by the Leukemia Research Fund (grant no. 04013), Deutsche Krebshilfe, European MCL Network (EU), Lymphoma Research Fund (New York), Hensel-Stiftung, and Schleswig-Holsteinische Krebsgesellschaft.
D.Y.M. and R.S. contributed equally to this work.
Supplemental material for this article can be found on http://jmd.amjpathol.org.
References
- Kearney L. The impact of the new fish technologies on the cytogenetics of haematological malignancies. Br J Haematol. 1999;104:648–658. doi: 10.1046/j.1365-2141.1999.01181.x. [DOI] [PubMed] [Google Scholar]
- Gozzetti A, Le Beau MM. Fluorescence in situ hybridization: uses and limitations. Semin Hematol. 2000;37:320–333. doi: 10.1016/s0037-1963(00)90013-1. [DOI] [PubMed] [Google Scholar]
- Spagnolo DV, Ellis DW, Juneja S, Leong AS, Miliauskas J, Norris DL, Turner J. The role of molecular studies in lymphoma diagnosis: a review. Pathology. 2004;36:19–44. doi: 10.1080/00313020310001648404. [DOI] [PubMed] [Google Scholar]
- Belaud-Rotureau MA, Parrens M, Dubus P, Garroste JC, de Mascarel A, Merlio JP. A comparative analysis of FISH, RT-PCR, PCR, and immunohistochemistry for the diagnosis of mantle cell lymphomas. Mod Pathol. 2002;15:517–525. doi: 10.1038/modpathol.3880556. [DOI] [PubMed] [Google Scholar]
- Cataldo KA, Jalal SM, Law ME, Ansell SM, Inwards DJ, Fine M, Arber DA, Pulford KA, Strickler JG. Detection of t(2;5) in anaplastic large cell lymphoma: comparison of immunohistochemical studies, FISH, and RT-PCR in paraffin-embedded tissue. Am J Surg Pathol. 1999;23:1386–1392. doi: 10.1097/00000478-199911000-00009. [DOI] [PubMed] [Google Scholar]
- Colleoni GW, Jhanwar SC, Ladanyi M, Chen B. Comparison of a multiplex reverse transcriptase-polymerase chain reaction for BCR-ABL to fluorescence in situ hybridization, Southern blotting, and conventional cytogenetics in the monitoring of patients with Ph1-positive leukemias. Diagn Mol Pathol. 2000;9:203–209. doi: 10.1097/00019606-200012000-00005. [DOI] [PubMed] [Google Scholar]
- Viardot A, Martin-Subero JI, Siebert R, Harder S, Gesk S, Bentz M, Schlegelberger B. Detection of secondary genetic aberrations in follicle center cell derived lymphomas: assessment of the reliability of comparative genomic hybridization and standard chromosome analysis. Leukemia. 2001;15:177–183. doi: 10.1038/sj.leu.2401969. [DOI] [PubMed] [Google Scholar]
- Shaminie J, Peh SC, Tan MJ. Improvement in the detection rate of t(14;18) translocation on paraffin-embedded tissue: a combination approach using PCR and FISH. Pathology. 2003;35:414–421. doi: 10.1080/00313020310001602620. [DOI] [PubMed] [Google Scholar]
- Janz S, Potter M, Rabkin CS. Lymphoma- and leukemia-associated chromosomal translocations in healthy individuals. Genes Chromosomes Cancer. 2003;36:211–223. doi: 10.1002/gcc.10178. [DOI] [PubMed] [Google Scholar]
- Cook JR. Paraffin section interphase fluorescence in situ hybridization in the diagnosis and classification of non-Hodgkin lymphomas. Diagn Mol Pathol. 2004;13:197–206. doi: 10.1097/01.pdm.0000135286.05198.89. [DOI] [PubMed] [Google Scholar]
- Martin-Subero JI, Gesk S, Harder L, Grote W, Siebert R. Interphase cytogenetics of hematological neoplasms under the perspective of the novel WHO classification. Anticancer Res. 2003;23:1139–1148. [PubMed] [Google Scholar]
- Heim S, Mitelman F. New York: Wiley-Liss,; Cancer Cytogenetics. (ed 2) 1995 [Google Scholar]
- Chaganti RS, Nanjangud G, Schmidt H, Teruya-Feldstein J. Recurring chromosomal abnormalities in non-Hodgkin’s lymphoma: biologic and clinical significance. Semin Hematol. 2000;37:396–411. doi: 10.1016/s0037-1963(00)90019-2. [DOI] [PubMed] [Google Scholar]
- Rowley JD. Cytogenetic analysis in leukemia and lymphoma: an introduction. Semin Hematol. 2000;37:315–319. doi: 10.1016/s0037-1963(00)90012-x. [DOI] [PubMed] [Google Scholar]
- Willis TG, Dyer MJ. The role of immunoglobulin translocations in the pathogenesis of B-cell malignancies. Blood. 2000;96:808–822. [PubMed] [Google Scholar]
- Siebert R, Rosenwald A, Staudt LM, Morris SW. Molecular features of B-cell lymphoma. Curr Opin Oncol. 2001;13:316–324. doi: 10.1097/00001622-200109000-00002. [DOI] [PubMed] [Google Scholar]
- Rabbitts TH. Chromosomal translocations in human cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- Gesk S, Martin-Subero JI, Harder L, Luhmann B, Schlegelberger B, Calasanz MJ, Grote W, Siebert R. Molecular cytogenetic detection of chromosomal breakpoints in T-cell receptor gene loci. Leukemia. 2003;17:738–745. doi: 10.1038/sj.leu.2402884. [DOI] [PubMed] [Google Scholar]
- Dierlamm J, Baens M, Wlodarska I, Stefanova-Ouzounova M, Hernandez JM, Hossfeld DK, De Wolf-Peeters C, Hagemeijer A, Van den Berghe H, Marynen P. The apoptosis inhibitor gene API2 and a novel 18q gene MLT, are recurrently rearranged in the t(11;18)(q21;q21) associated with mucosa-associated lymphoid tissue lymphomas. Blood. 1999;93:3601–3609. [PubMed] [Google Scholar]
- Horsman DE, Okamoto I, Ludkovski O, Le N, Harder L, Gesk S, Siebert R, Chhanabhai M, Sehn L, Connors JM, Gascoyne RD. Follicular lymphoma lacking the t(14;18)(q32;q21): identification of two disease subtypes. Br J Haematol. 2003;120:424–433. doi: 10.1046/j.1365-2141.2003.04086.x. [DOI] [PubMed] [Google Scholar]
- Johansson B, Mertens F, Mitelman F. Cytogenetic evolution patterns in non-Hodgkin’s lymphoma. Blood. 1995;86:3905–3914. [PubMed] [Google Scholar]
- Li JY, Gaillard F, Moreau A, Harousseau JL, Laboisse C, Milpied N, Bataille R, Avet-Loiseau H. Detection of translocation t(11;14)(q13;q32) in mantle cell lymphoma by fluorescence in situ hybridization. Am J Pathol. 1999;154:1449–1452. doi: 10.1016/S0002-9440(10)65399-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remstein ED, Kurtin PJ, Buno I, Bailey RJ, Proffitt J, Wyatt WA, Hanson CA, Dewald GW. Diagnostic utility of fluorescence in situ hybridization in mantle-cell lymphoma. Br J Haematol. 2000;110:856–862. doi: 10.1046/j.1365-2141.2000.02303.x. [DOI] [PubMed] [Google Scholar]
- Wilkens L, Gerr H, Gadzicki D, Kreipe H, Schlegelberger B. Standardised fluorescence in situ hybridisation in cytological and histological specimens. Virchows Arch. 2005;447:586–592. doi: 10.1007/s00428-005-1211-9. [DOI] [PubMed] [Google Scholar]
- Taji S, Nomura K, Matsumoto Y, Sakabe H, Yoshida N, Mitsufuji S, Nishida K, Horiike S, Nakamura S, Morita M, Taniwaki M. Trisomy 3 may predict a poor response of gastric MALT lymphoma to Helicobacter pylori eradication therapy. World J Gastroenterol. 2005;11:89–93. doi: 10.3748/wjg.v11.i1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubele M, Zitzelsberger H, Szucs S, Werner M, Braselmann H, Hutzler P, Rodenacker K, Lehmann L, Minkus G, Hofler H. Comparative FISH analysis of numerical chromosome 7 abnormalities in 5-micron and 15-micron paraffin-embedded tissue sections from prostatic carcinoma. Histochem Cell Biol. 1997;107:121–126. doi: 10.1007/s004180050096. [DOI] [PubMed] [Google Scholar]
- D’Alessandro I, Zitzelsberger H, Hutzler P, Lehmann L, Braselmann H, Chimenti S, Hofler H. Numerical aberrations of chromosome 7 detected in 15 microns paraffin-embedded tissue sections of primary cutaneous melanomas by fluorescence in situ hybridization and confocal laser scanning microscopy. J Cutan Pathol. 1997;24:70–75. doi: 10.1111/j.1600-0560.1997.tb01099.x. [DOI] [PubMed] [Google Scholar]
- Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961–1971. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Matthiesen K, Winkemann M, Muller-Hermelink A, Schlegelberger B, Grote W. Simultaneous fluorescence immunophenotyping and interphase cytogenetics: a contribution to the characterization of tumor cells. J Histochem Cytochem. 1992;40:171–175. doi: 10.1177/40.2.1552161. [DOI] [PubMed] [Google Scholar]
- Martin-Subero JI, Chudoba I, Harder L, Gesk S, Grote W, Novo FJ, Calasanz MJ, Siebert R. Multicolor-FICTION: expanding the possibilities of combined morphologic, immunophenotypic, and genetic single cell analyses. Am J Pathol. 2002;161:413–420. doi: 10.1016/S0002-9440(10)64197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewald G, Stallard R, Al Saadi A, Arnold S, Bader PI, Blough R, Chen K, Elejalde BR, Harris CJ, Higgins RR, Hoeltge GA, Hsu WT, Kubic V, McCorquodale DJ, Micale MA, Moore JW, Phillips RM, Scheib-Wixted S, Schwartz S, Siembieda S, Strole K, VanTuinen P, Vance GH, Wiktor A, Zinsmeister A. A multicenter investigation with interphase fluorescence in situ hybridization using X- and Y-chromosome probes. Am J Med Genet. 1998;76:318–326. [PubMed] [Google Scholar]
- Dewald G, Stallard R, Alsaadi A, Arnold S, Blough R, Ceperich TM, Rafael Elejalde B, Fink J, Higgins JV, Higgins RR, Hoeltge GA, Hsu WT, Johnson EB, Kronberger D, McCorquodale DJ, Meisner LF, Micale MA, Oseth L, Payne JS, Schwartz S, Sheldon S, Sophian A, Storto P, Van Tuinen P, Wenger GD, Wiktor A, Willis LA, Yung JF, Zenger-Hain J. A multicenter investigation with D-FISH BCR/ABL1 probes. Cancer Genet Cytogenet. 2000;116:97–104. doi: 10.1016/s0165-4608(99)00120-x. [DOI] [PubMed] [Google Scholar]
- Dewald G, Stallard R, Bader PI, Chen K, Zenger-Hain J, Harris CJ, Higgins R, Hirsch B, Hsu WT, Johnson E, Kubic V, Kurczynski TW, Malone JM, McCorquodale DJ, Meilinger K, Meisner LF, Moore JW, Schwartz S, Siembieda S, Storto PD, Vance G, Van Tuinen P, Wiktor A, Yung JF. Toward quality assurance for metaphase FISH: a multi-center experience. Am J Med Genet. 1996;64:539–545. doi: 10.1002/(SICI)1096-8628(19960906)64:4<539::AID-AJMG3>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Dewald GW, Stallard R, Bader PI, Chen K, Zenger-Hain J, Harris CJ, Higgins R, Hirsch B, Hsu WT, Johnson E, Kubic V, Kurczynski TW, Malone JM, McCorquodale DJ, Meilinger K, Meisner LF, Moore JW, Schwartz S, Siembieda S, Storto PD, Vance G, Van Tuinen P, Wiktor A, Yung JF. Toward quality assurance for metaphase FISH: a multicenter experience. Am J Med Genet. 1996;65:190–196. doi: 10.1002/(SICI)1096-8628(19961028)65:3<190::AID-AJMG4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mascarello JT, Brothman AR, Davison K, Dewald GW, Herrman M, McCandless D, Park JP, Persons DL, Rao KW, Schneider NR, Vance GH, Cooley LD. Proficiency testing for laboratories performing fluorescence in situ hybridization with chromosome-specific DNA probes. Arch Pathol Lab Med. 2002;126:1458–1462. doi: 10.5858/2002-126-1458-PTFLPF. [DOI] [PubMed] [Google Scholar]
- Gascoyne RD, Lamant L, Martin-Subero JI, Lestou VS, Harris NL, Muller-Hermelink HK, Seymour JF, Campbell LJ, Horsman DE, Auvigne I, Espinos E, Siebert R, Delsol G. ALK-positive diffuse large B-cell lymphoma is associated with Clathrin-ALK rearrangements: report of 6 cases. Blood. 2003;102:2568–2573. doi: 10.1182/blood-2003-03-0786. [DOI] [PubMed] [Google Scholar]
- Martinez-Ramirez A, Cigudosa JC, Maestre L, Rodriguez-Perales S, Haralambieva E, Benitez J, Roncador G. Simultaneous detection of the immunophenotypic markers and genetic aberrations on routinely processed paraffin sections of lymphoma samples by means of the FICTION technique. Leukemia. 2004;18:348–353. doi: 10.1038/sj.leu.2403230. [DOI] [PubMed] [Google Scholar]
- Andersen CL, Hostetter G, Grigoryan A, Sauter G, Kallioniemi A. Improved procedure for fluorescence in situ hybridization on tissue microarrays. Cytometry. 2001;45:83–86. doi: 10.1002/1097-0320(20011001)45:2<83::aid-cyto1149>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Grigoryan AM, Dougherty ER, Kononen J, Bubendorf L, Hostetter G, Kallioniemi O. Morphological spot counting from stacked images for automated analysis of gene copy numbers by fluorescence in situ hybridization. J Biomed Opt. 2002;7:109–122. doi: 10.1117/1.1428292. [DOI] [PubMed] [Google Scholar]
- Tai YC, Kim LH, Peh SC. Common ALK gene rearrangement in Asian CD30+ anaplastic large cell lymphoma: an immunohistochemical and fluorescence in situ hybridisation (FISH) study on paraffin-embedded tissue. Pathology. 2003;35:436–443. doi: 10.1080/00313020310001602594. [DOI] [PubMed] [Google Scholar]
- Tan LH, Do E, Chong SM, Koay ES. Detection of ALK gene rearrangements in formalin-fixed, paraffin-embedded tissue using a fluorescence in situ hybridization (FISH) probe: a search for optimum conditions of tissue archiving and preparation for FISH. Mol Diagn. 2003;7:27–33. doi: 10.1007/BF03260017. [DOI] [PubMed] [Google Scholar]
- Haralambieva E, Kleiverda K, Mason DY, Schuuring E, Kluin PM. Detection of three common translocation breakpoints in non-Hodgkin’s lymphomas by fluorescence in situ hybridization on routine paraffin-embedded tissue sections. J Pathol. 2002;198:163–170. doi: 10.1002/path.1197. [DOI] [PubMed] [Google Scholar]
- Haralambieva E, Banham AH, Bastard C, Delsol G, Gaulard P, Ott G, Pileri S, Fletcher JA, Mason DY. Detection by the fluorescence in situ hybridization technique of MYC translocations in paraffin-embedded lymphoma biopsy samples. Br J Haematol. 2003;121:49–56. doi: 10.1046/j.1365-2141.2003.04238.x. [DOI] [PubMed] [Google Scholar]
- Haralambieva E, Schuuring E, Rosati S, van Noesel C, Jansen P, Appel I, Guikema J, Wabinga H, Bleggi-Torres LF, Lam K, van den Berg E, Mellink C, van Zelderen-Bhola S, Kluin P. Interphase fluorescence in situ hybridization for detection of 8q24/MYC breakpoints on routine histologic sections: validation in Burkitt lymphomas from three geographic regions. Genes Chromosomes Cancer. 2004;40:10–18. doi: 10.1002/gcc.20009. [DOI] [PubMed] [Google Scholar]
- Dave BJ, Weisenburger DD, Higgins CM, Pickering DL, Hess MM, Chan WC, Sanger WG. Cytogenetics and fluorescence in situ hybridization studies of diffuse large B-cell lymphoma in children and young adults. Cancer Genet Cytogenet. 2004;153:115–121. doi: 10.1016/j.cancergencyto.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Zhang X, Karnan S, Tagawa H, Suzuki R, Tsuzuki S, Hosokawa Y, Morishima Y, Nakamura S, Seto M. Comparison of genetic aberrations in CD10+ diffused large B-cell lymphoma and follicular lymphoma by comparative genomic hybridization and tissue-fluorescence in situ hybridization. Cancer Sci. 2004;95:809–814. doi: 10.1111/j.1349-7006.2004.tb02186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Nomura K, Matsumoto S, Ueda K, Nakao M, Nishida K, Sakabe H, Yokota S, Horiike S, Nakamine H, Nakamura S, Taniwaki M. Detection of t(14;18) in follicular lymphoma by dual-color fluorescence in situ hybridization on paraffin-embedded tissue sections. Cancer Genet Cytogenet. 2004;150:22–26. doi: 10.1016/j.cancergencyto.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Masaki Y, Ozaki M. Fluorescence in situ hybridization detection of chromosome IGH/BCL2 translocations from paraffin-embedded tissue: evaluation in follicular lymphoma. Int J Hematol. 2003;78:154–159. doi: 10.1007/BF02983385. [DOI] [PubMed] [Google Scholar]
- Aguilera NS, Tomaszewski MM, Moad JC, Bauer FA, Taubenberger JK, Abbondanzo SL. Cutaneous follicle center lymphoma: a clinicopathologic study of 19 cases. Mod Pathol. 2001;14:828–835. doi: 10.1038/modpathol.3880398. [DOI] [PubMed] [Google Scholar]
- Kodet R, Mrhalova M, Krskova L, Soukup J, Campr V, Neskudla T, Szepe P, Plank L. Mantle cell lymphoma: improved diagnostics using a combined approach of immunohistochemistry and identification of t(11;14)(q13;q32) by polymerase chain reaction and fluorescence in situ hybridization. Virchows Arch. 2003;442:538–547. doi: 10.1007/s00428-003-0809-z. [DOI] [PubMed] [Google Scholar]
- Ye H, Gong L, Liu H, Hamoudi RA, Shirali S, Ho L, Chott A, Streubel B, Siebert R, Gesk S, Martin-Subero JI, Radford JA, Banerjee S, Nicholson AG, Ranaldi R, Remstein ED, Gao Z, Zheng J, Isaacson PG, Dogan A, Du MQ. MALT lymphoma with t(14;18)(q32;q21)/IGH-MALT1 is characterized by strong cytoplasmic MALT1 and BCL10 expression. J Pathol. 2005;205:293–301. doi: 10.1002/path.1715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.