Abstract
Two mutations in the MCOLN1 mucolipidosis IV (ML IV) gene represent ∼95% of the mutations in Ashkenazi-Jewish patients with ML IV. The mutations, a splice site mutation (IVS3-2A>G) and an ∼6.4-kb deletion (511del6434), account for 72% and 23% of ML IV alleles in this population, respectively. An automated high-throughput assay was developed using the 5′-nuclease (TaqMan) method for the simultaneous detection of both mutations in a single reaction. Three fluorescent probes specifically detected wild-type, IVS3-2A>G, and 511del6434 alleles in each reaction real-time. Data collected were automatically analyzed, and genotype results were uploaded into a laboratory information management system. The assay was validated using genomic controls, demonstrating high robustness and accuracy. Carrier screening of 10,527 samples revealed 77 heterozygote carriers of IVS3-2A>G, 25 heterozygote carriers of 511del6434, and two compound heterozygote of both mutant alleles. The frequency of mutated alleles was 0.73% for IVS3-2A>G and 0.24% for 511del6434. The combined carrier frequency was 1:103 with predicted disease incidence of 1:42,436 individuals in this population, slightly lower than previously described frequencies. This automated high-throughput assay is labor saving, because two mutations can be detected in a single reaction. The method has potential for use in other assays requiring simultaneous detection of two mutations.
Mucolipidosis IV (ML IV) is an autosomal recessive neurodegenerative lysosomal storage disease in which 80 to 90% of patients are of Ashkenazi-Jewish (AJ) descent.1,2 Published heterozygote carrier frequencies in the AJ population vary from 1:62 to 1:127 (Online Mendelian Inheritance in Man, no. 252650).3,4,5 Although there is some heterogeneity, affected individuals generally exhibit psychomotor retardation, and most patients do not progress beyond a developmental age of 1 to 2 years. Other symptoms include characteristic opthalmological abnormalities (Online Mendelian Inheritance in Man).3,5,6
ML IV is caused by loss of function mutations in the MCOLN1 gene.8 The gene has been mapped to chromosome 19 and codes for a potential membrane channel.1,3,8 Two predominant mutations were identified in ML IV patients of AJ descent, IVS3-2A>G and 511del6434, which together account for more than 95% of ML IV chromosomes. The IVS3-2A>G mutation causes aberrant mRNA splicing and skipping of exon 3 and accounts for ∼72% of AJ ML IV chromosomes. The 511del6434 is a large deletion that spans exon 1 through exon 7 and represents 23% of AJ ML IV chromosomes.1,8 Other rare mutations have been described in patients of Ashkenazi and non-Ashkenazi descent.1,8
Previous genotyping methods have relied on polymerase chain reaction-restriction fragment length polymorphism,4 polymerase chain reaction (PCR)-single strand conformational polymorphism,9 or allele-specific oligonucleotide hybridization.5 These techniques and methods, however, are not easily amenable to automation or high-throughput genotyping required in a commercial genotyping laboratory. A commercially available kit from Elucigene (Tepnel Diagnostics, Oxford, UK) using the amplification-refractory mutation system can be used to detect several mutations more prevalent in the AJ population, including both ML IV mutations. Recently, a method that relies on the Tag-It system was developed by Tm Bioscience (Toronto, Canada). Because of its high multiplexing capabilities, the Tag-It method for detecting ML IV mutations was developed as part of a panel for detection of mutations more prevalent in AJ, such as Tay-Sachs and Canavan diseases. The Tag-It AJ panel assay detects more than 33 mutations and, therefore, is much more demanding in terms of successful even multiplex amplification and allele-specific primer extension reactions.
We developed a one-step, single-tube method for the detection of wild-type and both AJ mutant alleles using the TaqMan platform. We describe the development and validation of the method and the initial results from screening >10,000 samples submitted for carrier testing. We also report on the results of a platform comparison of our method to the Tag-It method.
Materials and Methods
Oligonucleotides and Reagents
Table 1 shows primers and probes used in this study. TaqMan MGB probes and TaqMan Universal PCR Master Mix (2×) were from Applied Biosystems (Foster City, CA). An ABI 7900 with sequence detection system software (Applied Biosystems) was used for amplification and data analysis. DNA was extracted using Qiagen (Valencia, CA) or Genovision Robotic workstations according to the manufacturers’ instructions. Reagents and DNA were dispensed manually or by using an automated pipetting workstation. For the clinical laboratory assay, we used a Biomek2000 (Beckman Coulter, Fullerton, CA).
Table 1.
Primers and Probes Used in This Study (IDT, Purified by Desalting)
| Primer/probes | Sequence | Nucleotide position GB accession no. AF287270 |
|---|---|---|
| IVS3-2A>G-F | 5′-AGC GGG CCG GAC TCA-3′ | 5494 to 5509 |
| IVS3-2A>G-R | 5′-TAA CCA CCA TCG GAT CAA TGT C-3′ | 5671 to 5698 |
| PRI F1 | 5′-CTT GCT CTG TTG CCC AGG CT-3′ | 441 to 460 |
| PRI R2 | 5′-CTC ACC GTG CTG GAA GAC ACT-3′ | 7017 to 7037 |
| IVS3-2A>G WT probe | 5′-(VIC) CTG CCC ACA GTA CC-3′ | 5526 to 5539 |
| IVS3-2A>G MUT probe | 5′-(FAM) TGC CCA CGG TAC C-3′ | 5527 to 5539 |
| 511del6434 Probe | 5′-(TET) ACC CAG GCC CAC AT-3′ | 6984 to 6197 |
Underlined bases indicate wt/mutant allele.
Source of Genomic Controls
Cell lines from patients with ML IV mutations were purchased from Coriell Cell Repositories to validate the assay. Based on preliminary experiments, we identified the ML IV mutations present in these cell lines. The cell lines used were GM02525, a fibroblast cell line carrying a homozygous 511del6434 mutation; GM02526, a fibroblast cell line harboring compound heterozygous IVS3-2A>G/511del6434 mutations; GM02527, a fibroblast cell line carrying a homozygous IVS3-2A/G mutation; and GM02533, a B-lymphocyte cell line harboring compound heterozygous IVS3-2A>G/511del6434 mutations.
PCR Conditions
Reactions contained 25 μl of 2× TaqMan Universal PCR master mix, 0.9 μmol/L of each primer (Table 1), 0.2 μmol/L of IVS3-2A>G wild-type (wt) probe, 0.36 μmol/L IVS3-2A>G probe, 0.5 μmol/L 511del6434 probe (Table 1), 2× bovine serum albumin (NEB, Ipswich, MA), 4× Q solution (Qiagen), 0.5 μl of 25 mmol/L MgCl2 solution (Qiagen), 4 μl DNA, and H2O (Sigma Chemical Co., St. Louis, MO) to a final reaction volume of 50 μl. Master mix was aliquoted into optical reaction plates and sealed with an optical adhesive seal (Applied Biosystems). All plates included positive and negative DNA controls. Assays were run on the ABI 7900 instrument in the real-time mode using the following conditions: 50°C for 2 minutes, 95°C for 10 minutes, followed by 45 cycles of 95°C for 15 seconds, 60°C for 1 minute.
Data Analysis
Data contained in the results table [which displays cycle threshold (Ct) values] and in the multicomponent results table (which displays the signals generated in relative fluorescent units) were used. Proprietary software was developed to analyze the data and provide allele assignments based on the Ct value and signal generation. Using Ct alone can lead to misinterpretation of background signals in an empty well that did not receive reagents and can generate false values for Ct. Manual review of the data can prevent this error because the fluorescent signals generated using the multicomponent plot will indicate no amplification in the no reagent well. Calculations using both Ct and fluorescence provide quality assurance of the allele call. The automated allele calls were recorded as wt/wt, heterozygous for either mutation, compound heterozygous, or homozygous mutant for either allele.
Platform Comparison
We analyzed 583 patient samples and 55 wt/wt or positive control samples for the IVS3-2A>G and 511del6434 mutations using the Tag-It mutation detection kit for Ashkenazi-Jewish mutations from Tm Bioscience (Toronto, Ontario). This detection methodology utilizes multiplex PCR followed by allele-specific primer extension. The mutations are detected using the Luminex platform and Tm Bioscience’s proprietary universal tag system. Samples were analyzed on the Tag-It platform using the manufacturer’s protocol.
Results and Discussion
Experimental Design and Assay Validations
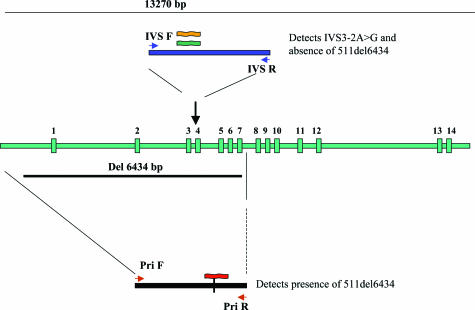
An assay was designed for detecting the IVS3-2A>G and 511del6434 mutations in MCOLN1 using 5′-exonuclease TaqMan real-time PCR. As the deletion encompasses exons 1 to 7, a probe to detect the presence of IVS3-2A>G wt allele would by default indicate the absence of the 511del6434 mutation in at least one allele. To detect the presence of the deletion, two PCR primers were designed that flank the deletion break points. If the deletion is absent, the conditions of the PCR will not allow amplification of a large 6.4-kb fragment. In the presence of a deletion, a 200-bp product will be generated, and a probe specific for that amplicon will hybridize, indicating the presence of the deletion. Thus, each reaction can have one or two amplicon products (depending on the absence or presence of the deletion). The strategy is shown in Figure 1.
Figure 1.
Design of single tube real-time TaqMan assay for the detection of two common ML IV mutations. Vertical green bars represent 14 MCOLN gene exons. A duplex PCR reaction amplifies a fragment from exon 3 or the junction fragment created by the 511del6434 mutation. Three probes are included per reaction. The VIC probe (orange) detects the WT IVS3 allele and absence of the 511del6434. The FAM probe (dark green) detects the presence of the mut IVS3-2A>G allele, whereas the TET probe (red) detects the 511del6434 allele when the PCR product using primers that flank the deletion is amplified.
Both cycle thresholds as well as fluorescence signals (in the multicomponent results) were used to generate automated allele calls after exporting the result tables into our laboratory information system. The signals generated from the wt/wt sample were used as a reference in performing the calculations, and the ratio of signals of the VIC (wt), FAM (IVS3-2A>G), and TET (511del6434) probes were used for automated calculations. Each assay included positive and negative controls. The total time of the assay setup (after DNA extraction) and detection on the ABI 7900, including automated allele calls, was 3 to 4 hours (total run time of plate on the ABI 7900 is ∼2.5 hours). This enables a rapid turn around time, as fast as 1-day turn-around time if extraction is performed on the same day.
To validate the assay, cell lines containing the following genotypes were used: homozygous IVS3-2A>G, homozygous 511del6434, and compound heterozygous IVS3-2A>G/511del6434. Heterozygote DNA samples were made by mixing equal concentrations of the homozygous mutant sample DNAs from Coriell Cell Repositories with a wt/wt sample. These controls were used for optimizing assay conditions. As shown in Figure 2, the assay was able to detect the presence of wt/wt, IVS3-2A>G/wt, and 511del6434/wt heterozygotes as well as the homozygous IVS3-2A>G/IVS3-2A>G and 511del6434/511del6434 mutations. Furthermore, the assay was able to detect the compound heterozygote IVS3-2A>G/511del6434. A validation panel consisting of 89 samples was assembled after anonymizing the samples, and two laboratory personnel performed the assay validations a total of three times using two different lots of reagents. All samples were detected and genotyped correctly, including all controls in all three setups with one exception in one validation experiment. In one sample, the Ct value was positive but the fluorescence units were very low, possibly because of a pipetting error. In a clinical setting this sample would be repeated because of very low relative fluorescent unit signals.
Figure 2.
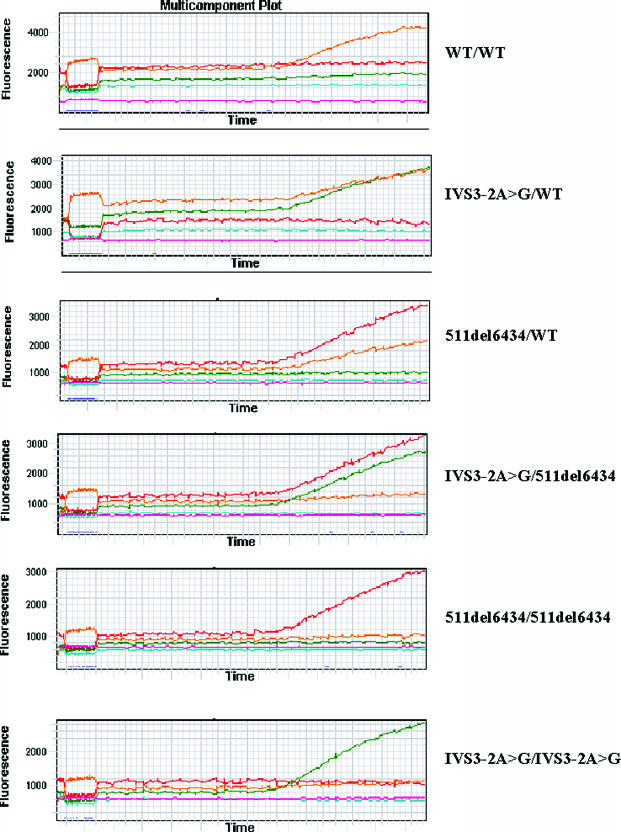
Example of all possible results obtained using the single tube multiplex ML IV assay. Orange, green, and red signals represent the VIC (WT), FAM (IVS3-2A>G), and TET (511del6434) alleles detected, respectively. Other colors in the figure reflect normalizing internal standard ROX (pink) present in 2× TaqMan master mix and background signals (light blue). Numbers on the left are relative fluorescence units.
Examples of the results are shown in Figure 2. In a wt/wt sample, only the VIC probe was detected, while the FAM and TET were not. In heterozygotes the VIC and either the FAM (wt/IVS3-2A>G) or the TET (wt/511del6434) signals were detected. Homozygous IVS3-2A>G samples showed only the FAM signals, and homozygous 511del6434 sample showed only TET signals. In a compound heterozygous IVS3-2A>G/511del6434 sample, the FAM and the TET signals were detected (Figure 2).
Allele Frequency in Our Tested Populations
Samples submitted to Quest Diagnostics for ML IV carrier testing were analyzed using the TaqMan method. In each assay setup, positive controls representing wt/wt, IVS3-2A>G/wt, 511del6434/wt, and IVS3-2A>G/511del6434 were included. We analyzed 10,527 consecutive samples submitted for carrier testing, and the results are shown in Table 2. The results for 2006 of these cases were published previously.10 Seventy-seven chromosomes of 21,054 carried the IVS3-2A>G mutation for a mutant allele frequency of 0.37% while 25 chromosomes carried the 511del6434 mutation for an allele frequency of 0.12%. The heterozygote carrier frequency was 0.73% for the IVS3-2A>G and 0.24% for the 511del6434; the combined carrier frequency was 0.97% or 1:103 individuals and a predicted disease incidence of ∼1:42,436. We also identified two cases of affected females. In these cases, the probands were 20 and 14 months old. Genotyping using the method we developed showed that the patients carried compound heterozygosity of the two mutated alleles. No cases harboring homozygous mutations of either allele were detected.
Table 2.
Carrier Frequency of the ML IV Mutations in Ashkenazi Jews
| Study | Study population | IVS3-2A>G
|
511del6434
|
Combined
|
||||
|---|---|---|---|---|---|---|---|---|
| Heterozygote carriers | Carrier % (frequency) | Heterozygote carriers | Carrier % (frequency) | Number of carriers | Carrier % (frequency) | Predicted disease incidence | ||
| Present study | United States | 77/10527 | 0.73 (1:137) | 25/10527 | 0.24 (1:416) | 102/10527 | 0.97 (1:103) | 1:42436 |
| Edelmann et al6 | New York | 11/2029 | 0.54 (1:184) | 5/2029 | 0.25 (1:406) | 16/2029 | 0.79 (1:127) | 1:64500 |
| Wang et al5 | New York | 2/123 | 1.62 (1:62) | 0 | 0 | 2/123 | 1.62 (1:62) | 1:15000 |
| Bargal et al | Israel | 17/2000 | 0.85 (1:117) | 1 | 0.05 (1:2000) | 18/2000 | 0.9 (1:111) | 1:49000 |
Platform Comparison
We analyzed 583 patient samples by the TaqMan assay and the Tag-It assays for concordance and platform comparison. The concordance between the real-time assay and the Tag-It assay was 100%, as both assays detected three carriers of the IVS3-2A>G and two carriers of the 511del6434 mutations. The repeat rate for the patient samples tested using the method we developed was 1.89% (11 of 583) whereas the repeat rate using the Tag-It system was 3.26% (19 of 583). Both platforms required repeat analysis of the same nine samples, suggesting DNA quality as the issue in these samples, while two extra samples were repeated on the TaqMan assay and 10 different samples required repeating on the Tag-It system. Therefore, the Tag-It method has a slightly higher repeat rate than the method we developed, likely due to the complexity of multiplexing the PCR and allele-specific primer extension reactions. In comparison to the Tag-It system, the TaqMan assay is a rapid method as patient samples are analyzed and detected in a single-step, single-tube assay. The Tag-It system utilizes several steps that include PCR, allele-specific primer extension reactions, purification, and analysis on the Luminex instrument, each step subsequent to PCR requiring opening of tubes for addition or aliquoting of reagents. As shown recently,11 the time for running the Tag-It system is ∼7 hours after PCR. The utility of the Tag-It system is for multiplex detection of several alleles. In summary, both the TaqMan and the Tag-It system detected the same number of mutations indicating the sensitivity of both methods.
In the expanding field of genetic testing, development of accurate high-throughput automated assays is desirable for the ease of use, minimization of human error, and rapid turn-around time. We chose the TaqMan assay platform to develop a rapid assay for the detection of the prevalent mutant alleles in the MCOLN1 gene in AJ. Our method combines a single tube assay for the detection of both alleles, IVS3-2A>G3 and 511del6434 mutations. No further manipulation, such as enzymatic digestion or allele-specific oligonucleotide, is needed as has been previously described.4,5 Additionally, automated PCR setup on a liquid handling robotic workstation minimizes hands-on manipulation, and the ability of our method for automated allele calls removes human bias in interpreting results. Automated uploading of the results into the laboratory information system eliminates transcription errors. Both the TaqMan assay and the Tag-It detection method performed well for mutation detection, with the TaqMan assay having the advantage of simplicity and ease of operation and the Tag-It system showing the advantage for multiplex detection of several alleles, but with the increase in the number of steps for sample analysis.
When examining the data generated from analysis of greater than 10,000 patient samples, our results are similar to those of Bargal and colleagues2 and Edelmann and colleagues5 with regard to overall carrier frequency (1:103 in our set versus 1:127 in Edelmann et al). The distribution of the mutations is also consistent with those of other reports in the literature (Table 2). We have no way of assuring all our samples are derived from AJ individuals. The frequency of the IVS3-2A>G is approximately three times that of the 511del6434. Previous data published by this laboratory10 reported a carrier frequency of 1:182, although the number of samples was lower (n = 2006). The larger number of samples tested in the current study yielded a carrier frequency more consistent with other reports (Table 2). The same is true for the proportion of the two mutated alleles tested in the current study, as our results here are consistent with other published studies.
The carrier frequency of 1:103 is not as high as that of Tay-Sachs (1:36) or Gaucher disease (1:19), but it is close to Niemann-Pick (1:125) and Fanconi anemia type C (1:108), and higher than Bloom syndrome (1:164, Strom et al10). Whether a recommendation is required for population-based screening for ML IV in AJs, as is the case for Tay-Sachs disease, is up for debate and will require evaluation of the potential benefits and costs involved.
Acknowledgments
We thank Christina Deeter and Elaina Kapoor for technical assistance; Joy B. Redman, M.S., Genetic Counselor, for clinical information on affected patients; and the reviewers for their helpful comments.
Footnotes
Current address of S.C.O.: Biocept, Inc., 5810 Nancy Ridge Dr., Suite 150, San Diego, CA 92121.
References
- Bassi MT, Manzoni M, Monti E, Pizzo MT, Ballabio A, Borsani G. Cloning of the gene encoding a novel integral membrane protein, mucolipidin—and identification of the two major founder mutations causing mucolipidosis type IV. Am J Hum Genet. 2000;67:1110–1120. doi: 10.1016/s0002-9297(07)62941-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargal R, Avidan N, Olender T, Ben Asher E, Zeigler M, Raas-Rothschild A, Frumkin A, Ben-Yoseph O, Friedlender Y, Lancet D, Bach G. Mucolipidosis type IV: novel MCOLN1 mutations in Jewish and non-Jewish patients and the frequency of the disease in the Ashkenazi Jewish population. Hum Mutat. 2001;17:397–402. doi: 10.1002/humu.1115. [DOI] [PubMed] [Google Scholar]
- Bargal R, Avidan N, Ben-Asher E, Olender Z, Zeigler M, Frumkin A, Raas-Rothschild A, Glusman G, Lancet D, Bach G. Identification of the gene causing mucolipidosis type IV. Nat Genet. 2000;26:118–123. doi: 10.1038/79095. [DOI] [PubMed] [Google Scholar]
- Wang ZH, Zeng B, Pastores GM, Raksadawan N, Ong E, Kolodny EH. Rapid detection of the two common mutations in Ashkenazi Jewish patients with mucolipidosis type IV. Genet Test. 2001;5:87–92. doi: 10.1089/109065701753145529. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Dong J, Desnick RJ, Kornreich R. Carrier screening for mucolipidosis type IV in the American Ashkenazi Jewish population. Am J Hum Genet. 2002;70:1023–1027. doi: 10.1086/339519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altarescu G, Sun M, Moore DF, Smith JA, Wiggs EA, Solomon BI, Patronas NJ, Frei KP, Gupta S, Kaneski CR, Quarrell OW, Slaugenhaupt SA, Goldin E, Schiffmann R. The neurogenetics of mucolipidosis type IV. Neurology. 2002;59:306–313. doi: 10.1212/wnl.59.3.306. [DOI] [PubMed] [Google Scholar]
- Goldin E, Cooney A, Kaneski CR, Brady RO, Schiffmann R. Mucolipidosis IV consists of one complementation group. Proc Natl Acad Sci USA. 1999;96:8562–8566. doi: 10.1073/pnas.96.15.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Goldin E, Stahl S, Falardeau JL, Kennedy JC, Acierno JS, Jr, Bove C, Kaneski CR, Nagle J, Bromley MC, Colman M, Schiffmann R, Slaugenhaupt SA. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum Mol Genet. 2000;9:2471–2478. doi: 10.1093/hmg/9.17.2471. [DOI] [PubMed] [Google Scholar]
- Bargal R, Goebel HH, Latta E, Bach G. Mucolipidosis IV: novel mutation and diverse ultrastructural spectrum in the skin. Neuropediatrics. 2002;33:199–202. doi: 10.1055/s-2002-34496. [DOI] [PubMed] [Google Scholar]
- Strom CM, Crossley B, Redman JB, Quan F, Buller A, McGinniss M, Sun W. Molecular screening for diseases frequent in Ashkenazi Jews: lessons learned from more than 100,000 tests performed in a commercial laboratory. Genet Medicine. 2004;6:145–152. doi: 10.1097/01.gim.0000127267.57526.d1. [DOI] [PubMed] [Google Scholar]
- Krafft AE, Lichy JH. Time-motion analysis of 6 cystic fibrosis mutation detection systems. Clin Chem. 2005;51:1116–1122. doi: 10.1373/clinchem.2004.047423. [DOI] [PubMed] [Google Scholar]