Abstract
Latent transforming growth factor (TGF)-β binding proteins (LTBPs) play important roles in the secretion and activation of TGF-β. We previously reported that LTBP-1L is overexpressed in some patients with ovarian cancer. To clarify the molecular mechanism of LTBP-1L regulation, we analyzed DNA sequences in the promoter region of LTBP-1L and identified two novel single nucleotide polymorphisms, −202G/C and +20A/C. While the alleles with −202C and +20C were initially reported, our data demonstrated that −202G and +20A are common in both ovarian cancer patients and healthy patients in the Japanese population. Luciferase reporter assays revealed that the G-A haplotype induced transcriptional activation in a Sp1-dependent manner. Electrophoretic mobility shift assays showed that increased binding affinity of Sp1 to the promoter with −202G and +20A. Interestingly, ovarian cancer patients (n = 42) with G-A/G-A homozygous genotype had increased expression of LTBP-1 and apparently poorer survival than those with other genotypes (P = 0.02). These findings suggest that the single nucleotide polymorphisms −202G/C and +20A/C on the LTBP-1L promoter may affect the clinical outcome of ovarian cancer patients, probably via up-regulating protein expression. Further studies using a larger number of samples will definitively determine the correlation between LTBP-1 haplotype and clinical behavior of ovarian cancer.
Transforming growth factor (TGF)-β is a potent growth inhibitor for most cell types, including epithelial cells.1,2 The secretion of TGF-β by tumor cells may contribute to tumor suppression by autocrine growth inhibition, but on the other hand, it may also promote tumor progression by stimulating tumor invasion and angiogenesis, and inhibiting immune response.3,4,5 TGF-β is synthesized as latent high-molecular weight complexes, composed of TGF-β, the NH2-terminal part of the TGF-β precursor, and the latent TGF-β binding protein (LTBP).6,7,8,9 LTBP is a glycoprotein with a molecular weight of more than 190 kd; it possesses 16 to 18 epidermal growth factor (EGF)-like domains and several repeats of a unique motif containing eight cysteine residues.10,11,12 Four isoforms of LTBP have been found in mammalian species (LTBP-1 to LTBP-4).13,14,15,16,17 LTBP-1 binds to TGF-β1 through one of the eight cysteine motifs and facilitates assembly and secretion18,19 and activation20,21,22 of TGF-β1. Immunoelectron-microscopic observations indicate that LTBP-1 is one of the extracellular microfibrillar components23; it targets TGF-β1 to extracellular structures and participates in the activation of latent TGF-β1, perhaps by concentrating latent TGF-β1 on the cell surface where activation occurs.9 LTBP-1S (short) and LTBP-1L (long) are derived from independent promoters and alternative splicing between codons 145 and 146 of LTBP-1S.24 LTBP-1L has a N-terminal extension of 346 amino acids that is not found in the LTBP-1S.25 The N-terminal extension contains an EGF-like domain that facilitates matrix incorporation, and LTBP-1L has been confirmed to associate more efficiently than LTBP-1S with the extracellular matrix.25
We previously demonstrated overexpression of LTBP-1 and TGF-β1 proteins in some ovarian cancer patients, predominantly because of up-regulation of LTBP-1L mRNA rather than LTBP-1S mRNA.26 This overexpression is thought to affect the function of TGF-β1 and its signaling, possibly contributing to ovarian carcinogenesis. However, little information is available concerning the transcriptional regulation of these genes. The present study therefore aimed to clarify the molecular mechanisms of LTBP-1L overexpression in ovarian cancers and to investigate the clinical significance of the overexpression. We found that two novel single nucleotide polymorphisms in the 5′-regulatory region of the LTBP-1L gene critically contribute to the overexpression of this gene.
Materials and Methods
Samples and DNA Preparation
Tissue specimens were surgically obtained from a consecutive series of 42 patients aged 23 to 77 with ovarian cancer in Kanazawa University Hospital and stored at −80°C for DNA analysis or fixed with formaldehyde for immunohistochemical analysis. Patients gave written informed consent for the use of their tissues in this study. All of the patients were newly diagnosed, previously untreated (chemotherapy or radiotherapy), and histologically confirmed without family history of ovarian cancer. Healthy control DNA samples were obtained from buccal epithelial cells of 156 Japanese individuals (80 males and 76 females) age 20 to 60 years, including staff and students in Kanazawa University, with written informed consent. Buccal epithelial cells were collected by modifying the mouthwash method described previously.27,28 Briefly, at least 3 hours after brushing their teeth, participants swished 10 ml of mouthwash, Mondahmin (Earth Chemical, Tokyo, Japan), vigorously in their mouths for 30 seconds and then spat it into a conical tube. The buccal cells were harvested by centrifugation at 3000 × g for 10 minutes. Total genomic DNA was purified using a Wizard genomic DNA purification kit (Promega, Madison, WI).
Sequencing of LTBP-1L Promoter
The LTBP-1L promoter region spanning −2164 and +130 was analyzed by direct sequencing. The polymerase chain reaction (PCR) primer sequences were F(forward)-1: 5′-CGTCGACTCGATCTCAAAGTGTTGC-3′ and R(reverse)-1: 5′-GAGGATTGAGGTGAGTCACAAGG-3′, F-2: 5′-GTAGAACAAGGAATTGGATCCGT-3′ and R-2: 5′-TTGATTTGGCAGGCAGGGCCTC-3′, F-3: 5′-GTTCTCACAAGCAGC-TAGTGCT-3′ and R-3: 5′-GAAAGTCCACAGTCATAGCA-GTC-3′, F-4: 5′-CAAAGCCTTGGAAACACACCATC-3′ and R-4: 5′-TTAGGGTAGGACTAGAGTTCA-3′, F-5: 5′-GTCGGATTACGGTCCCGTGA-3′ and R-5: 5′-TTACTGAACGA-TCCTGTCCTTTC-3′, F-6: 5′-AGTATCACAGCAAACACGGAT-3′; and R-6: 5′-GGTGCACCACGTAGGTGATCCTCC-3′. All PCR products were purified and subsequently directly sequenced from both ends. All sequencing reactions were performed using dye-terminator chemistry (Applied Biosystems, Foster City, CA) in an ABI 310 DNA analyzer.
Haplotype Analysis
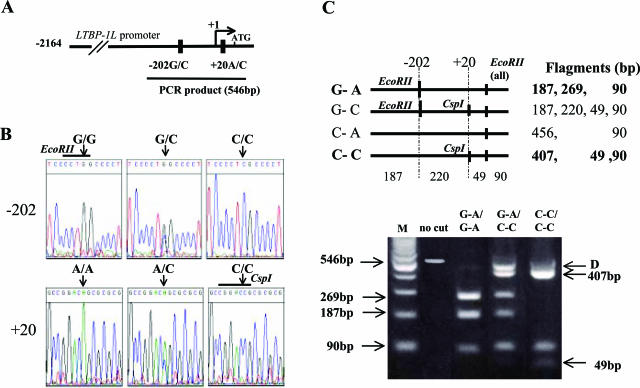
The haplotypes of −202G/C and +20A/C were analyzed by PCR-based restriction fragment length polymorphism (RFLP) methods. The LTBP-1L promoter region spanning −392 and +154 was amplified using F-7: 5′-TTGGCTGCTCAGGTCTGACA-3′ and R-6 primers. The PCR cycling conditions were 2 minutes at 94°C followed by 31 cycles of 1 minute at 94°C, 1 minute at 56°C, and 1 minute at 72°C, with a final step at 72°C for 10 minutes to allow for the complete extension of all PCR fragments. The number of cycles was within the exponential region of the PCR reaction. The 546-bp PCR products were subjected to EcoRII (Toyobo, Osaka, Japan) and CspI (Toyobo) digestion, and the DNA fragments were separated on a 3% agarose gel. The haplotypes were determined as follows: three fragments of 187, 269, and 90 bp represent the G-A haplotype; three fragments of 407, 49, and 90 bp represent the C-C haplotype; four fragments of 187, 220, 49, and 90 bp represent the G-C haplotype; and two fragments of 456 and 90 bp represent the C-A haplotype. Hetero-duplex fragments of 456 bp, which could not be digested by the restriction enzymes, were also produced because of mismatched annealing of heterogeneous alleles. We confirmed that such 456-bp products detected in patients with heterozygous genotypes were not due to the C-A allele by means of DNA sequencing of the purified bands.
Cell Culture
The RMUG-S cell line was derived from ovarian mucinous adenocarcinoma (JCRB, Osaka, Japan). The cells were maintained in Ham’s F12 medium (Sigma-Aldrich Corp., St. Louis, MO) containing 10% fetal bovine serum (JRH Bioscience, Lenexa, KS), 100 U/ml penicillin, and 100 μg/ml streptomycin.
Preparation of the Plasmids
The LTBP-1L promoter-luciferase reporter plasmid was a generous gift from Dr. Jorma Keski-Oja (University of Helsinki, Helsinki, Finland).24 It contains the promoter fragment spanning −2211 to +54 upstream of the initiating ATG codon, inserted into luciferase reporter vector pGL3-Basic (Promega). The mutated plasmids were constructed using the GeneTailor site-directed mutagenesis system (Invitrogen Life Technologies, Carlsbad, CA). The forward and reverse mutagenic primers for −202C to G were 5′-TGCGCGGCCCGCTCCCCTGGCCCCTCCCCGCTCCC-3′ and 5′-AGGGGAGCGGGCCGCGCAAGGTGAGGGTCC-3′, respectively. The forward and reverse mutagenic primers for +20C to A were 5′-GGCCGGGGGAGGGGGCCGGACAGCGCGCGACC-3′ and 5′-GTCCGGCCCCCTCCCCCGGCCGTGCGGCTCGCCT-3′, respectively (mutated sites are underlined). Nucleotide sequences were checked by DNA sequencing analyses. The plasmid constructs pM, pM-Sp1, and pCMV-DNSp3 were kind gifts from Dr. Yoshihiro Sowa (Kyoto Prefectural University of Medicine, Kyoto, Japan). pM-Sp1 is an expression vector for Sp1 protein. pCMV-DNSp3 is an expression vector for the dominant-negative form of Sp3, in which the function of Sp1 is abrogated.29,30
Luciferase Assays
RMUG-S cells (5 × 104) were seeded on 24-well plates 1 day before transfection. The cells were transfected with 0.4 μg of each reporter plasmid alone or together with the same amount of effector plasmid using FuGENE6 transfection reagent (Roche Molecular Biochemicals, Mannheim, Germany). Renilla luciferase plasmid phRL-tk was co-transfected to normalize the transfection efficiency. After 48 hours, the cells were lysed using a dual-luciferase reporter assay system (Promega) and the firefly luciferase and Renilla luciferase activities were measured with a Lumat LB9507 (Berthold Technologies, Tokyo, Japan). All experiments were performed at least three times for each reporter plasmid and the relative luciferase activity was calculated. Results are mean ± SD (n = 3).
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed as described previously.31 Briefly, synthetic double-stranded oligonucleotides 5′-GAGAATGCGGACCCTCCTGGGAGT-3′ and 5′-GAGAATGCGGACTCTCCTGGGAGT-3′ corresponding to the −202C and −202G sequences of the LTBP-1L promoter region were labeled with [γ-32P]ATP (Amersham Pharmacia Biotech, Piscataway, NJ) using the MEGALABEL kit (Takara, Shiga, Japan). One μg of recombinant human Sp1 protein (Promega) was incubated with 1 μg of poly(dI-dC) (Amersham Biosciences) in the presence or absence of a 100-fold molar excess of unlabeled competitor DNA on ice for 20 minutes in a 25-μl reaction mixture containing 10% glycerol, 25 mmol/L HEPES (pH 7.9), 50 mmol/L KCl, 0.5 mmol/L phenylmethyl sulfonyl fluoride, and 1 mmol/L dithiothreitol. For supershift assay, specific antibodies against Sp1 (Santa Cruz Biotechnologies, Santa Cruz, CA) or estrogen receptor (Santa Cruz Biotechnologies) were preincubated with recombinant Sp1 proteins (Promega) on ice for 60 minutes. After incubation, an aliquot of labeled oligonucleotide (>30,000 cpm) was added, and the reaction mixture was incubated at room temperature for an additional 20 minutes. The DNA-protein complexes were then separated from free probes by electrophoresis on a 5% polyacrylamide gel. The gel was dried and subjected to autoradiography using a Fuji BAS-III bioimaging analyzer (Fuji Photo Film, Kanagawa, Japan). Consensus oligonucleotides for Sp1 (5′-ATTCGATCGGGGCGGGGCGAGC-3′; Promega), or AP2 (5′-GATCGAACTGACCGCCCGCGGCCCGT-3′; Promega) were also used as competitors to confirm the specific binding of Sp1.
Immunohistochemistry
Immunohistochemical assays for LTBP-1 were performed on formalin-fixed, paraffin-embedded specimens of ovary tissues, using the Vectastain ABC Elite kit (Vector Laboratories, Burlingame, CA) as described previously.32 Antigen retrieval was performed for 10 minutes in 1× antigen retrieval solution (Biogenex, San Ramon, CA), and endogenous peroxidase was quenched in methanol with 3% H2O2. Then, the tissue sections were incubated for 16 hours at 4°C with mouse monoclonal anti-LTBP-1 antibody (R& D Systems, Minneapolis, MN) at a dilution of 1:1000 or with nonimmune whole mouse serum. After washing the sections were incubated with a biotinylated secondary antibody, followed by detection by sequential reaction with a streptavidin-biotin-horseradish peroxidase complex, biotinylated tyramide, streptavidin peroxidase, and 3,3′-diaminobenzidine (Dako Cytomation, Carpinteria, CA). Sections were lightly counterstained with Mayer’s hematoxylin, and then mounted. Staining was evaluated as negative 0 (no staining), low 1+ (positive in 5 to 25% cells or staining intensity was extremely low), intermediate 2+ (positive in 25 to 75% cells, intensely positive), high 3+ (positive in >75% cells and staining intensity was extremely high). Two independent pathologists judged the results and cases with discordant results were excluded from the analyses.
Statistical Analysis
Deviations of the genotype frequencies in the cancer cases and controls from those expected under Hardy-Weinberg equilibrium were assessed with the χ2 test. The luciferase assay data were analyzed by analysis of variance followed by Fisher’s protected least significant difference (PLSD) test. Immunohistochemical staining was analyzed by Fisher’s exact probability test. Overall survival was measured from primary surgery to death or the date of last follow-up. Patient follow-up was updated through December 1, 2004. Survival curves were plotted by using the Kaplan-Meier method,33 and compared using the log-rank test.34
Results
Identification of Novel SNPs in the LTBP-1L Upstream Region
We compared the LTBP-1L promoter sequences in patients with ovarian cancers and in healthy controls with those registered in GenBank (accession no. AF171934) and identified two novel SNPs, −202G/C and +20A/C (Figure , 1Aand 1B). Because −202G generates an EcoRII site and +20C generates a CspI site (Figure 1B), the haplotypes between −202G/C and +20A/C were distinguished by RFLP analysis (Figure 1C). Of four predicted haplotypes, only two types, G-A and C-C were identified in both ovarian cancer patients and healthy controls and therefore three genotypes, G-A/G-A, G-A/C-C, and C-C/C-C, were detected.
Figure 1.
Screening for the LTBP-1L genotypes. A: Schematic representation of the two novel SNPs in the LTBP-1L promoter. The transcriptional start site is indicated as +1. B: Representative results of nucleotide sequence analysis. Arrows indicate the location of SNPs. Restriction enzymes used in PCR-restriction fragment length polymorphism (RFLP) analysis are indicated above the sequences. C: Representative results of PCR-RFLP analysis. Numbers in parentheses are the size of each fragment (bp). Arrow D indicates the heteroduplex complexes produced by mismatched annealing of heterogeneous alleles (see Materials and Methods). M, 100-bp DNA ladder marker.
The −202C and +20C, which were originally registered in GenBank, were rather rare in both ovarian cancer patients and healthy Japanese controls (Table 1). The allele frequency in the LTBP-1L promoter SNPs in ovarian cancer patients was not significantly different from that in healthy controls (Table 1). Thus, we newly identified the G-A allele in the LTBP-1L promoter, and showed that it is frequently detected in the Japanese population.
Table 1.
Genotyping of LTBP-1LPromoter in Ovarian Cancers and Healthy Controls
| Genotyping of LTBP-1Lpromoter
|
Allele frequency
|
||||
|---|---|---|---|---|---|
| G-A/G-A | G-A/C-C | C-C/C-C | G-A | C-C | |
| Ovarian cancer (n = 42) | 15 (35.7%) | 18 (42.9%) | 9 (21.4%) | 0.571 | 0.429 |
| Healthy control (n = 156) | 49 (31.4%) | 84 (53.9%) | 23 (14.7%) | 0.583 | 0.417 |
Hardy-Weinberg equilibrium test indicates that both populations were in equilibrium.
P= 0.391 by χ 2 test
Impact of LTBP-1L SNPs on the Transcriptional Activity of the Promoter
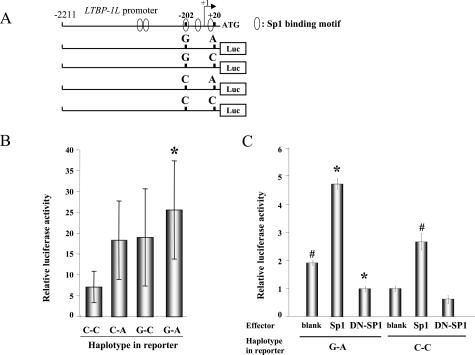
To examine the effect of the SNPs at −202 and +20 on the transcriptional activity of the LTBP-1L promoter, luciferase reporter plasmids containing the LTBP-1L promoter region spanning −2211 to +54 with four haplotypes (G-A, G-C, C-A, C-C) were prepared (Figure 2A) and transfected into ovarian cancer RMUG-S cells, and luciferase assays were performed. Transcriptional activity of the G-A haplotype was the highest, being 3.6-fold higher than that of the C-C haplotype (Figure 2B) (25.7 ± 11.7 versus 7.17 ± 3.77, P < 0.001), whereas the G-C and C-A haplotypes exhibited intermediate activity.
Figure 2.
Luciferase assays with LTBP-1L promoter reporter constructs having the four haplotypes, using ovarian cancer RMUG-S cells. A: Schematic representation of luciferase reporter constructs containing a −2211 to +54 promoter region with different haplotypes at the SNP sites. B: The relative luciferase activities of reporter plasmids with the different haplotypes. Each column represents the mean of three independent experiments, each done in triplicate; bars, ±SD (*P < 0.01 compared with the C-C construct). C: Luciferase assays to examine the role of Sp1 in LTBP-1L promoter. Luciferase plasmids with G-A or C-C haplotype were co-transfected with blank vector, Sp1, or dominant-negative Sp3 expression vector to inhibit Sp1 activity. The relative luciferase activities in each reporter plasmid are shown, based on the luciferase activity in C-C reporter plasmid transfected with blank vector, normalized to 1. Each column represents the mean of three independent experiments, each done in triplicate; bars, ±SD (*P < 0.0001 compared with the G-A blank, #P < 0.0001 compared with the C-C blank).
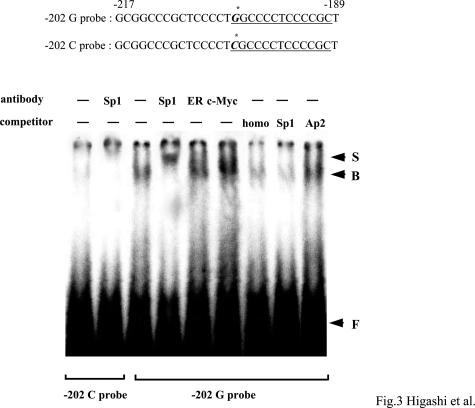
The LTBP-1L promoter contains GC-boxes, which are reminiscent of a Sp1 binding motif. Computer-assisted homology search identified five putative Sp1 motifs in the LTBP-1L promoter (Figure 2A), and two of them overlapped with, or were adjacent to, SNP sites. We therefore tested whether Sp1 regulates the LTBP-1L promoter. Co-transfection of reporter plasmids having the G-A haplotype with Sp1 expression vectors markedly increased the transcriptional activity (Figure 2C). In contrast, co-transfection with a dominant-negative Sp3 expression vector, which is known to inhibit the function of Sp1, significantly abrogated the transcriptional activity. Similar effects of Sp1 were observed with promoter having the C-C haplotype as well, but with lower efficiency. These data suggest a potential role of Sp1 in activating the LTBP-1L promoter, especially in the case of the G-A haplotype. To further clarify the functional impact of these SNPs on the promoter activity, EMSAs were performed. Recombinant Sp1 proteins were incubated with promoter fragments spanning −217 to −189, with G or C at −202, respectively (Figure 3A). No band or only faint bands were formed with probes with −202C, whereas probes with −202G clearly generated binding complexes. To confirm the binding specificity, supershift assays were performed. The binding complexes were supershifted by the addition with Sp1 antibody, but not by unrelated estrogen receptor or c-myc antibody. Furthermore, these bands were eliminated by the addition of homologous competitors, as well as Sp1 consensus oligonucleotides, but not by unrelated Ap2 oligonucleotides. Similar findings were observed using probes containing the SNP site at +20: probes with +20A formed definite binding complexes, whereas those with C did not. However, the binding affinity of Sp1 with the probe containing +20 was weaker than that with the probe containing −202G (data not shown). Taken together, these results clearly demonstrate that the SNPs at −202 and +20 in the LTBP-1L promoter contribute to increased binding affinity of Sp1, probably leading to transcriptional activation of this promoter.
Figure 3.
EMSAs for Sp1 binding to the LTBP-1L promoter. The nucleotide sequences of the probes with SNP at −202 are shown. Underlining indicates the Sp1 binding motif. The asterisk shows the SNP site. Recombinant Sp1 proteins were incubated with 32P-labeled probes with G or C at −202, respectively, and separated by electrophoresis. Antibodies and competitors used are shown. Homo, homologous competitors of −202G probe. Arrow B represents specific binding complexes of Sp1 and arrow S exhibits the supershifted bands. Arrow F shows free probes.
Immunohistochemical Analyses of LTBP-1 Expression in Ovarian Cancers
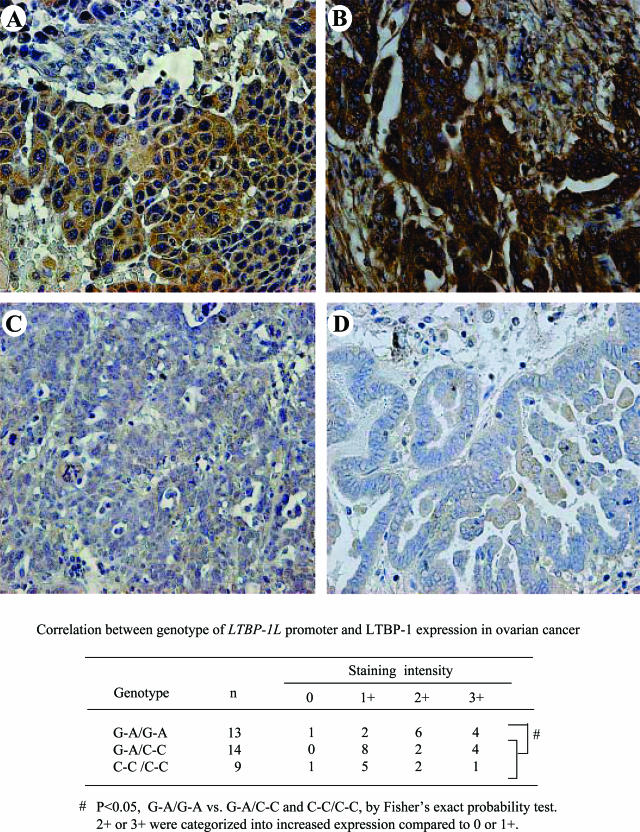
The effect of these two SNPs on the LTBP-1 protein expression level was further examined by immunohistochemical analysis using surgical specimens obtained from 36 patients with different genotypes. Immunostaining of LTBP-1 was detected in both cancer and stromal cells in all tissue samples examined (Figure 4). Of 13 ovarian cancers with G-A homozygous alleles, 4 exhibited high and 6 showed intermediate expression (Figure , 4Aand 4B). Of 14 cancer samples with heterozygous alleles, 4 exhibited high expression and 2 showed intermediate expression, and 8 had low expression (Figure 4C). Of nine cancers with C-C homozygous alleles, one exhibited high expression and two showed intermediate expression, and five had low and one had no expression (Figure 4D). When patients with intermediate or high expression were categorized as having increased expression, G-A homozygous alleles were significantly associated with increased expression of LTBP-1, compared with heterozygous and C-C homozygous alleles (P < 0.05).
Figure 4.
Immunohistochemical staining of LTBP-1 protein in ovarian cancer tissues. Representative results of immunohistochemistry of LTBP-1 are shown in ovarian cancers with different genotypes of LTBP-1L. Tissues from patients with G-A/G-A homozygous (A, B), those with G-A/C-C heterozygous (C) and C-C/C-C homozygous (D) LTBP-1L promoter are shown. Cancer tissues with G-A/G-A genotypes exhibited high levels of LTBP-1 staining. Original magnifications, ×200.
Impact of LTBP-1L SNPs on Clinical Features and Outcomes of Patients with Ovarian Cancer
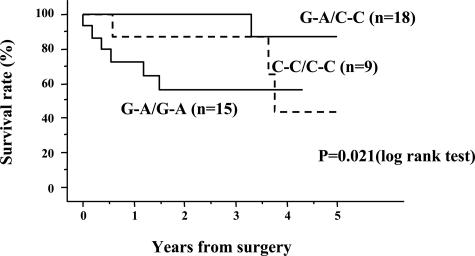
We next examined the relationship between genotype of LTBP-1L and clinicopathological characteristics of ovarian cancers, including age, histological types, tumor grading, and FIGO clinical staging. There was no significant correlation between LTBP-1L genotype and such parameters. Survival analyses were then performed. In univariate analyses, we found that the 5-year overall survival rate was significantly lower in patients with G-A homozygous alleles than in patients with the heterozygous or C-C homozygous alleles (P = 0.021) (Figure 5).
Figure 5.
Survival curves according to LTBP-1L promoter genotype in ovarian cancer patients. Patients with G-A/G-A homozygous genotype had a significantly lower survival rate than those with G-A/C-C heterozygous or C-C/C-C homozygous genotype. The P value was determined using the log-rank test.
Discussion
We identified two novel SNPs, −202G/C and +20A/C, in the LTBP-1L promoter and 5′-UTR regions. The LTBP-1L promoter was initially cloned from a human placental library,24 and −202C and +20C were registered in GenBank. However, we found that G at −202 and A at +20 are commonly observed in the Japanese population. Haplotype analyses between −202G/C and +20A/C revealed that there are only G-A and C-C haplotypes in the LTBP-1 promoter, so −202G is tightly linked to +20A. No difference of allele frequency was observed between male and female healthy controls (data not shown). At present, we do not know the ethnic differences in allele frequencies or haplotypes of these SNPs, and this would be a worthwhile direction for future investigation. There was no difference in the allele frequency between patients and healthy controls. These findings indicate that the SNPs are not involved in susceptibility to ovarian cancer.
It has been reported that SNPs in promoter regions can modulate gene transcription by affecting DNA binding of trans-acting elements.35,36,37,38,39 Luciferase reporter assays revealed that the G-A haplotype caused transcriptional activation more efficiently than other haplotypes. The LTBP-1L promoter has a high GC content, but lacks a TATA box. Such promoters are often activated by Sp family proteins.40,41 In fact, there are five putative Sp1 motifs in the LTBP-1L promoter, and interestingly, one overlaps with the SNP site at −202, while another is adjacent to the SNP site at +20. Luciferase reporter assays revealed that overexpression of Sp1 significantly activated the LTBP-1L promoter, with higher efficiency in G-A haplotype than C-C haplotype. EMSAs showed increased binding affinity of Sp1 to the promoter with −202G and +20A. These findings suggest that a change in Sp1 binding affinity by SNPs leads to increased transcriptional activity. It is well known that Sp1 is constitutively overexpressed in a variety of human cancers.42,43,44,45 In ovarian cancers, such overexpression may facilitate LTBP-1 expression in patients with G-A alleles, as was observed in the present immunohistochemical analyses. Tightly linked SNPs within or adjacent to Sp1 binding sites are also known in the promoters of other cancer-related genes, such as EGFR, MMP-2, and vascular endothelial growth factor.37,38,41 Most of them affect the transcriptional activity of the promoters by interacting with Sp1. The SNPs we identified in the LTBP-1 promoter may be categorized as such functional SNPs. Thus, the Sp1 site may be a hot spot of functional SNPs in GC-rich promoters, and it might be interesting to screen these regions in a variety of cancer-related genes. We conclude that the newly identified SNPs in the LTBP-1L promoter may play roles in up-regulation of LTBP-1L.
One interesting finding in the present study is that ovarian cancer patients with G-A/G-A alleles had significantly poorer survival although analysis using a larger number of samples is required to obtain a definite conclusion. How can we explain such a correlation? TGF-β can act both as a tumor suppressor and as a stimulator of tumor progression. At an early stage of carcinogenesis, when growth-inhibitory responses to TGF-β are maintained in cells, TGF-β directly suppresses tumor growth.46,47 However, after cancer cells become resistant to TGF-β growth inhibition at the later stages of carcinogenesis, TGF-β contributes to tumor invasion and metastasis by stimulating tumor cell motility, angiogenesis, remodeling of the extracellular matrix protein by stromal cells, and even by suppressing the immune system.48,49,50,51,52,53 Because LTBP-1 may be an important molecule for activation of TGF-β1,18,19,20,21,22 one possible explanation of the poor prognosis of patients with G-A/G-A alleles may be that increased expression of LTBP-1L enhances TGF-β1 activity and facilitates tumor progression. We are currently investigating the possible correlation between elevated levels of TGF-β1 activity and LTBP-1L expression both in vitro and in vivo to test this hypothesis.
In summary, we have identified novel SNPs in the LTBP-1L promoter, and shown that they contribute to transcriptional activation, leading to up-regulation of LTBP-1L expression and poorer survival in ovarian cancer patients. Further studies using a larger number of samples will definitively determine the correlation between LTBP-1 haplotype and clinical behavior of ovarian cancer. Analysis of the roles and mechanisms of LTBP-1L expression in ovarian cancer may improve our understanding of the molecular mechanisms of ovarian carcinogenesis, as well as assisting in the development of novel molecular-based therapies.
Acknowledgments
We thank Dr. Jorma Keski-Oja and Dr. Yoshihiro Sowa for generously providing LTBP-1 promoter construct and Sp1 expression vector, respectively; Dr. Yasuhito Ishigaki of Kanazawa Medical University and Dr. Miki Nakajima of Kanazawa University for helpful advice; the staff and students in Kanazawa University for cooperation as healthy volunteers; and all of the members of Departments of Hygiene and Obstetrics and Gynecology.
Footnotes
Supported by the Ministry of Education, Science, Sports, and Culture of Japan (grant-in-aid 15790283); and the Honjin Foundation.
References
- Masui T, Wakefield LM, Lechner JF, LaVeck MA, Sporn MB, Harris CC. Type beta transforming growth factor is the primary differentiation-inducing serum factor for normal human bronchial epithelial cells. Proc Natl Acad Sci USA. 1986;83:2438–2442. doi: 10.1073/pnas.83.8.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipley GD, Pittelkow MR, Wille JJ, Jr, Scott RE, Moses HL. Reversible inhibition of normal human prokeratinocyte proliferation by type beta transforming growth factor-growth inhibitor in serum-free medium. Cancer Res. 1986;46:2068–2071. [PubMed] [Google Scholar]
- Taipale J, Saharinen J, Keski-Oja J. Extracellular matrix-associated transforming growth factor-beta: role in cancer cell growth and invasion. Adv Cancer Res. 1998;75:87–134. doi: 10.1016/s0065-230x(08)60740-x. [DOI] [PubMed] [Google Scholar]
- Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- Roberts AB, Wakefield LM. The two faces of transforming growth factor beta in carcinogenesis. Proc Natl Acad Sci USA. 2003;100:8621–8623. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Hellman U, Wernstedt C, Heldin CH. Latent high molecular weight complex of transforming growth factor beta 1. Purification from human platelets and structural characterization. J Biol Chem. 1988;263:6407–6415. [PubMed] [Google Scholar]
- Olofsson A, Miyazono K, Kanzaki T, Colosetti P, Engstrom U, Heldin CH. Transforming growth factor-beta 1, -beta 2, and -beta 3 secreted by a human glioblastoma cell line. Identification of small and different forms of large latent complexes. J Biol Chem. 1992;267:19482–19488. [PubMed] [Google Scholar]
- Dallas SL, Park-Snyder S, Miyazono K, Twardzik D, Mundy GR, Bonewald LF. Characterization and autoregulation of latent transforming growth factor beta (TGF beta) complexes in osteoblast-like cell lines. Production of a latent complex lacking the latent TGF beta-binding protein. J Biol Chem. 1994;269:6815–6821. [PubMed] [Google Scholar]
- Taipale J, Miyazono K, Heldin CH, Keski-Oja J. Latent transforming growth factor-beta 1 associates to fibroblast extracellular matrix via latent TGF-beta binding protein. J Cell Biol. 1994;124:171–181. doi: 10.1083/jcb.124.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saharinen J, Taipale J, Keski-Oja J. Association of the small latent transforming growth factor-beta with an eight cysteine repeat of its binding protein LTBP-1. EMBO J. 1996;15:245–253. [PMC free article] [PubMed] [Google Scholar]
- Gleizes PE, Beavis RC, Mazzieri R, Shen B, Rifkin DB. Identification and characterization of an eight-cysteine repeat of the latent transforming growth factor-beta binding protein-1 that mediates bonding to the latent transforming growth factor-beta1. J Biol Chem. 1996;271:29891–29896. doi: 10.1074/jbc.271.47.29891. [DOI] [PubMed] [Google Scholar]
- Saharinen J, Keski-Oja J. Specific sequence motif of 8-Cys repeats of TGF-beta binding proteins, LTBPs, creates a hydrophobic interaction surface for binding of small latent TGF-beta. Mol Biol Cell. 2000;11:2691–2704. doi: 10.1091/mbc.11.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki T, Olofsson A, Moren A, Wernstedt C, Hellman U, Miyazono K, Claesson-Welsh L, Heldin CH. TGF-beta 1 binding protein: a component of the large latent complex of TGF-beta 1 with multiple repeat sequences. Cell. 1990;61:1051–1061. doi: 10.1016/0092-8674(90)90069-q. [DOI] [PubMed] [Google Scholar]
- Moren A, Olofsson A, Stenman G, Sahlin P, Kanzaki T, Claesson-Welsh L, ten Dijke P, Miyazono K, Heldin CH. Identification and characterization of LTBP-2, a novel latent transforming growth factor-beta-binding protein. J Biol Chem. 1994;269:32469–32478. [PubMed] [Google Scholar]
- Yin W, Smiley E, Germiller J, Mecham RP, Florer JB, Wenstrup RJ, Bonadio J. Isolation of a novel latent transforming growth factor-beta binding protein gene (LTBP-3). J Biol Chem. 1995;270:10147–10160. doi: 10.1074/jbc.270.17.10147. [DOI] [PubMed] [Google Scholar]
- Giltay R, Kostka G, Timpl R. Sequence and expression of a novel member (LTBP-4) of the family of latent transforming growth factor-beta binding proteins. FEBS Lett. 1997;411:164–168. doi: 10.1016/s0014-5793(97)00685-6. [DOI] [PubMed] [Google Scholar]
- Saharinen J, Taipale J, Monni O, Keski-Oja J. Identification and characterization of a new latent transforming growth factor-beta-binding protein, LTBP-4. J Biol Chem. 1998;273:18459–18469. doi: 10.1074/jbc.273.29.18459. [DOI] [PubMed] [Google Scholar]
- Miyazono K, Olofsson A, Colosetti P, Heldin CH. A role of the latent TGF-beta 1-binding protein in the assembly and secretion of TGF-beta 1. EMBO J. 1991;10:1091–1101. doi: 10.1002/j.1460-2075.1991.tb08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Thyberg J, Heldin CH. Retention of the transforming growth factor-beta 1 precursor in the Golgi complex in a latent endoglycosidase H-sensitive form. J Biol Chem. 1992;267:5668–5675. [PubMed] [Google Scholar]
- Flaumenhaft R, Abe M, Sato Y, Miyazono K, Harpel J, Heldin CH, Rifkin DB. Role of the latent TGF-beta binding protein in the activation of latent TGF-beta by co-cultures of endothelial and smooth muscle cells. J Cell Biol. 1993;120:995–1002. doi: 10.1083/jcb.120.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes I, Gleizes PE, Metz CN, Rifkin DB. Latent transforming growth factor-beta binding protein domains involved in activation and transglutaminase-dependent cross-linking of latent transforming growth factor-beta. J Cell Biol. 1997;136:1151–1163. doi: 10.1083/jcb.136.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annes JP, Chen Y, Munger JS, Rifkin DB. Integrin alphaVbeta6-mediated activation of latent TGF-beta requires the latent TGF-beta binding protein-1. J Cell Biol. 2004;165:723–734. doi: 10.1083/jcb.200312172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoi T, Ohtani H, Miyazono K, Miyazawa M, Matsuno S, Nagura H. Immunoelectron microscopic localization of transforming growth factor beta 1 and latent transforming growth factor beta 1 binding protein in human gastrointestinal carcinomas: qualitative difference between cancer cells and stromal cells. Cancer Res. 1993;53:183–190. [PubMed] [Google Scholar]
- Koski C, Saharinen J, Keski-Oja J. Independent promoters regulate the expression of two amino terminally distinct forms of latent transforming growth factor-beta binding protein-1 (LTBP-1) in a cell type-specific manner. J Biol Chem. 1999;274:32619–32630. doi: 10.1074/jbc.274.46.32619. [DOI] [PubMed] [Google Scholar]
- Olofsson A, Ichijo H, Moren A, ten Dijke P, Miyazono K, Heldin CH. Efficient association of an amino-terminally extended form of human latent transforming growth factor-beta binding protein with the extracellular matrix. J Biol Chem. 1995;270:31294–31297. doi: 10.1074/jbc.270.52.31294. [DOI] [PubMed] [Google Scholar]
- Higashi T, Sasagawa T, Inoue M, Oka R, Shuangying L, Saijoh K. Overexpression of latent transforming growth factor-beta 1 (TGF-beta 1) binding protein 1 (LTBP-1) in association with TGF-beta 1 in ovarian carcinoma. Jpn J Cancer Res. 2001;92:506–515. doi: 10.1111/j.1349-7006.2001.tb01123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lench N, Stanier P, Williamson R. Simple non-invasive method to obtain DNA for gene analysis. Lancet. 1988;1:1356–1358. doi: 10.1016/s0140-6736(88)92178-2. [DOI] [PubMed] [Google Scholar]
- Lum A, Le Marchand L. A simple mouthwash method for obtaining genomic DNA in molecular epidemiological studies. Cancer Epidemiol Biomarkers Prev. 1998;7:719–724. [PubMed] [Google Scholar]
- Sowa Y, Orita T, Minamikawa-Hiranabe S, Mizuno T, Nomura H, Sakai T. Sp3, but not Sp1, mediates the transcriptional activation of the p21/WAF1/Cip1 gene promoter by histone deacetylase inhibitor. Cancer Res. 1999;59:4266–4270. [PubMed] [Google Scholar]
- Takakura M, Kyo S, Sowa Y, Wang Z, Yatabe N, Maida Y, Tanaka M, Inoue M. Telomerase activation by histone deacetylase inhibitor in normal cells. Nucleic Acids Res. 2001;29:3006–3011. doi: 10.1093/nar/29.14.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takakura M, Kyo S, Kanaya T, Hirano H, Takeda J, Yutsudo M, Inoue M. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 1999;59:551–557. [PubMed] [Google Scholar]
- Kyo S, Masutomi K, Maida Y, Kanaya T, Yatabe N, Nakamura M, Tanaka M, Takarada M, Sugawara I, Murakami S, Taira T, Inoue M. Significance of immunological detection of human telomerase reverse transcriptase: re-evaluation of expression and localization of human telomerase reverse transcriptase. Am J Pathol. 2003;163:859–867. doi: 10.1016/S0002-9440(10)63446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;54:457–481. [Google Scholar]
- Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50:163–170. [PubMed] [Google Scholar]
- Baseggio L, Bartholin L, Chantome A, Charlot C, Rimokh R, Salles G. Allele-specific binding to the −308 single nucleotide polymorphism site in the tumour necrosis factor-promoter. Eur J Immunogenet. 2004;31:15–19. doi: 10.1111/j.1365-2370.2004.00440.x. [DOI] [PubMed] [Google Scholar]
- Gazzoli I, Kolodner RD. Regulation of the human MSH6 gene by the Sp1 transcription factor and alteration of promoter activity and expression by polymorphisms. Mol Cell Biol. 2003;23:7992–8007. doi: 10.1128/MCB.23.22.7992-8007.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harendza S, Lovett DH, Panzer U, Lukacs Z, Kuhnl P, Stahl RA. Linked common polymorphisms in the gelatinase A promoter are associated with diminished transcriptional response to estrogen and genetic fitness. J Biol Chem. 2003;278:20490–20499. doi: 10.1074/jbc.M211536200. [DOI] [PubMed] [Google Scholar]
- Stevens A, Soden J, Brenchley PE, Ralph S, Ray DW. Haplotype analysis of the polymorphic human vascular endothelial growth factor gene promoter. Cancer Res. 2003;63:812–816. [PubMed] [Google Scholar]
- Mann V, Hobson EE, Li B, Stewart TL, Grant SF, Robins SP, Aspden RM, Ralston SH. A COL1A1 Sp1 binding site polymorphism predisposes to osteoporotic fracture by affecting bone density and quality. J Clin Invest. 2001;107:899–907. doi: 10.1172/JCI10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Chang LS. The human POLD1 gene. Identification of an upstream activator sequence, activation by Sp1 and Sp3, and cell cycle regulation. J Biol Chem. 1997;272:4869–4882. [PubMed] [Google Scholar]
- Liu W, Innocenti F, Wu MH, Desai AA, Dolan ME, Cook EH, Jr, Ratain MJ. A functional common polymorphism in a Sp1 recognition site of the epidermal growth factor receptor gene promoter. Cancer Res. 2005;65:46–53. [PubMed] [Google Scholar]
- Shi Q, Le X, Abbruzzese JL, Peng Z, Qian CN, Tang H, Xiong Q, Wang B, Li XC, Xie K. Constitutive Sp1 activity is essential for diff-erential constitutive expression of vascular endothelial growth factor in human pancreatic adenocarcinoma. Cancer Res. 2001;61:4143–4154. [PubMed] [Google Scholar]
- Zannetti A, Del Vecchio S, Carriero MV, Fonti R, Franco P, Botti G, D’Aiuto G, Stoppelli MP, Salvatore M. Coordinate up-regulation of Sp1 DNA-binding activity and urokinase receptor expression in breast carcinoma. Cancer Res. 2000;60:1546–1551. [PubMed] [Google Scholar]
- Kitadai Y, Yasui W, Yokozaki H, Kuniyasu H, Haruma K, Kajiyama G, Tahara E. The level of a transcription factor Sp1 is correlated with the expression of EGF receptor in human gastric carcinomas. Biochem Biophys Res Commun. 1992;189:1342–1348. doi: 10.1016/0006-291x(92)90221-6. [DOI] [PubMed] [Google Scholar]
- Chiefari E, Brunetti A, Arturi F, Bidart JM, Russo D, Schlumberger M, Filetti S. Increased expression of AP2 and Sp1 transcription factors in human thyroid tumors: a role in NIS expression regulation? BMC Cancer. 2002;2:35–38. doi: 10.1186/1471-2407-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick AB, Kulkarni AB, Tennenbaum T, Hennings H, Flanders KC, O’Reilly M, Sporn MB, Karlsson S, Yuspa SH. Loss of expression of transforming growth factor beta in skin and skin tumors is associated with hyperproliferation and a high risk for malignant conversion. Proc Natl Acad Sci USA. 1993;90:6076–6080. doi: 10.1073/pnas.90.13.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engle SJ, Hoying JB, Boivin GP, Ormsby I, Gartside PS, Doetschman T. Transforming growth factor beta1 suppresses nonmetastatic colon cancer at an early stage of tumorigenesis. Cancer Res. 1999;59:3379–3386. [PubMed] [Google Scholar]
- Tang B, Vu M, Booker T, Santner SJ, Miller FR, Anver MR, Wakefield LM. TGF-beta switches from tumor suppressor to prometastatic factor in a model of breast cancer progression. J Clin Invest. 2003;112:1116–1124. doi: 10.1172/JCI18899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel PM, Shu W, Cardiff RD, Muller WJ, Massague J. Transforming growth factor beta signaling impairs Neu-induced mammary tumorigenesis while promoting pulmonary metastasis. Proc Natl Acad Sci USA. 2003;100:8430–8435. doi: 10.1073/pnas.0932636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka RS, Koh Y, Roebuck LR, Sanders ME, Brantley-Sieders D, Gorska AE, Moses HL, Arteaga CL. Increased malignancy of Neu-induced mammary tumors overexpressing active transforming growth factor beta1. Mol Cell Biol. 2003;23:8691–8703. doi: 10.1128/MCB.23.23.8691-8703.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Fowlis DJ, Bryson S, Duffie E, Ireland H, Balmain A, Akhurst RJ. TGFbeta1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell. 1996;86:531–542. doi: 10.1016/s0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- Torre-Amione G, Beauchamp RD, Koeppen H, Park BH, Schreiber H, Moses HL, Rowley DA. A highly immunogenic tumor transfected with a murine transforming growth factor type beta 1 cDNA escapes immune surveillance. Proc Natl Acad Sci USA. 1990;87:1486–1490. doi: 10.1073/pnas.87.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka RS, Dumont N, Ritter CA, Dugger TC, Brantley DM, Chen J, Easterly E, Roebuck LR, Ryan S, Gotwals PJ, Koteliansky V, Arteaga CL. Blockade of TGF-beta inhibits mammary tumor cell viability, migration, and metastases. J Clin Invest. 2002;109:1551–1559. doi: 10.1172/JCI15234. [DOI] [PMC free article] [PubMed] [Google Scholar]