Abstract
Methacarn and RCL2, a new noncrosslinking fixative, were compared to formalin-fixed or frozen tissue samples of the same invasive breast carcinoma and were evaluated for their effects on tissue morphology and immunohistochemistry as well as DNA and RNA integrity. The histomorphology of methacarn- or RCL2-fixed paraffin-embedded tumors was similar to that observed with the matched formalin-fixed tissues. Immunohistochemistry using various antibodies showed comparable results with either fixative, leading to accurate breast tumor diagnosis and determination of estrogen and progesterone receptors, and HER2 status. Methacarn and RCL2 fixation preserved DNA integrity as demonstrated by successful amplification and sequencing of large DNA amplicons. Similarly, high-quality RNA could be extracted from methacarn- or RCL2-fixed paraffin-embedded MCF-7 cells, whole breast tumor tissues, or microdissected breast tumor cells, as assessed by electropherogram profiles and real-time reverse transcriptase-polymerase chain reaction quantification of various genes. Moreover, tissue morphology and RNA integrity were preserved after 8 months of storage. Altogether, these results indicate that methacarn, as previously shown, and RCL2, a promising new fixative, have great potential for performing both morphological and molecular analyses on the same fixed tissue sample, even after laser-capture microdissection, and can open new doors for investigating small target lesions such as premalignant breast lesions.
In the last few years, gene and protein expression profiling have been widely developed in pathological tissues to better understand the molecular events leading to diseases and to identify new prognostic and therapeutic markers for treatment of patients. Such approaches were permitted by rapid technical progress in profiling methods,1,2,3,4 mRNA quantification,5 or microdissection.6 However, these approaches still remain highly dependent on the quality of the tissue analyzed, which varies according to tissue acquisition, fixation, and preservation.7,8
Formalin-fixed, paraffin-embedded tissues represent the most abundant supply of archival material for clinical and molecular analyses. Although formalin is adapted to morphological examination of tissues, it is a crosslinking agent that induces RNA chemical modifications and fragmentation, impairing quantification of gene expression.8,9,10 The gold standard for molecular analyses remains unfixed fresh or snap-frozen tissues. Unfortunately, these treatments cannot be used for routine laboratory samples because they do not provide accurate morphological details and may impair histological diagnosis. Molecular analyses of small target lesions such as benign or premalignant lesions thus become challenging as the whole surgical tissue is usually fixed and paraffin-embedded according to the routine procedures for histological diagnosis, most often using formalin, so that no fresh or frozen tissue sample remains available.
New tissue fixation procedures for these small lesions are therefore critically needed, providing preservation of both tissue morphology for accurate diagnosis and RNA, DNA, and proteins for further molecular analysis. Recently, some fixatives with such properties have been described. Methacarn, a solution of methanol, chloroform, and acetic acid, is a noncrosslinking organic solvent that was shown to maintain tissue morphology and to preserve DNA, RNA, and protein integrity.11,12,13 Interestingly, methacarn-fixed tissues have been successfully used for quantitative expression analysis of mRNAs after tissue microdissection.14 Similarly, Morales and colleagues15 and Vincek and colleagues16 recently demonstrated that UMFIX, a mixture of methanol and polyethylene glycol, is a simple valuable fixative, when combined with a rapid processing technique, for histomorphology and for analysis of DNA, RNA, and protein in clinical samples.
To facilitate analysis of premalignant breast lesions, we searched for new fixation protocols that would allow both morphological and molecular analyses on the same fixed tissue sample. First, we evaluated methacarn and a promising new noncrosslinking fixative, RCL2, because of their performance regarding nucleic acid and protein preservation (M. Lacroix-Triki, L. Lamant, B. A. Cuider, A. Decha, J.-J. Voigt, P. Rochaix, submitted manuscript). Both fixatives were used to test invasive breast carcinoma according to tissue morphology, breast tumor staging and diagnosis, as well as DNA and RNA integrity. Because premalignant breast lesions are small lesions that need microdissection before molecular analyses, we also analyzed the effect of these fixatives on RNA integrity after laser-capture microdissection (LCM) of breast tumor cells. Our data indicate that methacarn and the new fixative RCL2 give accurate morphological and immunohistochemical results for breast tumor diagnosis and allow extraction of high-quality DNA and RNA, including RNA extracted from laser-captured breast tumor cells.
Materials and Methods
Fixatives and Paraffin
Neutral buffered formaldehyde 4% (NBF) stored at room temperature or FAAM (formaldehyde, 40%, 0.1:1, v/v; acetic acid, 100%, 0.1:1, v/v; and methanol, 100%, 0.4:1, v/v, in distilled water), routinely used in the Department of Pathology (Centre Hospitalier Universitaire, Montpellier, France) for diagnosis, were considered as gold standards for morphological and immunohistochemistry studies. The two fixatives tested in this report were a methacarn solution, consisting of 60% (v/v) methanol, 30% chloroform, and 10% glacial acetic acid, and RCL2 solution (patent application no. WO 2004/083369) stored at 4°C. Both fixatives were freshly prepared before use. Paraffin (Paraplast Plus) was from Tyco Heathcare Group (Mansfield, MA). All solvents used in this study were RNase-free and of molecular biology grade.
Cell and Breast Tumor Preparation
MCF-7 cells were cultured at confluence in T175 vials in Dulbecco’s modified Eagle’s medium/F12 medium supplemented with 10% fetal calf serum. After two washes in phosphate-buffered saline, the cell pellet was either immediately snap-frozen in liquid nitrogen and stored at −80°C or fixed in methacarn or RCL2 (10 ml per cell pellet) overnight at 4°C, dehydrated, and then embedded in paraffin for 1 hour at 58°C. For estradiol (E2)-stimulation experiments, MCF-7 cells were cultured for 5 days in 10% charcoal dextran-fetal calf serum phenol red-free medium before E2 treatment (10−8 mol/L E2 for 24 hours).
The effects of methacarn or RCL2 fixation on tissues were analyzed using residual samples from large surgical specimens of invasive breast carcinoma sent to the Department of Pathology for histological diagnosis. The time delay between surgery and tissue processing for the six invasive breast tumors analyzed was ∼15 minutes. Among the six tumors tested, five were invasive ductal carcinoma and one was diagnosed as a mammary carcinoma with giant osteoclast-like cells. One of these tumors contained in situ ductal breast carcinoma adjacent to the invasive tumor. On arrival in the laboratory, the tumors were immediately processed according to the various procedures described below. One part of the tumor was fixed in NBF or FAAM for 24 hours at room temperature and then embedded in paraffin using a TissueTek VIP automated processor (Sakura Finetek, Torrance, CA) according to the standard protocol used for breast tumor diagnosis and staging in the laboratory. Briefly, this protocol was performed overnight at room temperature and consisted of dehydration in absolute ethanol (four baths for a total of 3 hours), followed by butanol for 30 minutes, toluene (three baths for a total of 2 hours), and paraffin-immersion (four baths at 58°C for 3 hours). The remaining tissue was divided in three samples (∼2 × 1 × 0.5 cm thick). One sample was immediately snap-frozen in liquid nitrogen and stored at −80°C. The other samples were fixed overnight at 4°C in methacarn or RCL2. Optimization of the paraffin-embedding protocol using these latter fixatives led to the following steps: dehydration at 4°C with absolute ethanol (1 hour, ×3) and acetone (1 hour, ×3) and immersion in soluble paraffin at 58°C for 1 hour. The methacarn paraffin (MP) or RCL2 paraffin (RCL2P) blocks were then kept at −20°C until histological or molecular studies.
Histology, Immunohistochemistry, and Chromogenic in Situ Hybridization (CISH) Analyses
Three-μm-thick sections of paraffin-embedded tissues from either NBF-, FAAM-, methacarn-, or RCL2-fixed tumors were deparaffinized before hematoxylin and eosin (H&E) staining or immunohistochemical analyses. Immunostaining was performed automatically with a DAKO Autostainer (Universal staining system; DakoCytomation, Trappes, France) using the streptavidin-peroxidase method. Whenever needed, antigen retrieval was performed in a water bath, according to standard procedures, using citrate or ethylenediaminetetraacetic acid buffers.
For all breast tumors, immunostaining was performed for estrogen receptor (ER)-α (6F11 antibody, 1/50; Novocastra, Newcastle, UK), progesterone receptor (PR) (PgR 636 antibody, 1/50; DakoCytomation), and HER2 (CB11 antibody, 1/250; Novocastra). ER-α and PR status of breast tumors was determined as previously described17 with a positivity cutoff of ≥10% of stained nuclei. For HER2 immunostaining, results (0, 1+, 2+, 3+) were expressed according to the Herceptest scoring system.18
For some tumors, the following antibodies were also tested and the immunostaining intensity (none, +, ++, +++ for strong intensity) was compared according to the fixatives used (NBF, methacarn, or RCL2): P63 (4A4, 1/200; DakoCytomation), S100 protein (polyclonal rabbit antibody, 1/200; DakoCytomation), smooth muscle actin (1A4, 1/100; DakoCytomation), E-cadherin (36B5, 1/100; TEBU), cytokeratin 5/6 (D5/16B4, 1/50; DakoCytomation), cytokeratin 7 (OVTL12/30, 1/25; DakoCytomation), and pan-cytokeratin (AE1/AE3, 1/50; DakoCytomation). To evaluate HER2 gene amplification, CISH experiments were performed according to Tanner and colleagues19 (Spot-Light; Zymed, San Francisco, CA) on two invasive breast carcinomas harboring HER2 protein overexpression by immunohistochemistry (3+ status).
DNA and RNA Extraction
For whole tissue sections, two to three sections 7 μm in thickness, from either snap-frozen tissues (control) or paraffin-embedded tissues (fixed in NBF, methacarn, or RCL2), were loaded on SuperFrost slides (Menzel GmbH, Germany). Before nucleic acid extraction, paraffin-embedded samples were deparaffinized in 100% RNase-free ethanol for 5 minutes at 60°C in a water bath. DNA or RNA extraction buffer was then loaded directly on the slides to homogenize the tissue sections.
For DNA extraction, tissue sections were homogenized in 180 μl of proteinase K digestion buffer (50 mmol/L Tris, pH 8.1, 1 mmol/L ethylenediaminetetraacetic acid, and 0.5% Tween 20) containing 20 μl of proteinase K (2 mg/ml); digestion was performed for 2 to 3 hours at 55°C. After digestion, proteinase K was inactivated by heating at 70°C for 10 minutes and DNA was further purified using the QIAmp minikit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. DNA was eluted in a final volume of 50 μl of water.
RNA was extracted using the RNeasy minikit (Qiagen) including a DNase I digestion step. Total RNA was finally eluted in 20 μl of RNase-free water loaded twice on the column. One μl of RNA solution was used to assess RNA integrity using the RNA 6000 Pico assay kit with Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).20 Total RNA from the MCF-7 cell pellets was extracted using the same protocol. For microdissected samples, total RNA was extracted using the RNeasy microkit (Qiagen).
Laser Capture Microdissection
LCM of frozen or paraffin-embedded breast tumors was performed as previously described using the PixCell II LCM system (Arcturus Engineering, Inc., Mountain View, CA).20 Briefly, tissue samples were sectioned at 7 μm, and tissue sections were stained according to the manufacturer’s protocol using H&E RNase-free reagents. For paraffin-embedded tissues, the tissue sections were deparaffinized before staining in 100% RNase-free ethanol as already indicated. After staining tissue sections were stored in a desiccated container for at least 15 minutes before LCM. For each analysis, ∼1500 tumor cells were laser-captured from the same breast tumor, either frozen or fixed in methacarn or RCL2. Efficiency of microdissection was assessed by examining the cells present on the LCM cap. For each breast tumor and for each condition of tissue processing (freezing or fixation in methacarn or RCL2), LCM was performed in duplicates or triplicates, and all these samples were used for RNA extraction and real-time reverse transcriptase-polymerase chain reaction (RT-PCR) analysis.
PCR and Real-Time RT-PCR
The quality of DNA extracted from methacarn- or RCL2-fixed breast tumors was tested by performing amplification and sequencing of a 250-bp fragment of the human Gs α gene, as described in Lumbroso and colleagues,21 or amplification of an 850-bp fragment of the human FSH receptor (FSH-R). Primers for the FSH-R were as follow: FSHR-A-FW 5′-ACCCTGCACAAAGACAGTGA-3′ and FSHR-A-R 5′-AGCTGTGACAAAGGGCTGTC-3′. For this purpose, 2 μl of extracted DNA was amplified (Gs α and FSH-R) and sequenced (Gs α). The sequences obtained with methacarn- or RCL2-fixed, paraffin-embedded tissues were compared to the sequences obtained with DNA extracted from the same frozen tissue (control) or with DNA extracted from NBF-fixed tissue.
For real-time RT-PCR, 9 μl of total RNA was subjected to a reverse transcription step using oligo dT primers and either the Omniscript (whole tissue section or MCF-7 pellets) or the Sensiscript (microdissected cells) reverse transcriptase kit (Qiagen). Real-time PCR was performed using a SYBR Green approach (Light Cycler Fast DNA Masterplus SYBR Green kit and Light Cycler technology; Roche, Mannheim, Germany). Primers and PCR conditions for ER-α, PR, and TBP,22,23 cyclin D1 (CCDN1) and β2m,24 HER2,25 and HPRT20 have been published elsewhere. mRNA levels for the various parameters (ER-α, PR, CCDN1, or TBP) were expressed relative to the HPRT housekeeping gene.
Results
Effects of Methacarn and RCL2 Fixatives on Breast Tumor Morphology, Immunohistochemistry, and HER2 CISH Analysis
Tissue morphology and immunohistochemistry were analyzed on six invasive breast carcinomas after tissue fixation and paraffin-embedding using either routine fixatives for breast tumor diagnosis and staging (NBF, FAAM) or methacarn or RCL2 fixatives. Methacarn and RCL2 preserved tissue integrity, giving morphological information similar to that obtained using the reference fixative FAAM with respect to cytoplasmic and nuclear details (Figure 1). Some tissue retraction was observed using FAAM, methacarn, RCL2, or NBF fixation, but this did not impair the global tissue architecture nor the cellular details in the tumor analyzed.
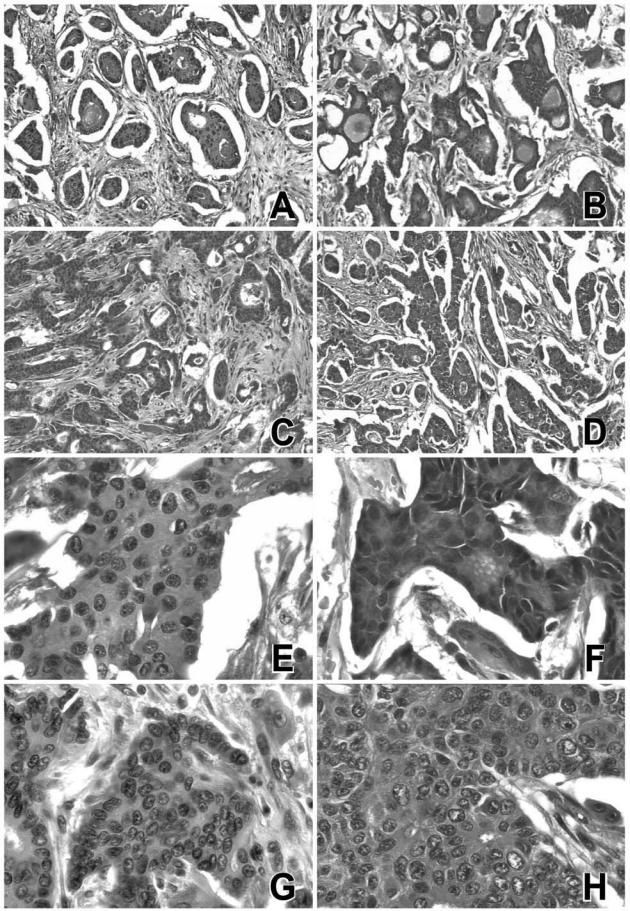
Figure 1.
Morphology of breast tumors exposed to methacarn or RCL2 fixatives. A breast carcinoma sample was either fixed in FAAM (A, E), NBF (B, F), methacarn (C, G), or RCL2 (D, H) before paraffin-embedding. A representative view of H&E-stained sections is presented. Similar views were obtained for five additional invasive breast carcinomas. Original magnifications: ×100 (A–D); ×400 (E–H).
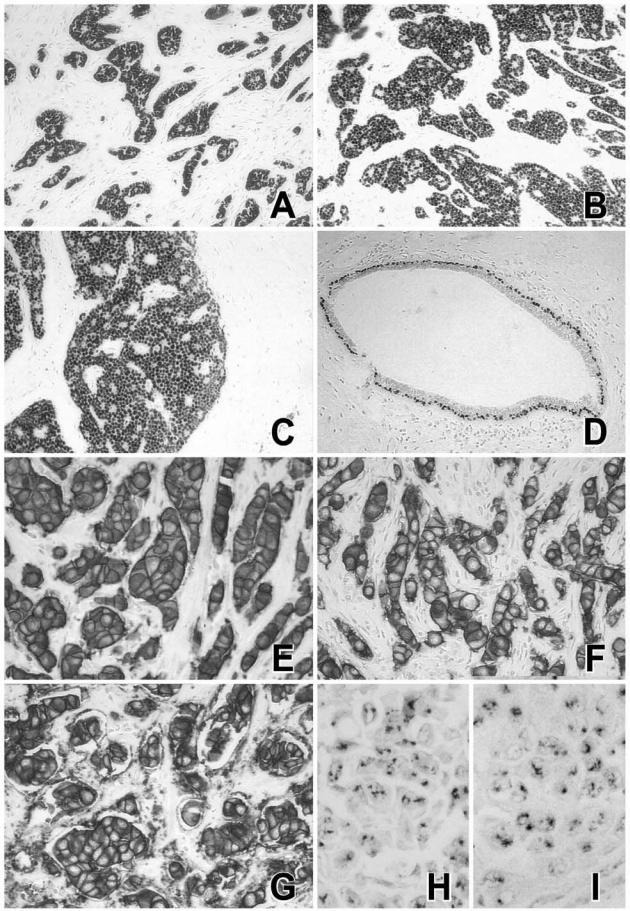
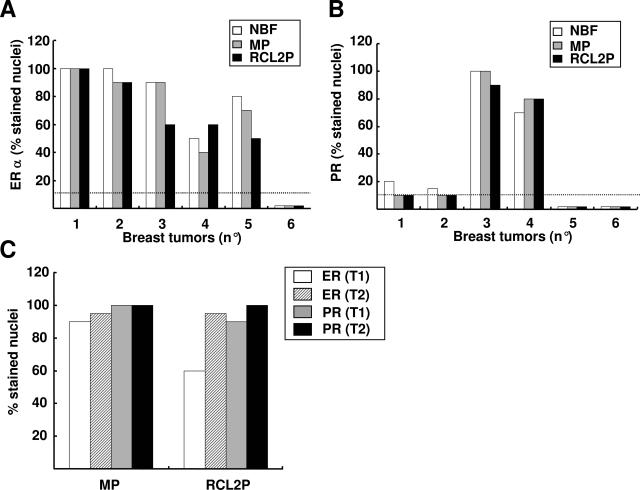
The effects of methacarn and RCL2 fixation on immunostaining were tested on the same invasive breast tumors (Figures 2and 3). Although both fixatives required some optimization of the immunostaining procedures including antigen retrieval, the percentage of stained cells was similar for ER-α, PR, or HER2 antigens as compared to NBF fixative (Figures 2and 3). Five breast tumors were diagnosed as ER-α-positive and one tumor as ER-α-negative regardless of the tissue fixative used (NBF, MP, or RCL2P) (Figure 3A). Similar results were obtained for PR status showing two tumors with high PR expression levels, two tumors with no PR expression, and two tumors with weak PR expression (Figure 3B). Analysis of the same breast tumor at the 8-month interval indicated good stability of tissue morphology as well as ER-α and PR immunostaining after methacarn and RCL2 fixation (Figure 3C). Of the six breast tumors tested, two tumors were scored 3+ for HER2 by immunohistochemistry and the other cases were negative (0 or 1+) (Figure 2E–G); the same results were obtained using NBF, methacarn, or RCL2 fixation procedures. HER2 gene amplification was confirmed on the HER2-positive tumors by CISH experiments using the reference NBF-fixed tumor tissues as well as the methacarn- and RCL2-fixed samples (Figure 2, H and I). Finally, immunoreactivity for various antibodies used for breast tumor diagnosis, including P63 (Figure 2D), S100 protein, smooth muscle actin, E-cadherin, cytokeratin 5/6, cytokeratin 7, and cytokeratin AE1/AE3 was similar in methacarn- or RCL2-fixed tissue samples as compared to the reference NBF-fixed tissue (data not shown).
Figure 2.
Immunohistochemical staining of ER-α, P63, and HER2, and CISH detection of HER2 gene. A breast carcinoma sample was fixed in NBF (A, E, H), methacarn (B, F), or RCL2 (C, D, G, I) before paraffin-embedding. Immunoreactivity for ER-α (A–C), P63 (D), HER2 (E–G), and CISH detection of HER2 gene (H, I) are shown. Similar views were obtained for five additional invasive breast carcinomas. Original magnifications: ×100 (A–D); ×400 (E–G); ×600 (H, I).
Figure 3.
Quantification of ER-α and PR in NBF-, methacarn-, and RCL2-fixed breast tumors. Six breast tumors (numbers 1 to 6) were processed according to routine procedures (NBF) or fixed in methacarn (MP) or RCL2 (RCL2P) before paraffin-embedding. For each condition of fixation, immunostaining of ER-α (A) and PR (B) was performed and quantified as percentage of stained nuclei. The cutoff value (10% stained nuclei) is indicated by a dotted line. C: The same breast tumor was fixed using methacarn (MP) or RCL2 (RCL2P), and immunostaining analysis of ER-α (ER) and PR was performed either immediately after surgical excision (T1) or 8 months later (T2).
Quality of DNA Extracted from Methacarn- and RCL2-Fixed Breast Tumors
Total DNA extracted from NBF-fixed breast tumor was significantly degraded as suggested by the smear observed on agarose gel and by the failure to amplify a large amplicon (FSH receptor; FSH-R) (Figure 4, A and B). By contrast, intact genomic DNA was extracted from the same frozen, methacarn (MP)- or RCL2 (RCL2P)-fixed tissue (Figure 4A) allowing amplification of an 850-bp FSH-R PCR product (Figure 4B). Amplification followed by sequencing of a 250-bp amplicon of the human Gs α gene was also performed, giving similar results with no nucleotide alterations for methacarn- and RCL2-fixed tissues as compared to the frozen control, contrasting with poor sequencing results using the matched NBF-fixed tissue sample (Figure 4C). Similarly, we could successfully amplify and sequence 825 nucleotides of the human ER gene using DNA extracted from the same frozen, methacarn- or RCL2-fixed paraffin-embedded breast tumor (data not shown).
Figure 4.
Quality of DNA extracted from methacarn- or RCL2-fixed breast tumors. Invasive breast tumors were processed as described in Materials and Methods, and total DNA was extracted from whole tissue sections using proteinase K digestion. A: Four μl of total DNA extracted from formalin-fixed (NBF), methacarn-fixed (MP), or RCL2-fixed (RCL2P) paraffin-embedded tissues or DNA extracted from the same frozen tissue (frozen) was submitted to electrophoresis on a 2% agarose gel after ethidium bromide staining. L: Ladder. B: An 850-bp fragment of the human FSH receptor (FSH-R) was amplified using DNA extracted from the tissue samples described in A and visualized on an agarose gel. The 850-bp amplicon is indicated by an arrow. NC: Negative PCR control; L: ladder. Similar results were obtained for two different breast tumors. C: Representative results of sequences obtained for a 250-bp amplicon of the human Gs α gene amplified from the various processed tissues. These results are representative of experiments performed on two different breast tumors.
Methacarn, RCL2, and RNA Quality
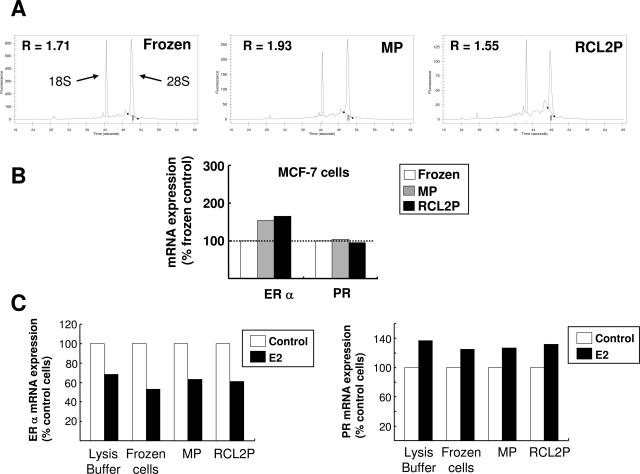
The effect on RNA quality of methacarn and RCL2 fixation followed by paraffin-embedding was first tested using MCF-7 breast tumor cells and compared to the results obtained from the same snap-frozen cells (Figure 5). High-quality total RNA was extracted from MCF-7 cells prepared in either condition (frozen, MP, and RCL2P) as judged from the electropherogram profiles shown in Figure 5A. RNA integrity was confirmed by real-time RT-PCR, showing similar ER-α and PR mRNA levels in methacarn- or RCL2-fixed paraffin-embedded MCF-7 cells as compared to frozen cells (Figure 5B). Because ER-α mRNA levels seemed slightly higher in methacarn- or RCL2-fixed cells than their frozen counterparts (Figure 5B, see also Figures 6Band 7C), the effect of both fixatives on dynamic mRNA variations was evaluated by measuring ER-α and PR mRNA levels in MCF-7 cells upon estradiol (E2) treatment (Figure 5C). As expected from previous reports,25,26,27,28 a significant decrease in ER-α and a weak increase in PR mRNA levels were observed in E2-treated MCF-7 cells, and those results were obtained regardless of the fixation procedure used.
Figure 5.
Quality of total RNA extracted from breast tumor MCF-7 cells after methacarn or RCL2 fixation and paraffin-embedding. Total RNA was extracted from MCF-7 cell pellets either frozen or fixed in methacarn (MP) or RCL2 (RCL2P) before paraffin-embedding as described in Materials and Methods. A: One μl of extracted RNA was run on Agilent RNA 6000 Picochip. The 18S and 28S ribosomal RNAs are indicated by an arrow and the 28S/18S ratio (R) is indicated for each sample. B: The same RNA was used for ER-α and PR mRNA quantification using real-time RT-PCR. Results represent the mean of duplicate samples. ER-α and PR mRNA levels are normalized to HPRT mRNA levels (housekeeping gene), and results for methacarn- and RCL2-fixed MCF-7 cells are expressed relative to the MCF-7 frozen cells control. C: MCF-7 cells were treated (E2) or not treated (control) with 10−8 mol/L estradiol (E2) for 24 hours, and MCF-7 cells were directly processed for RNA extraction (lysis buffer) or snap-frozen in liquid nitrogen (frozen cells) or fixed in methacarn (MP) or RCL2 (RCL2P) before paraffin-embedding. Total RNA was then extracted before real-time RT-PCR for ER-α and PR mRNA quantification. ER-α and PR mRNA levels are normalized to HPRT mRNA levels (housekeeping gene). Results are expressed relative to the nontreated MCF-7 cells (control cells).
Figure 6.
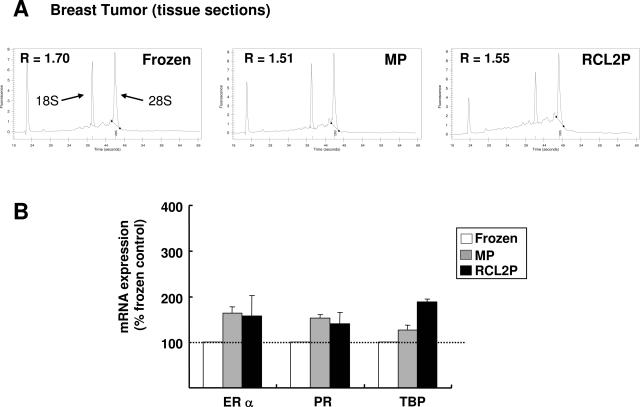
Quality of RNA extracted from methacarn- or RCL2-fixed breast tumors. Breast tumors were processed as indicated in Materials and Methods, and whole tissue sections were used for total RNA extraction. A: Electropherogram profiles of RNA extracted from frozen, methacarn-fixed (MP), and RCL2-fixed (RCL2P) tissue samples. The 28S/18S ratio (R) is indicated for each sample. B: Real-time RT-PCR quantification of ER-α, PR, and TBP mRNA was performed using the same RNA as presented in A. Results are normalized to the HPRT mRNA levels and expressed relative to the frozen tissue control. The results represent mean and standard deviations of three different sections from the same tissue sample. These results are representative of those obtained for three different breast tumors.
Figure 7.
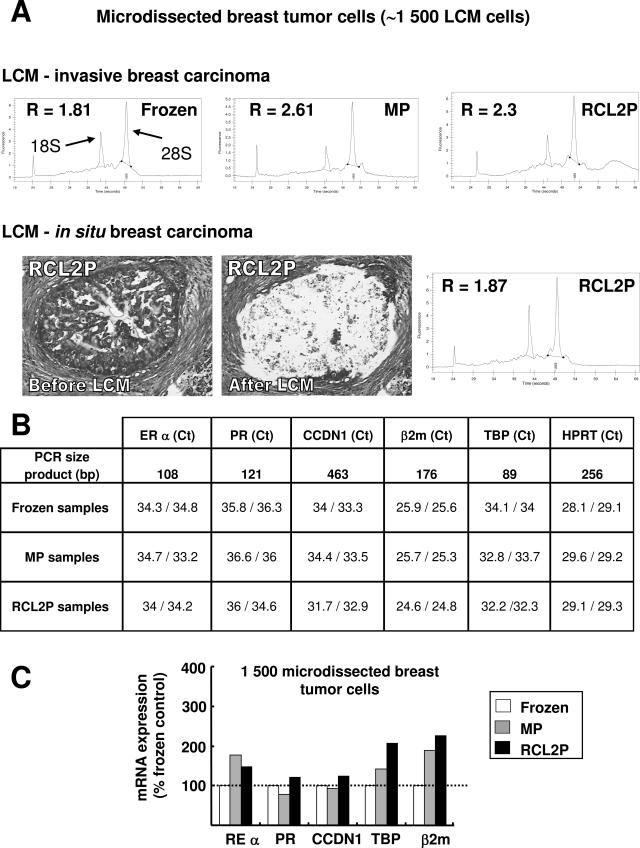
RNA and LCM of breast tumors after various fixation procedures. Breast tumors were processed as indicated in Materials and Methods, and total RNA was extracted from ∼1500 laser-captured breast tumor cells. A: Electropherogram profiles of total RNA extracted after LCM of an invasive breast carcinoma that was frozen or fixed in methacarn (MP) or RCL2 (RCL2P) and paraffin-embedded. The 28S/18S ratio (R) is presented for each electropherogram profile. Similar results were obtained after LCM of two additional invasive breast carcinomas. LCM of in situ breast carcinoma, adjacent to an invasive tumor and fixed in RCL2, was also performed, and the electropherogram profile of the corresponding extracted RNA is presented. Similar results were obtained from in situ breast carcinoma fixed in methacarn. B: Real-time RT-PCR quantification of ER-α, PR, CCDN1, β2m, TBP, and HPRT genes using RNA extracted from 1500 laser-captured breast tumor cells. The Ct values obtained for duplicate LCM samples from the invasive breast tumor presented in A are shown. C: ER-α, PR, CCDN1, and TBP mRNA levels for the same tumor as in B are normalized to the HPRT mRNA levels and expressed relative to the frozen tissue control. Similar results were obtained for the two other invasive breast carcinomas tested.
Tumor tissues are more difficult to handle than cultured cells because RNA may be adversely affected by the time delay between surgery and tissue processing. However, similar results in terms of RNA quality were obtained using total RNA extracted from breast tumor tissue sections. According to the electropherogram profiles (Figure 6A) or the quantification of specific mRNA (Figures 6B), intact RNA could be extracted from frozen, methacarn- or RCL2-fixed paraffin-embedded breast tumors, although we still noticed higher levels of ER-α, PR, or TBP mRNA using methacarn- or RCL2-fixed tissues as compared to frozen tissues (Figure 6B). Conversely, RNA extracted from NBF-fixed paraffin-embedded tissues was found to be repeatedly degraded, even when the dehydration and paraffin-embedding steps were performed at 4°C as for methacarn and RCL2 protocols (data not shown).
RNA Quality and LCM of Breast Tumors
LCM is a powerful approach for analyzing gene expression in specific cells from complex tissue structures and has been widely used in cancer research. However, mRNA quantification from laser-captured cells is challenged by the small amounts and by the quality of total RNA extracted, with the latter being highly dependent on the quality of the tissue used before microdissection. We thus evaluated the quality of RNA extracted from laser-captured breast tumor cells from methacarn- and RCL2-fixed paraffin-embedded tissues. Previous studies in our laboratory indicated that a minimum of 1500 microdissected breast tumor cells was necessary to allow reproducible mRNA quantification using real-time RT-PCR (data not shown).
High-quality total RNA could be extracted from 1500 laser-captured breast tumor cells from either processed tissues, as shown by electropherogram profiles (Figure 7A). This result was obtained after laser-capture microdissection of invasive breast carcinoma as well as in situ breast carcinoma (Figure 7A). Real-time RT-PCR experiments using the same RNA gave similar cycle threshold (Ct) measurements and reproducible mRNA quantifications for various genes such as ER-α, PR, CCDN1, β2m, HPRT, TBP (Figure 7, B and C) or HER2 (data not shown). Moreover, real-time RT-PCR quantification was possible even with amplicons larger than 200 bp (CCDN1, HPRT; Figure 7B).
The effect of long-term storage on RNA quality from methacarn- or RCL2-fixed tumors was evaluated by performing LCM and real-time RT-PCR on a breast tumor after surgical excision and 8 months later using the same tissue samples stored either at −80°C (frozen sample) or at −20°C (methacarn-paraffin or RCL2-paraffin samples). Analysis of the Ct measured for ER-α, PR, CCDN1, β2m, and HPRT genes in the same tumor between the two experiments (Table 1) indicated limited variations in mRNA levels [ΔCt (T1 to T2)], in the range of the interassay variations (Ct between 29 to 34). This suggested that, in addition to immunostaining (Figure 2), methacarn and RCL2 allow a good preservation of mRNA integrity throughout several months.
Table 1.
Effect of Long-Term Storage on RNA Quality
| ΔCt (T1 to T2) | ER-α | PR | CCDN1 | β2m | HPRT |
|---|---|---|---|---|---|
| Frozen samples | −0.4 | −1.3 | −0.8 | −1.2 | −0.6 |
| MP samples | −0.9 | −2 | −2 | 0.2 | −0.9 |
| RCL2P samples | −0.3 | 1.4 | 0.6 | 1.6 | 1.1 |
Laser capture microdissection of ∼1500 tumor cells was performed from the same breast tumor either frozen or fixed in methacarn (MP) or RCL2 (RCL2P) and paraffin-embedded. Total RNA was extracted before real-time RT-PCR quantification of ER-α, PR, CCDN1, β2m, and HPRT mRNA. This experiment was performed with the same tissue samples once (T1), and 8 months later (T2). The difference in Ct values obtained for the various genes at T1 and T2 are presented [ΔCt (T1 to T2)]. For each condition, laser capture microdissection was performed in duplicates or triplicates and the ΔCt values represent the differences between the mean Ct values obtained for each condition.
Discussion
Molecular analysis of premalignant lesions, such as preinvasive breast lesions, is an important step to better understand the early events of tumor progression and to identify new prognostic markers. Analysis of these microscopic lesions has recently gained advantage from the development of microdissection, allowing specific analysis of pathological cells without contamination by surrounding tissue.29,30 Unfortunately, these molecular approaches have been hampered by the limited availability of fresh or frozen clinical samples, as these small target lesions are usually entirely fixed in formalin before paraffin-embedding for accurate diagnosis and staging of the lesion. Formalin is a crosslinking fixative that is known to alter and fragment nucleic acids, thus impairing extraction efficiency and quality of DNA and, more strikingly, RNA.9,10,31,32 Although recent studies have reported an improved protocol for gene expression analysis from formalin-fixed tissues, including formalin-fixed breast tumors,33,34,35,36,37 this approach has technical limitations linked to the lower yield of RNA recovered, to the need for small amplicon size (<100 bp) and to further RNA degradation with increased archival storage.8,10,33 These limitations may be critical when working with genes harboring weak expression levels or with small amounts of RNA such as RNA extracted from a few microdissected cells.
Molecular analysis of small target lesions thus requires new fixation procedures allowing accurate histomorphological diagnosis as well as DNA and RNA preservation on the same fixed tissue sample. Based on these requirements, we evaluated two noncrosslinking fixatives on invasive breast tumors: the methacarn solution because of its known protecting properties on DNA, RNA, and proteins and RCL2, a new promising fixative that was recently described to preserve both nucleic acids and proteins (M. Lacroix-Triki, L. Lamant, B. A. Cuider, A. Decha, J.-J. Voigt, P. Rochaix; submitted manuscript).
Because histomorphological diagnosis of breast tumors is critical for patient treatment and prognosis, we first performed a careful evaluation of the quality of tissue morphology and immunostaining for the standard therapeutic markers ER-α, PR, and HER2 in methacarn- and RCL2-fixed paraffin-embedded breast tumors. Methacarn has previously been shown to be a valuable fixative for tissue morphology and immunohistochemical studies.8,38 Similarly, in preliminary experiments, RCL2 gave interesting histomorphological results on various tissues. In this report, we confirm that both methacarn and RCL2 are suitable for morphological diagnosis of breast tumors.
Although some optimization of the immunostaining procedures was needed, similar quantification of ER-α, PR, and HER2 antigens were obtained for breast tumors fixed in methacarn or RCL2, as compared to the standard NBF-fixed tumors. Furthermore, HER2 gene amplification could be confirmed by CISH experiments using either methacarn- or RCL2-fixed tumor samples, suggesting DNA preservation. Successful immunostaining was also obtained for various antigens currently tested for breast tumor diagnosis. Finally, methacarn and RCL2 were found to preserve tissue morphology and immunostaining after several months of tissue storage. Altogether, these data validate methacarn and RCL2 for routine breast tumor histomorphological diagnosis.
Both fixatives were previously validated for DNA integrity using fixed and paraffin-embedded tissues by showing high yields of extracted DNA and amplification of large DNA fragments (P. Rochaix, unpublished data).12 We confirmed these findings using human breast tumors, and further demonstrated that both fixatives allowed accurate DNA sequencing of more than 800 bp. This is an important point because DNA isolated from formalin-fixed tissues has been shown to exhibit a high frequency of nonreproducible sequence alterations, up to 1 mutation artifact per 500 bases, which may alter sequencing results in archival formalin-fixed material.39 Thus, although numerous studies have used formalin-fixed tissues for DNA amplification, methacarn or RCL2 may have higher performances for fine DNA analysis such as DNA sequencing and may give similar results as frozen tissues.
Methacarn is a noncrosslinking organic fixative that was shown to preserve RNA, possibly by causing precipitation and inactivation of endogenous tissue RNases.8 Recent studies by Shibutani and colleagues11 confirmed that high-quality RNA could be extracted from methacarn-fixed paraffin-embedded rat liver sections, with efficiency similar to that with unfixed cryosections. Moreover, methacarn preserved the relative abundance of amplifiable mRNA in microdissected tissue specimens.13,14 Similarly, RCL2 has been shown to preserve RNA integrity on mouse liver tissue sections (P. Rochaix et al, unpublished data). To validate both fixatives at the RNA level in human breast tumors, we evaluated RNA integrity by analyzing ribosomal RNA profiles using miniaturized capillary electrophoresis, by measuring specific mRNA expression, including genes with low expression levels (such as ER-α, PR, or HPRT genes) and by quantifying various mRNAs in a limited number of laser-captured breast tumor cells. This latter condition is particularly stringent because additional steps from human tissue processing to mRNA quantification, including tissue section and staining as well as LCM, may alter RNA integrity. Using these different approaches, we found that methacarn and RCL2 were excellent fixatives for preserving RNA from human surgical samples. Interestingly, we could validate these new fixation procedures to analyze RNA extracted from in situ ductal breast carcinoma. Moreover, we found reproducible mRNA quantification using microdissected methacarn- and RCL2-fixed paraffin-embedded breast tumors after 8 months of tissue storage at −20°C.
Regarding mRNA quantification, we surprisingly found that some genes, including ER-α or TBP genes, had higher relative expression in methacarn- or RCL2-fixed breast tumors as compared to their frozen counterparts. However, in our MCF-7 tumor cell models, the dynamic expression of ER-α upon estradiol treatment was similar regardless of the fixative used when compared to the results obtained when total RNA was immediately extracted from cells. The reason for this higher expression of ER-α or TBP genes in methacarn- and RCL2-fixed tissues is unclear. This is probably not due to breast tumor heterogeneity because similar observations were made using MCF-7 cultured cells. ER-α mRNA was shown to be unstable, especially under estradiol conditions.40 It is thus possible that ER-α mRNA is not entirely protected from degradation in snap-frozen tissues, in contrast to methacarn- or RCL2-fixed tissues. In agreement with this hypothesis, we repeatedly observed lower relative expression of ER-α gene using total RNA extracted from snap-frozen cultured breast tumor cells as compared to total RNA immediately extracted from the same fresh cultured cells (data not shown). Whatever the reason for this observation, this suggests that one may be cautious when analyzing gene expression in tissue specimens either frozen or fixed using various procedures.
In this study, breast tumor samples were fixed overnight at 4°C in methacarn or RCL2 and then dehydrated at the same temperature in ethanol and acetone. The requirement for dehydration steps at 4°C is not convenient for a routine laboratory use, hampering automation of the fixation procedure. The use of methacarn may also be limited by its poor stability and its potential toxicity. In contrast, preliminary results indicate that RCL2 is as efficient in protecting RNA as previously shown when performing the dehydration steps at room temperature, which makes its use in routine practice likely in the future. Moreover, this new fixative is devoid of toxicity with easy processing steps (P. Rochaix, unpublished data). In this respect, RCL2, and to a lesser extent methacarn, belong to a new generation of tissue fixatives, including UMFIX described by Vincek and colleagues,16 which could be used in routine laboratory for pathological diagnosis and molecular analysis of the same tissue samples. Although the question of long-term tissue storage should now be addressed, these new fixation procedures will undoubtedly make molecular analysis of human clinical samples easier, including analysis of small premalignant lesions in which microdissection approaches are needed.
In conclusion, we made a careful evaluation of methacarn and RCL2 fixatives using paraffin-embedded human breast tumor samples and found that both fixatives had high performance, even after several months of tissue storage, regarding tissue histomorphology and DNA and RNA preservation, including RNA extracted from microdissected tissue samples. These fixatives will be helpful in the future for investigating small target lesions such as premalignant breast lesions.
Acknowledgments
We thank Mrs. Marie-Christine Lecq and Nadia Chapelle for their careful technical assistance and Pr. T. Lavabre-Bertrand for critical reading of the manuscript.
Footnotes
Supported by the “Ligue Nationale Contre le Cancer” (Comité de l’Hérault), the “Centre Hospitalier Universitaire de Montpellier,” the “Institut National de la Santé et de la Recherché Médicale,” and the “Cancéropôle du Grand Sud Ouest.”
C.D. and P. Roger contributed equally to this work.
References
- Schena M, Shalon D, Davis R, Brown P. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Velculescu V, Zhang L, Volgelstein B, Kinzler K. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Emmert-Buck M, Gillespie J, Paweletz C, Ornstein D, Berserk V, Appella E, Wang Q-H, Huang J, Hu N, Taylor P, Petricoin E., III An approach to proteomic analysis of human tumors. Mol Carcinog. 2000;27:158–165. [PubMed] [Google Scholar]
- Lim M, Elenitoba-Johnson K. Proteomics in pathology research. Lab Invest. 2004;84:1227–1244. doi: 10.1038/labinvest.3700167. [DOI] [PubMed] [Google Scholar]
- Bustin S. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Emmert-Buck M, Bonner R, Smith P, Chuaqui R, Zhuang Z, Goldstein S, Weiss R, Liotta L. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- Goldsworthy S, Stockton P, Trempus C, Foley J, Maronpot R. Effects of fixation on RNA extraction and amplification from laser capture microdissected tissue. Mol Carcinog. 1999;25:86–91. [PubMed] [Google Scholar]
- Srinivasan M, Sedmark D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961–1971. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27:4436–4443. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis F, Maughan N, Smith V, Hillan K, Quirke P. Unlocking the archive-gene expression in paraffin-embedded tissue. J Pathol. 2001;195:66–71. doi: 10.1002/1096-9896(200109)195:1<66::AID-PATH921>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Shibutani M, Uneyama C, Miyazaki K, Toyoda K, Hirose M. Methacarn fixation: a novel tool for analysis of gene expressions in paraffin-embedded tissue specimens. Lab Invest. 2000;80:199–208. doi: 10.1038/labinvest.3780023. [DOI] [PubMed] [Google Scholar]
- Uneyama C, Shibutani M, Masutomi N, Takagi H, Hirose M. Methacarn fixation for genomic DNA analysis in microdissected, paraffin-embedded tissue specimen. J Histochem Cytochem. 2002;52:903–913. doi: 10.1177/002215540205000911. [DOI] [PubMed] [Google Scholar]
- Kim J-O, Kim H-N, Hwang M-H, Shin H-I, Kim S-Y, Park R-W, Park E-Y, Kim I-S, van Wijnen A, Stein J, Lian J, Stein G, Choi J-Y. Differential gene expression analysis using paraffin-embedded tissues after laser microdissection. J Cell Biochem. 2003;90:998–1006. doi: 10.1002/jcb.10680. [DOI] [PubMed] [Google Scholar]
- Takagi H, Shibutani M, Kato N, Fujita H, Lee K-Y, Takigami S, Mitsumori K, Hirose M. Microdissected region-specific gene expression analysis with methacarn-fixed, paraffin-embedded tissues by real-time RT-PCR. J Histochem Cytochem. 2004;52:903–913. doi: 10.1369/jhc.3A6215.2004. [DOI] [PubMed] [Google Scholar]
- Morales A, Essenfeld H, Essenfeld E, Duboue M, Vincek V, Nadji M. Continuous-specimen-flow, high-throughput, 1-hour tissue processing: a system for rapid diagnostic tissue preparation. Arch Pathol Lab Med. 2002;126:583–590. doi: 10.5858/2002-126-0583-CSFHTH. [DOI] [PubMed] [Google Scholar]
- Vincek V, Nassiri M, Nadji M, Morales A. A tissue fixative that protects macromolecules (DNA, RNA and protein) and histomorphology in clinical samples. Lab Invest. 2003;83:1427–1435. doi: 10.1097/01.lab.0000090154.55436.d1. [DOI] [PubMed] [Google Scholar]
- Roger P, Esslimani Sahla M, Mäkelä S, Gustafsson J, Baldet P, Rochefort H. Decreased expression of estrogen receptor β protein in proliferative preinvasive mammary tumors. Cancer Res. 2001;61:2537–2541. [PubMed] [Google Scholar]
- Vincent-Salomon A, MacGrogan G, Couturier J, Arnould L, Denoux Y, Fiche M, Jacquemier J, Mathieu M, Penault-Llorca F, Rigaud C, Roger P, Treilleux I, Vilain M, Mathoulin-Pelissier S, Le Doussal V. Calibration of immunohistochemistry for assessment of HER2 in breast cancer: results of the French multicentre GEFPICS study. Histopathology. 2003;42:337–347. doi: 10.1046/j.1365-2559.2003.01598.x. [DOI] [PubMed] [Google Scholar]
- Tanner M, Gancberg D, Di Leo A, Larsimont D, Rouas G, Piccart M, Isola J. Chromogenic in situ hybridization. A practical alternative for fluorescence in situ hybridization to detect Her-2/neu oncogene amplification in archival breast cancer samples. Am J Pathol. 2000;157:1467–1472. doi: 10.1016/S0002-9440(10)64785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copois V, Bret C, Bibeau F, Brouillet J, del Rio M, Berthe M-L, Maudelonde T, Boulle N. Assessment of RNA quality extracted from laser-captured tissues using miniaturized capillary electrophoresis. Lab Invest. 2003;83:599–602. doi: 10.1097/01.lab.0000062853.92828.20. [DOI] [PubMed] [Google Scholar]
- Lumbroso S, Paris F, Sultan C. Activating Gs alpha mutations: analysis of 113 patients with signs of McCune-Albright syndrome—a European collaborative study. J Clin Endocrinol Metab. 2004;89:2107–2113. doi: 10.1210/jc.2003-031225. [DOI] [PubMed] [Google Scholar]
- Bièche I, Onody P, Laurendeau I, Olivi M, Vidaud D, Lidereau R, Vidaud M. Real-time reverse transcription-PCR assay for future management of ERBB2-based clinical applications. Clin Chem. 1999;45:1148–1156. [PubMed] [Google Scholar]
- Bieche I, Parfait B, Tozlu S, Lidereau R, Vidaud M. Quantitation of androgen receptor gene expression in sporadic breast tumors by real-time RT-PCR: evidence that MYC is an AR-regulated gene. Carcinogenesis. 2001;22:1521–1526. doi: 10.1093/carcin/22.9.1521. [DOI] [PubMed] [Google Scholar]
- Rey J, Pujol P, Callier C, Cavailles V, Freiss G, Maudelonde T, Brouillet J. Semiquantitative reverse transcription-polymerase chain reaction to evaluate the expression patterns of genes involved in the oestrogen pathway. J Mol Endocrinol. 2000;24:433–440. doi: 10.1677/jme.0.0240433. [DOI] [PubMed] [Google Scholar]
- de Cremoux P, Tran-Perennou C, Brockdorff B, Boudou E, Brünner N, Magdelenat H, Lykkesfeldt A. Validation of real-time RT-PCR for analysis of human breast cancer cell lines resistant or sensitive to treatment with antiestrogens. Endocr Relat Cancer. 2003;10:409–418. doi: 10.1677/erc.0.0100409. [DOI] [PubMed] [Google Scholar]
- May F, Johnson M, Wiseman L, Wakeling A, Kastner P, Westley B. Regulation of progesterone receptor mRNA by oestradiol and antioestrogens in breast cancer cell lines. J Steroid Biochem. 1989;33:1035–1041. doi: 10.1016/0022-4731(89)90406-8. [DOI] [PubMed] [Google Scholar]
- Pink J, Jordan V. Models of estrogen receptor regulation by estrogens and antiestrogens in breast cancer cell lines. Cancer Res. 1996;56:2321–2330. [PubMed] [Google Scholar]
- Bentrem D, Dardes R, Liu H, Macgregor-Schafer J, Zapf J, Jordan V. Molecular mechanism of action at estrogen receptor a of a new clinically relevant antiestrogen (GW7604) related to tamoxifen. Endocrinology. 2001;142:838–846. doi: 10.1210/endo.142.2.7932. [DOI] [PubMed] [Google Scholar]
- Wulfkuhle J, Sgroi D, Krutzsch H, McLean K, McGarvey K, Knowlton M, Chen S, Sahin A, Kurek R, Wallwiener D, Merino M, Petricoin E, Zhao Y, Steeg P. Proteomics of human breast ductal carcinoma in situ. Cancer Res. 2002;62:6740–6749. [PubMed] [Google Scholar]
- Ma X-J, Salunga R, Tuggle J, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang B, Zhou Y-X, Varnholt H, Smith B, Gadd M, Chatfield E, Kessler J, Baer T, Erlander M, Sgroi D. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata D. Extraction of DNA from paraffin-embedded tissue for analysis by polymerase: new tricks from an old friend. Hum Pathol. 1994;25:561–563. doi: 10.1016/0046-8177(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Frank T, Svoboda-Newman S, Hsi E. Comparison of methods for extracting DNA from formalin-fixed, paraffin sections for nonisotopic PCR. Diagn Mol Pathol. 1996;5:220–224. doi: 10.1097/00019606-199609000-00012. [DOI] [PubMed] [Google Scholar]
- Specht K, Richter T, Müller U, Walch A, Werner M, Höfler H. Quantitative gene expression analysis in microdissected archival formalin-fixed and paraffin-embedded tumor tissue. Am J Pathol. 2001;158:419–429. doi: 10.1016/S0002-9440(10)63985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann U, Kreipe H. Real-time analysis of DNA and RNA extracted from formalin-fixed and paraffin embedded biopsies. Methods. 2001;25:409–418. doi: 10.1006/meth.2001.1263. [DOI] [PubMed] [Google Scholar]
- Cohen C, Gröne H-J, Gröne E, Nelson P, Schlöndorff D, Kretzler M. Laser microdissection and gene expression analysis on formaldehyde-fixed archival tissue. Kidney Int. 2002;61:125–132. doi: 10.1046/j.1523-1755.2002.00112.x. [DOI] [PubMed] [Google Scholar]
- Gjerdrum L, Sorensen B, Kjeldsen E, Sorensen F, Nexo E, Hamilton-Dutoit S. Real-time quantitative PCR of microdissected paraffin-embedded breast carcinoma. J Mol Diag. 2004;6:42–51. doi: 10.1016/S1525-1578(10)60490-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin M, Pho M, Dutta D, Stephans J, Shak S, Kiefer M, Esteban J, Baker J. Measurement of gene expression in archival paraffin-embedded tissues. Am J Pathol. 2004;164:35–42. doi: 10.1016/S0002-9440(10)63093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D, Ibrahim S, Gusterson B. Improved immunohistochemical localization of tissue antigens using modified methacarn fixation. J Histochem Cytochem. 1985;33:491–495. doi: 10.1177/33.5.3921605. [DOI] [PubMed] [Google Scholar]
- Williams C, Ponten F, Moberg C, Soderkvist P, Uhlen M, Ponten J, Sitbon G, Lundeberg J. A high frequency of sequence alterations is due to formalin fixation of archival specimen. Am J Pathol. 1999;155:1467–1471. doi: 10.1016/S0002-9440(10)65461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saceda M, Lindsey R, Solomon H, Angeloni S, Martin M. Estradiol regulates estrogen receptor mRNA stability. J Steroid Biochem Mol Biol. 1998;66:113–120. doi: 10.1016/s0960-0760(98)00049-1. [DOI] [PubMed] [Google Scholar]