Abstract
Monitoring breakpoint cluster region-Abelson kinase (BCR-ABL) levels in patients treated for chronic myelogenous leukemia (CML) has become an integral part of patient management. Real-time reverse transcriptase-polymerase chain reaction is the method of choice for this purpose because of its high analytical sensitivity and reproducibility. Given the variation of RNA quality and quantity in clinical specimens, accurate quantitative assessment of BCR-ABL depends on normalization of the BCR-ABL signal to an appropriate internal reference. However, the controls used by different laboratories vary, and there is no clear consensus on an ideal reference due to limited investigations. In this study, we compared nine commonly used control genes for three criteria: mRNA abundance, levels in CML and non-CML cells, and their degradation kinetics in comparison with BCR-ABL. We found that β-glucuronidase (GUSB) is the most suitable among the nine genes tested. Although ABL is most widely used, our data suggest that the amount of ABL is different in CML and non-CML cells. Moreover, ABL levels are regulated by cellular stress. These findings have a direct impact on current clinical laboratory practice and patient care because the use of a proper control gene affects the reported levels of BCR-ABL transcripts used for patient management decisions.
Chromosomal translocation t(9;22) is a hallmark of chronic myelogenous leukemia (CML).1,2 It can be found in 95% of patients with CML. At the molecular level, the translocation joins the 5′ segment of the breakpoint cluster region (BCR) gene on chromosome 22 to the 3′ portion of the Abelson kinase (ABL) gene on chromosome 9. Reverse transcriptase-polymerase chain reaction (RT-PCR) detection of BCR-ABL has long been used to aid in the diagnosis of CML and monitoring of residual leukemia after therapy.3
With the recent advent of newer therapies, such as the tyrosine kinase inhibitor Gleevec (also known as STI-571 or imatinib mesylate), molecular monitoring has become indispensable for assessment of patients’ therapeutic response and early detection of relapse.4,5,6,7,8 For patients who develop acquired resistance to Gleevec, therapeutic strategies have been developed to overcome such resistance.9,10 Conventional RT-PCR that generates only positive or negative results does not allow timely assessment of therapeutic response because many patients remain positive for a long period even after they achieve a cytogenetic response.11 In contrast, quantitative assessment of BCR-ABL transcripts using real-time technology has become the method of choice. It has been proven as a clinically useful test because patients with high or increasing levels of BCR-ABL over the disease course have a greater probability of relapse than those with steady-state or decreasing levels of BCR-ABL.3 Essential to accurate determination of BCR-ABL is the application of an appropriate internal normalization control because RNA derived from the clinical samples varies a great deal in both quality and quantity.
Review of the literature and a survey conducted by the Association for Molecular Pathology in 2002 showed that internal control genes that are widely used in Europe and North America for BCR-ABL quantitative RT-PCR include ABL,12,13,14,15,16 BCR,5,17 glucose 6-phosphate dehydrogenase (G6PD),7,18 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH).19,20 These were used as internal controls mainly because they had been historically used in conventional RT-PCR assays to assess the quality of cDNA. So far, only one study has been performed by the Europe Against Cancer (EAC) Program to examine suitability of different controls for BCR-ABL quantification. The study has proposed using ABL as the internal control after comparing several commonly used internal control genes.21 However, the primers/probe for ABL used in the study detect not only ABL wild-type allele but also the BCR-ABL fusion gene. The ratio of BCR-ABL to control then becomes BCR-ABL/(BCR-ABL+ABL). Although the authors were concerned that the detected level of ABL may be affected by the level of BCR-ABL that would lead to an underestimation of the tumor load, the authors stated that the ABL assay by this design led to a limited inaccuracy for diagnostic specimens expressing high levels of BCR-ABL transcripts. Because the assay is primarily used for CML monitoring after therapy, the impact of the ABL primers/probe design on minimal residual disease assessment need to be addressed.
In this study, we investigated nine commonly used control genes for BCR-ABL quantification, including β-actin (ACTB), β2-microglobulin (B2M), GAPDH, G6PD, GUSB, hypoxanthine phosphoribosyltransferase (HPRT), phosphoglycerate kinase 1 (PGK), TATA-box binding protein (TBP), and ABL, according to the following criteria: 1) suitable control genes are expressed at similar level to BCR-ABL, 2) suitable control genes are expressed in CML cells at similar level to that in non-CML cells, and 3) degradation kinetics of suitable control genes parallels that of the BCR-ABL transcripts. We have found that ABL does not serve as an appropriate control gene regardless of how primers/probe are designed. Among genes studied, G6PD and GUSB meet all criteria as appropriate controls for BCR-ABL quantification. We recommend using GUSB as the control gene of choice because mutations or variations occur at a much rarer frequency in the gene than in the G6PD locus.
Materials and Methods
Specimens
A total of 21 patient specimens from 19 patients were studied under an Institutional Review Board protocol. These include 12 specimens from newly diagnosed CML patients, 5 from patients who had been treated, 1 from a patient in blast crisis, 1 from a patient in accelerated phase of disease, and 2 different types of specimens from a patient with minimal residual disease. Specimen types included 15 bone marrow aspirates and 6 peripheral blood samples (Table 1).
Table 1.
Sample Description
| Specimen | Gender | Age | Specimen type | Stage of disease |
|---|---|---|---|---|
| 1 | M | 60 | BM | CP/diagnosis |
| 2 | M | 59 | BM | CP/diagnosis |
| 3 | F | 52 | BM | CP/diagnosis |
| 4 | M | 72 | BM | CP/diagnosis |
| 5 | F | 33 | BM | CP/diagnosis |
| 6 | M | 42 | BM | CP/diagnosis |
| 7* | F | 68 | BM | CP/diagnosis |
| 8 | M | 34 | PB | CP/diagnosis |
| 9 | M | 68 | PB | CP/diagnosis |
| 10 | M | 50 | BM | CP/diagnosis |
| 11 | M | 56 | PB | CP/diagnosis |
| 12 | M | 35 | BM | CP/diagnosis |
| 13 | F | 65 | BM | CP/treated |
| 14 | M | 43 | PB | CP/treated |
| 15 | F | 61 | PB | CP/treated |
| 16 | F | 31 | BM | CP/treated |
| 17 | M | 58 | BM | BP |
| 18† | F | 55 | PB | Post BMT/MRD |
| 19† | F | 55 | BM | Post BMT/MRD |
| 20* | F | 68 | BM | CP/treated |
| 21 | M | 63 | BM | AP |
Specimens 7 and 20 were obtained from the same patient at different times.
Specimens 18 and 19 were from the same patient.
M, male; F, female; BM, PB, peripheral blood; bone marrow; BMT/MRD, bone marrow transplantation/ minimal residual disease; CP, chronic phase; BP, blastic phase; AP, accelerated phase.
RNA Isolation, Quantification, and Reverse Transcription
Total cellular RNA was isolated from patient samples using RNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. RNA was eluted from the RNeasy column in 30 μl of RNase-free water. The amount of total RNA isolated from the cells was quantified using spectrophotometric measurements. Four micrograms of RNA was reverse-transcribed in an 80-μl reaction volume using a Reverse Transcription System (Promega, Madison, WI) according to the manufacturer’s protocol.
Real-Time PCR
Real-time PCR was conducted in an ABI PRISM 7000 Sequence Detection System (Applied Biosystems [ABI], Foster City, CA). cDNA made from 100 ng of total RNA was added to 25 μl of 1× Taqman Universal PCR master mix. The reaction contains 300 nmol/L of primers and 200 nmol/L probe. PCR was conducted using the following default TaqMan PCR conditions: 50°C for 2 minutes, 95°C for 10 minutes, followed by 50 cycles of 95°C for 15 seconds and 60°C for 60 seconds. Triplicate PCR reactions were conducted for each sample. Water instead of cDNA was included as a blank sample to control for PCR contamination.
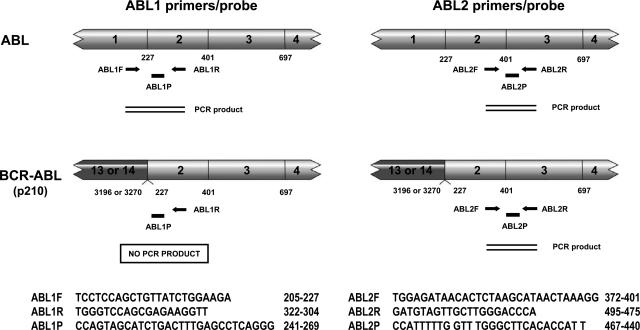
Except for G6PD and two ABL primer/probe sets, all other primer/probe sets, including ACTB, B2M, GAPDH, GUSB, HPRT, PGK, and TBP, were TaqMan Pre-developed Assay Reagents from ABI. G6PD was obtained from ABI as an Assay-on-Demand gene expression product. Two ABL primer/probe sets were reported previously21,22 and were custom-made by ABI. Their sequences and relative positions to ABL and BCR-ABL genes are illustrated in Figure 1. As shown in the left panel of Figure 1, the upstream primer of the ABL1 set hybridizes to exon 1, and the downstream primer and probe hybridize to exon 2 of the ABL gene. Because the breakpoints mostly occur in the intron between exons 1 and 2, the ABL1 set therefore detects only the wild-type allele of the ABL gene. In comparison, the upstream primer of ABL2 set hybridizes to exon 2, and the downstream primer and probe hybridize to exon 3 of the ABL gene (Figure 1, right). It therefore detects both the wild-type ABL and translocated BCR-ABL messages. Real-time PCR results were analyzed with ABI Prism 7000 SDS software, and autothresholds and autobaselines determined by the software for each individual gene target were applied to generate values of corresponding threshold cycles (Ct).
Figure 1.
Schematic diagram of the two different sets of primers/probes for ABL quantification (left, ABL1; right, ABL2). Lightly shaded boxes represent ABL cDNA and darkly shaded boxes represent BCR cDNA with exons indicated. Numbers below cDNAs indicate nucleotide positions at exon boundaries. Arrows represent PCR primers and their relative positions to ABL and BCR-ABL cDNAs. Black bars represent the TaqMan probes and their positions. Sequences of primers and probes and their locations are shown under each diagram.
Degradation Kinetics
To study the degradation kinetics of BCR-ABL and control genes, mononuclear cells from the bone marrow of two CML patients were placed on the bench to let cells die and RNA degrade. Aliquots of cells were collected at different time points, followed immediately by RNA extraction. RNA was reverse-transcribed, and levels of BCR-ABL and control genes were determined simultaneously by real-time RT-PCR in triplicate reactions. Levels of BCR-ABL and control genes at each time point after day 0 are expressed as a percentage of the initial time point calculated by the ΔCt method using the following formula: % = 2−ΔCt × 100%, where ΔCt = Ctday x − Ctday 0. For example, if on day x, Ct is one cycle higher than on day 0, then ΔCt = 1 and 2−ΔCt × 100% = 50%.
Results
mRNA Abundance of Commonly Used Control Genes Compared with BCR-ABL
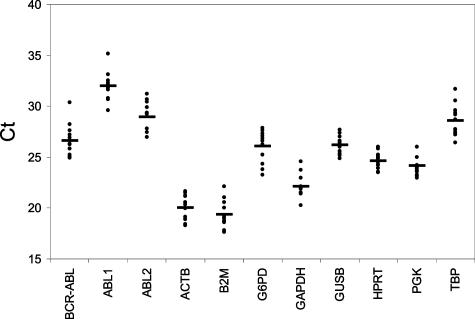
Suitable normalization control genes should be expressed at similar levels to the BCR-ABL gene so that they would be similarly sensitive to variations in the amount of RNA in test samples. To select for such control genes, we compared the expression levels of nine commonly used control genes, including ABL, ACTB, B2M, G6PD, GAPDH, GUSB, HPRT, PGK, TBP, and BCR-ABL in patient samples using TaqMan technology on an ABI PRISM 7000 Sequence Detection System. Except for ABL, none of the selected genes are located on chromosomes 8, 17, 19, or 22, which are frequently subject to rearrangements in CML. Among them, ABL was assessed using two sets of differently designed primer/probe sets (see Material and Methods; Figure 1). ABL1 primers flank the major breakpoint region in the ABL gene; they therefore only amplify cDNA derived from the wild-type ABL transcript. In contrast, both ABL2 primers are located downstream from the translocation breakpoints; ABL2 therefore amplified cDNA from both fusion and wild-type ABL transcript.23 Ct values of the BCR-ABL and the nine control genes were determined in 12 pretreatment patient samples (Figure 2; Table 1, patients 1 to 12). For each gene, mean Ct values of 12 samples and their differences from that of BCR-ABL are listed in Table 2. The mean Ct value of BCR-ABL in the 12 patients was 26.58. Among the 10 control genes, ACT, B2M, and GAPDH had mean Ct values more than four cycles lower than BCR-ABL. On the other hand, ABL level as assessed by ABL1 primer/probe set was more than five cycles higher than BCR-ABL. mRNA levels of the remaining genes including G6PD, GUSB, HPRT, PGK, TBP, and ABL by ABL2 primers/probe were less than 2.5 cycles different from the BCR-ABL; they therefore meet these criteria as suitable control genes.
Figure 2.
Threshold cycles of the BCR-ABL and control genes in 12 pretreatment patient samples. Triplicate PCR reactions were performed for each patient sample. The mean of Ct values of 12 samples are represented by horizontal bars.
Table 2.
Threshold Cycles of the Control Genes and BCR-ABL in CML Samples (n = 12)
| Genes | Ct (mean ± 2SD)* | ΔCt (BCR-ABL − control) |
|---|---|---|
| BCR-ABL | 26.58 ± 3.25 | — |
| ABL1 | 31.96 ± 2.75 | −5.38 |
| ABL2 | 28.94 ± 3.07 | −2.36 |
| ACTB | 20.02 ± 2.43 | +6.56 |
| B2M | 19.39 ± 2.65 | +7.19 |
| G6PD | 26.07 ± 3.09 | +0.51 |
| GAPDH | 22.12 ± 2.30 | +4.46 |
| GUSB | 26.32 ± 1.71 | +0.26 |
| HPRT | 24.66 ± 1.62 | +1.92 |
| PGK | 24.14 ± 2.05 | +2.44 |
| TBP | 28.57 ± 3.11 | −1.99 |
The Ct values for individual specimen are calculated based on six replicate reactions from two experiments. Data shown are means ± 2SD for 12 CML samples.
mRNA Levels of the Control Genes in CML and Non-CML Cells
Because quantitative BCR-ABL is primarily used in patients who have been treated for CML to monitor their response to therapy, the bone marrow or peripheral blood from these patients typically contains normal hematopoietic cells in addition to residual CML cells. A key criterion for a suitable control gene should be that it is expressed in CML cells at a comparable level to that in non-CML cells, so that the level of the control gene ultimately reflects the amount of total RNA being analyzed irrespective of the CML-to-non-CML cell ratio in the mixture. To study which control genes meet these criteria, we first analyzed the control genes in well-characterized leukemic cell lines. To mimic samples from treated patients, we mixed one part of K562 cells, a CML cell line bearing BCR-ABL, in nine parts of HL60 cells, a promyelocytic cell line that lacks BCR-ABL. We compared the levels of the control genes in K562 cells with no dilution and K562 diluted with HL60. It is expected that the Ct values of BCR-ABL differ by 3.32 (equivalent to a 10-fold difference in the amount of initial BCR-ABL cDNA). It is also expected that Ct values of a suitable control gene are essentially the same in equal amounts of pure K562 and mixed K562/HL60 cells if the control gene is expressed at comparable levels in both cell types. As shown in Table 3, among the nine genes tested by 10 primer/probe sets, there was a difference of less than 0.5 cycles in the Ct values of ACTB, GAPDH, GUSB, and PGK between K562 and the K562/HL60 mixture (ΔCt column in Table 3), suggesting that their levels are similar in both CML and non-CML cells. The Ct of B2M, G6PD, HPRT, TBP, and ABL levels by both ABL1 and ABL2 differ between the two samples from 0.76 cycle for HPRT to 2.51 cycles for ABL2, suggesting that K562 and HL60 cells contain significantly different amounts of the respective control gene mRNA. B2M, HPRT, and TBP were not further studied for this reason as well as the fact that they are not widely used controls for BCR-ABL quantification. Besides having a Ct that is more than six cycles lower than BCR-ABL, the amplification plot of ACTB is shaped differently from other control genes, because the plateau phase was reached much earlier, suggesting that some PCR components are limiting in the reaction (data not shown). ACTB, therefore, was also not chosen for further study.
Table 3.
Threshold Cycles of the Control Genes and BCR-ABL in K562 and K562/HL60 Mixtures
| Genes | Ct (mean ± 2SD)*
|
ΔCt† | P value‡ | |
|---|---|---|---|---|
| K562 | K562/HL60 | |||
| BCR-ABL | 21.50 ± 0.34 | 24.97 ± 0.08 | 3.47 | <0.001 |
| ABL1 | 29.45 ± 0.92 | 27.56 ± 0.94 | −1.89 | <0.001 |
| ABL2 | 22.90 ± 0.26 | 25.41 ± 0.50 | 2.51 | <0.001 |
| ACTB | 18.90 ± 0.16 | 18.98 ± 0.20 | 0.08 | 0.176 |
| B2M | 20.93 ± 0.18 | 18.52 ± 0.20 | −2.41 | <0.001 |
| G6PD | 22.87 ± 0.07 | 24.98 ± 0.02 | 2.12 | <0.001 |
| GAPDH | 18.99 ± 0.24 | 18.96 ± 0.08 | −0.03 | 0.618 |
| GUSB | 23.84 ± 0.26 | 23.72 ± 0.12 | −0.12 | 0.062 |
| HPRT | 21.59 ± 0.80 | 22.35 ± 0.72 | 0.76 | 0.006 |
| PGK | 21.97 ± 0.06 | 21.50 ± 0.50 | −0.47 | 0.001 |
| TBP | 23.61 ± 0.14 | 24.99 ± 0.08 | 1.38 | <0.001 |
The Ct values are calculated based on six replicate reactions from two experiments.
ΔCt = Ct (K562/HL60) − Ct (K562).
Analyzed by Student’s t-test.
Notably, the Ct of ABL assayed by ABL1 or ABL2 was significantly different in pure K562 and K562/HL60 mixtures (Table 3, P value), suggesting that ABL amounts are different in K562 and HL60 cell lines. Intriguingly, ABL detected by ABL1 and ABL2 changed unexpectedly in the opposite direction comparing K562/HL60 cell mixtures with pure K562 cells. Specifically, the Ct by ABL1 in the cell mixture was 1.89 cycles lower than pure K562 cells, whereas Ct by ABL2 was 2.51 cycles higher (Table 3). These apparently paradoxical changes probably pertain to the distinct design of the two ABL primer/probe sets (Figure 1; see Discussion).
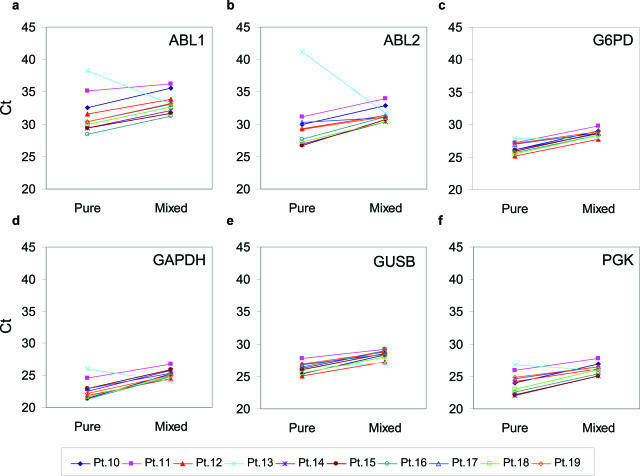
To further examine whether mRNA levels of the control genes vary in CML and non-CML cells taken from patients and to exclude that differences in expression were due to peculiarities of the cell lines tested, we mixed CML patient and normal specimens and compared Ct of mixed samples with pure patient samples. As with the study on cell lines, we expected no significant change in Ct on mixing if CML and normal peripheral blood mononuclear cells have similar levels of a control gene. Among six commonly used controls, Ct values of G6PD (Figure 3c) and GUSB (Figure 3e) had the least change between pure and mixed samples for all 10 specimens, although levels of G6PD appeared to be different in K562 and HL60 cells. GAPDH (Figure 3d) and PGK (Figure 3f) had intermediate differences between the pair of pure and mixed samples and were therefore not further analyzed. Notably, both ABL1 and ABL2 varied to a large degree between the sample pairs (Figure 3, a and b). The most dramatic change occurred in paired samples of patient 13. The Ct of both ABL1 and ABL2 dramatically decreased on mixing pure patient samples with the normal. Apparently, in this patient, using ABL as a normalization control for BCR-ABL quantification would definitely lead to erroneous results. Taken together with the study on cell lines, we conclude that ABL is not expressed at a similar level in CML and other hematopoietic cells and that ABL levels change as the ratio of CML to non-CML cells changes, even when total numbers of cells remain constant. In serial follow-up of treated patients, the BCR-ABL-to-ABL ratio would be misleading when used as a parameter to monitor residual disease.
Figure 3.
Threshold cycle difference of six internal control genes in 10 pairs of pure patient samples and mixed samples. cDNA from 10 patient specimens (Table 1, patients 10 to 19) were mixed with cDNA from normal peripheral blood mononuclear cells at 1:16 ratio. Levels of six different control genes as indicated in the graphs were determined in the 10 pairs of pure and mixed samples. Mean Ct values of triplicate real-time PCR reactions were plotted. Lines connect pure and mixed patient samples to show pairwised relationship.
Degradation Kinetics of BCR-ABL and Control Genes
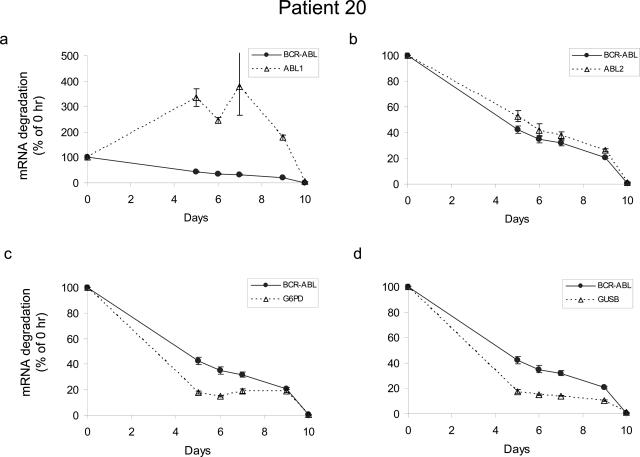
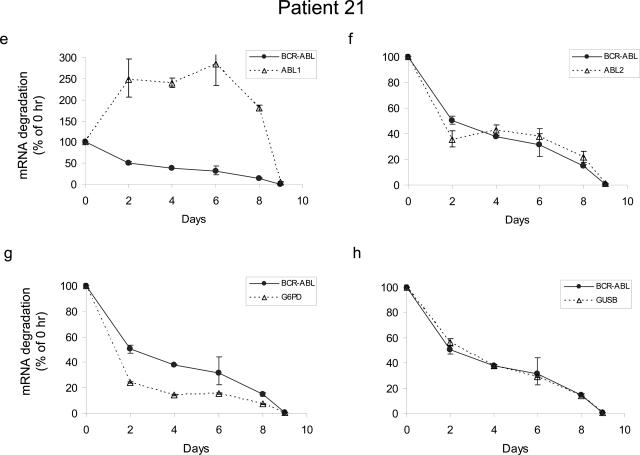
To serve as a legitimate control, the degradation kinetics of a gene should ideally parallel that of BCR-ABL transcripts so that the BCR-ABL-to-control gene ratio remains the same even when RNA is partially degraded. To study the degradation kinetics of BCR-ABL and control genes, mononuclear cells from the bone marrow of two CML patients were placed at room temperature to let cells die and RNA degrade. Levels of BCR-ABL, ABL by ABL1 and ABL2, G6PD, and GUSB were determined by real-time RT-PCR in aliquots of cells collected at different time points. Expression levels relative to the immediate sample (day 0) were analyzed and plotted in Figure 4. Apparently, patient-to-patient variation in the degradation of BCR-ABL and the four control genes existed. Nevertheless, of the four controls studied, G6PD, GUSB, and ABL2 showed similar degradation kinetics to BCR-ABL in both patients. Although degradation of ABL by ABL2 primer/probe followed almost perfectly with that of BCR-ABL (Figure 4,b and f), this finding is not unexpected, because ABL2 detects the ABL portion of the BCR-ABL fusion transcripts. As for ABL levels measured by ABL1, we observed unexpected upward changes of ABL after the initial time point (Figure 4, a and e). Interestingly, this upward change in ABL level was seen in both patient samples, suggesting that the finding was not incidental. It is possible that transcription of ABL kinase is up-regulated in response to cellular stress, likely nutrient/growth factor deficiency experienced by cells placed on the bench. ABL is implicated in various signaling pathways initiated by growth factor, DNA damage, oxidative stress, and integrin stimulation.24,25,26,27 No matter what causes this unexpected change in the ABL level, these results demonstrate that ABL is regulated by cellular conditions and cannot serve as a reference for BCR-ABL quantification, although it is widely used in current clinical practice.
Figure 4.
The degradation kinetics of four internal control transcripts in comparison with BCR-ABL. RNA was extracted from mononuclear cells from the bone marrow of two CML patients (Table 1, patients 20 and 21), placed at the bench for various periods of time. Real-time RT-PCR reactions were performed simultaneously for BCR-ABL and four different control genes as indicated in the legend boxes. Data at each time point after day 0 are expressed as a percentage of the initial time point calculated by the ΔCt method (for details, see Materials and Methods). Means ± SD are plotted. Note different y-axis scale is used in a and e.
Discussion
Recommendation of the Internal Control Gene for BCR-ABL Quantification
After applying three criteria to select the most suitable control genes, we have found that among nine genes tested with 10 primer/probe sets, only G6PD and GUSB meet all three criteria. Reasons for exclusion of other genes are summarized in Table 4. G6PD, as part of a commercially available kit for BCR-ABL quantification, is one of the most commonly used internal control genes. However, we do not recommend using G6PD for the following reasons. First, G6PD is located on the X chromosome. X chromosome location is normally avoided because there might be sex difference in expression levels.23 Second, G6PD deficiency is a fairly common genetic abnormality leading to anemia. Up to 5% of Chinese, 20% of Italians, 32% of Greeks, and 65% of Saudis are affected by G6PD deficiency.28 Furthermore, in the Online Mendelian Inheritance in Man database of the National Center for Biotechnology Information, 56 molecularly characterized variants of G6PD are currently documented together with a very long list of mutants that have not been characterized. We are concerned that frequent sequence variations may affect the binding of PCR primers and/or probe, leading to false-negative results.
Table 4.
Reasons for Exclusion
| Genes | Main reasons for exclusion |
|---|---|
| ABL1 | Much higher Ct than BCR-ABL, different levels in CML and non-CML cells, different degradation kinetics from BCR-ABL, and being a translocation partner of BCR-ABL |
| ABL2 | Different levels in CML and non-CML cells, and ability to detect the ABL portion of BCR-ABL |
| ACTB | More abundant than BCR-ABL and unusual amplification curve |
| B2M | More abundant than BCR-ABL and different levels in CML and non-CML cells |
| GAPDH | More abundant than BCR-ABL and different levels in CML and non-CML cells |
| G6PD | Presence of molecular variants and X chromosome location |
| HPRT | Different levels in CML and non-CML cells |
| PGK | Different levels in CML and non-CML cells |
| TBP | Different levels in CML and non-CML cells |
In contrast, GUSB is located on the long arm of chromosome 7. Mutations in GUSB cause mucopolysaccharidosis (MPS) VII, also known as Sly syndrome. MPS is currently known to consist of 13 subclasses.29 These are rare genetic disorders with a combined frequency of approximately 1 in 20,000.28 Moreover, among the 13 subclasses, MPS VII is the rarest of all forms of MPS.30 We therefore recommend GUSB over G6PD as the most suitable internal control for BCR-ABL quantification.
ABL as the Internal Control Gene
ABL is probably the most widely used normalization control for BCR-ABL quantification in Europe and North America. The EAC study has evaluated several commonly used control genes and concluded that ABL is the most suitable one.23 However, the primer/probe set used to assay ABL level (designated as ABL2 in this study) also detects the ABL portion of the BCR-ABL transcript. The ratio of BCR-ABL to control then becomes BCR-ABL/(BCR-ABL+ABL) with a changing denominator. We question whether a translocation partner is qualified to serve as an internal control gene for normalization because different gene structures in the malignant and nonmalignant cells may lead to different gene expression levels. It is also of our concern that ABL assayed this way would change along with the BCR-ABL during the leukemia/treatment course, giving rise to an inaccurate BCR-ABL-to-control ratio. We therefore designed our study using different criteria for control gene selection. We mimicked diagnostic and residual disease specimens using pure samples and samples mixed with non-CML hematopoietic cells. We selected control genes to ensure that for the same amount of RNA input, the level of a particular control gene does not change significantly between the pure and mixed samples. We found that ABL assayed by two differently designed primer/probe sets failed to meet this criterion. Lower Ct by ABL1 and higher Ct by ABL2 was observed in K562 cells mixed with HL60 cells (Table 3). Because a lower Ct value represents a higher level of mRNA, the lower Ct by ABL1 in the cell mixture (K562/HL60 = 1:9) suggests that a higher amount of ABL transcript is present in HL60 cells. This is not unexpected because HL60 cells contain two wild-type alleles of ABL, whereas K562 cells contains only one, and the other allele is disrupted by the translocation. Alternatively, expression of BCR-ABL may suppress the transcription of the wild-type ABL in K562, making it lower than HL60 cells. In contrast, ABL2 detects both wild-type ABL and translocated ABL in the BCR-ABL fusion transcript. The lower Ct value by ABL2 in K562 may simply reflect that the BCR-ABL plus ABL transcripts in K562 are much more abundant than ABL in HL60 cells. More importantly, in the mixing study performed with patient samples, ABL levels by ABL1 and ABL2 are different between pure and mixed samples, suggesting that the amounts of ABL message are different in CML and non-CML cells.
We have also applied another criterion that was not applied by the EAC study. In our opinion, degradation of the control gene should be proportional to degradation of the BCR-ABL transcripts. A control gene that is not degraded in the same fashion as BCR-ABL may lead to under- or overestimation of the BCR-ABL. This criterion is particularly important to diagnostic laboratories because varying degrees of degradation exist in clinical samples. We examined the degradation kinetics of several control genes and found that wild-type ABL assayed by ABL1 does not meet this criterion. Although degradation of ABL assayed by ABL2 paralleled that of the BCR-ABL, it is expected because ABL2 detects the ABL portion of the BCR-ABL.
An additional concern for use of ABL as an internal control is that the EAC study found that ABL2 amplifies genomic DNA in 7% of 150 samples tested. Ct values resulting from genomic amplification ranged from 35 to 45 cycles. As stated in the article, these high Ct values were far away from the Ct values obtained from good-quality RNA samples. However, RNA samples of variable quality are seen in routine clinical practice. In a partially degraded clinical specimen with high ABL Ct, contribution from amplification of ABL genomic locus would lead to a falsely low BCR-ABL result.
GUSB as the Internal Control Gene
By our three criteria plus the rare presence of sequence variations, we recommend GUSB as the most suitable control gene for BCR-ABL quantification. However, the EAC study found that GUSB levels are different between normal and leukemia samples at diagnosis. The leukemia samples used combined CML with acute lymphocytic leukemia and acute myelogenous leukemia samples in the analysis. Separate comparison of normal and CML samples was not provided. In addition, the primer/probe set of GUSB used in the EAC study was designed by the group and differs in nucleotide sequences from the commercially available set that we used in the current analysis, potentially accounting for the different results in the two studies. Of note, our studies were conducted using ABI primer/probe sets on ABI Prism 7000 instrument only. Whether similar results and conclusion can be obtained using other primer/probe sets for the same control genes or using other real-time instruments remains to be determined.
Conclusions
In conclusion, we recommend using GUSB assayed by the primer/probe set from ABI as the control gene for determination of BCR-ABL level in CML patients. Using a commercially available source of primer/probe facilitates standardization of reagents among different laboratories. It remains to be determined whether GUSB can be used as the control gene for quantification of fusion genes found in other types of leukemia. Because the degradation kinetics of each fusion gene is likely different, the control gene may need to be evaluated and selected on a target-by-target basis.
Note Added in Proof
After acceptance of the current article, a related manuscript31 was accepted for publication and will be appearing in an upcoming issue of The Journal of Molecular Diagnostics. This study applied additional clinically relevant criteria for further evaluation of the control genes.
Acknowledgments
We thank Wayne Tam, M.D., Ph.D., and Botond Timar, M.D., for critical reading of the manuscript.
References
- Nowell PC, Hungerford DA. A minute chromosome in human chronic granulocytic leukemia. Science. 1960;132:1497–1499. [Google Scholar]
- Rowley JD. Letter: a new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- Hochhaus A, Weisser A, La Rosee P, Emig M, Muller MC, Saussele S, Reiter A, Kuhn C, Berger U, Hehlmann R, Cross NC. Detection and quantification of residual disease in chronic myelogenous leukemia. Leukemia. 2000;14:998–1005. doi: 10.1038/sj.leu.2401811. [DOI] [PubMed] [Google Scholar]
- Kantarjian H, Talpaz M, O’Brien S, Garcia-Manero G, Verstovsek S, Giles F, Rios MB, Shan J, Letvak L, Thomas D, Faderl S, Ferrajoli A, Cortes J. High-dose imatinib mesylate therapy in newly diagnosed Philadelphia chromosome-positive chronic phase chronic myeloid leukemia. Blood. 2004;103:2873–2878. doi: 10.1182/blood-2003-11-3800. [DOI] [PubMed] [Google Scholar]
- Hughes TP, Kaeda J, Branford S, Rudzki Z, Hochhaus A, Hensley ML, Gathmann I, Bolton AE, van Hoomissen IC, Goldman JM, Radich JP. Frequency of major molecular responses to imatinib or interferon alfa plus cytarabine in newly diagnosed chronic myeloid leukemia. N Engl J Med. 2003;349:1423–1432. doi: 10.1056/NEJMoa030513. [DOI] [PubMed] [Google Scholar]
- Rosti G, Martinelli G, Bassi S, Amabile M, Trabacchi E, Giannini B, Cilloni D, Izzo B, De Vivo A, Testoni N, Cambrin GR, Bonifazi F, Soverini S, Luatti S, Gottardi E, Alberti D, Pane F, Salvatore F, Saglio G, Baccarani M. Molecular response to imatinib in late chronic-phase chronic myeloid leukemia. Blood. 2004;103:2284–2290. doi: 10.1182/blood-2003-07-2575. [DOI] [PubMed] [Google Scholar]
- Mauro MJ, Druker BJ, Maziarz RT. Divergent clinical outcome in two CML patients who discontinued imatinib therapy after achieving a molecular remission. Leuk Res. 2004;28(Suppl 1):S71–S73. doi: 10.1016/j.leukres.2003.10.017. [DOI] [PubMed] [Google Scholar]
- Cortes J, O’Brien S, Kantarjian H. Discontinuation of imatinib therapy after achieving a molecular response. Blood. 2004;104:2204–2205. doi: 10.1182/blood-2004-04-1335. [DOI] [PubMed] [Google Scholar]
- Hochhaus A, La Rosee P. Imatinib therapy in chronic myelogenous leukemia: strategies to avoid and overcome resistance. Leukemia. 2004;18:1321–1331. doi: 10.1038/sj.leu.2403426. [DOI] [PubMed] [Google Scholar]
- Druker BJ. Overcoming resistance to imatinib by combining targeted agents. Mol Cancer Ther. 2003;2:225–226. [PubMed] [Google Scholar]
- Lee M, Khouri I, Champlin R, Kantarjian H, Talpaz M, Trujillo J, Freireich E, Deisseroth A, Stass S. Detection of minimal residual disease by polymerase chain reaction of bcr/abl transcripts in chronic myelogenous leukaemia following allogeneic bone marrow transplantation. Br J Haematol. 1992;82:708–714. doi: 10.1111/j.1365-2141.1992.tb06948.x. [DOI] [PubMed] [Google Scholar]
- Hochhaus A, Reiter A, Saussele S, Reichert A, Emig M, Kaeda J, Schultheis B, Berger U, Shepherd PC, Allan NC, Hehlmann R, Goldman JM, Cross NC. Molecular heterogeneity in complete cytogenetic responders after interferon-alpha therapy for chronic myelogenous leukemia: low levels of minimal residual disease are associated with continuing remission. German CML Study Group and the UK MRC CML Study Group. Blood. 2000;95:62–66. [PubMed] [Google Scholar]
- Paschka P, Muller MC, Merx K, Kreil S, Schoch C, Lahaye T, Weisser A, Petzold A, Konig H, Berger U, Gschaidmeier H, Hehlmann R, Hochhaus A. Molecular monitoring of response to imatinib (Glivec) in CML patients pretreated with interferon alpha. Low levels of residual disease are associated with continuous remission. Leukemia. 2003;17:1687–1694. doi: 10.1038/sj.leu.2403033. [DOI] [PubMed] [Google Scholar]
- Emig M, Saussele S, Wittor H, Weisser A, Reiter A, Willer A, Berger U, Hehlmann R, Cross NC, Hochhaus A. Accurate and rapid analysis of residual disease in patients with CML using specific fluorescent hybridization probes for real time quantitative RT-PCR. Leukemia. 1999;13:1825–1832. doi: 10.1038/sj.leu.2401566. [DOI] [PubMed] [Google Scholar]
- Schoch C, Schnittger S, Bursch S, Gerstner D, Hochhaus A, Berger U, Hehlmann R, Hiddemann W, Haferlach T. Comparison of chromosome banding analysis, interphase- and hypermetaphase-FISH, qualitative and quantitative PCR for diagnosis and for follow-up in chronic myeloid leukemia: a study on 350 cases. Leukemia. 2002;16:53–59. doi: 10.1038/sj.leu.2402329. [DOI] [PubMed] [Google Scholar]
- Preudhomme C, Revillion F, Merlat A, Hornez L, Roumier C, Duflos-Grardel N, Jouet JP, Cosson A, Peyrat JP, Fenaux P. Detection of BCR-ABL transcripts in chronic myeloid leukemia (CML) using a ‘real time’ quantitative RT-PCR assay. Leukemia. 1999;13:957–964. doi: 10.1038/sj.leu.2401426. [DOI] [PubMed] [Google Scholar]
- Branford S, Hughes TP, Rudzki Z. Monitoring chronic myeloid leukaemia therapy by real-time quantitative PCR in blood is a reliable alternative to bone marrow cytogenetics. Br J Haematol. 1999;107:587–599. doi: 10.1046/j.1365-2141.1999.01749.x. [DOI] [PubMed] [Google Scholar]
- Neumann F, Herold C, Hildebrandt B, Kobbe G, Aivado M, Rong A, Free M, Rossig R, Fenk R, Schneider P, Gattermann N, Royer-Pokora B, Haas R, Kronenwett R. Quantitative real-time reverse-transcription polymerase chain reaction for diagnosis of BCR-ABL positive leukemias and molecular monitoring following allogeneic stem cell transplantation. Eur J Haematol. 2003;70:1–10. doi: 10.1034/j.1600-0609.2003.02811.x. [DOI] [PubMed] [Google Scholar]
- Jones CD, Yeung C, Zehnder JL. Comprehensive validation of a real-time quantitative bcr-abl assay for clinical laboratory use. Am J Clin Pathol. 2003;120:42–48. doi: 10.1309/60A9-C8WG-EGHR-NXEE. [DOI] [PubMed] [Google Scholar]
- Eder M, Battmer K, Kafert S, Stucki A, Ganser A, Hertenstein B. Monitoring of BCR-ABL expression using real-time RT-PCR in CML after bone marrow or peripheral blood stem cell transplantation. Leukemia. 1999;13:1383–1389. doi: 10.1038/sj.leu.2401489. [DOI] [PubMed] [Google Scholar]
- Gabert J, Beillard E, van der Velden VH, Bi W, Grimwade D, Pallisgaard N, Barbany G, Cazzaniga G, Cayuela JM, Cave H, Pane F, Aerts JL, De Micheli D, Thirion X, Pradel V, Gonzalez M, Viehmann S, Malec M, Saglio G, van Dongen JJ. Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia: a Europe Against Cancer program. Leukemia. 2003;17:2318–2357. doi: 10.1038/sj.leu.2403135. [DOI] [PubMed] [Google Scholar]
- Zanella I, Rossi G, Finazzi D, Capucci A, Albertini A, Cariani E. Quantification of bcr/abl mRNA expression by a rapid real-time reverse transcription-polymerase chain reaction assay in patients with chronic myeloid leukemia. Haematologica. 2001;86:318–319. [PubMed] [Google Scholar]
- Beillard E, Pallisgaard N, van der Velden VH, Bi W, Dee R, van der Schoot E, Delabesse E, Macintyre E, Gottardi E, Saglio G, Watzinger F, Lion T, van Dongen JJ, Hokland P, Gabert J. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using ‘real-time’ quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR): a Europe against cancer program. Leukemia. 2003;17:2474–2486. doi: 10.1038/sj.leu.2403136. [DOI] [PubMed] [Google Scholar]
- Van Etten RA. c-Abl regulation: a tail of two lipids. Curr Biol. 2003;13:R608–R610. doi: 10.1016/s0960-9822(03)00528-1. [DOI] [PubMed] [Google Scholar]
- Hernandez SE, Krishnaswami M, Miller AL, Koleske AJ. How do Abl family kinases regulate cell shape and movement? Trends Cell Biol. 2004;14:36–44. doi: 10.1016/j.tcb.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Wange RL. TCR signaling: another Abl-bodied kinase joins the cascade. Curr Biol. 2004;14:R562–R564. doi: 10.1016/j.cub.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Shaul Y, Ben-Yehoyada M. Role of c-Abl in the DNA damage stress response. Cell Res. 2005;15:33–35. doi: 10.1038/sj.cr.7290261. [DOI] [PubMed] [Google Scholar]
- Connor M, Ferguson-Smith M. Oxford: Blackwell Science Ltd; Medical Genetics, (ed 5) 1997 [Google Scholar]
- Goetz CG. St. Louis, MO: W.B. Saunders Company; Textbook of Clinical Neurology, (ed 2) 2003 [Google Scholar]
- Behrman RE, Kliegman RM, Jenson HB. St. Louis, MO: W.B. Saunders Company; Nelson Textbook of Pediatrics, (ed 17) 2004 [Google Scholar]
- Lee JW, Chen Q, Knowles DM, Cesarman E, Wang YL: β-Glucoronidase is an optimal normalization control gene for molecular monitoring of chronic myelogenous leukemian. J Mol Diagn 2006, (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]