Abstract
It has been demonstrated that lung cancers, specifically a subset of pulmonary adenocarcinomas, with epidermal growth factor receptor (EGFR) mutation are highly sensitive to EGFR-targeted drugs. Therefore, a rapid, sensitive assay for mutation detection using routine pathological specimens is demanded in clinical practice to predict the response. We therefore developed a new assay for detecting EGFR mutation using only a paraffin section of a small biopsy specimen. The method was very sensitive, detecting as few as 5% cancer cells in a background of normal cells, the results usually being obtained within 4 hours. Furthermore, it was accurate, as shown by the high concordance with reverse transcriptase-polymerase chain reaction-coupled direct sequencing (186 of 195, 95%). The practical application of this assay to 29 cases treated with gefitinib resulted in a high prediction rate: 10 of the 11 responders were shown to be positive for the mutation, and all patients with progressive disease were negative. In addition, a mutation at codon 790, conferring gefitinib resistance, was successfully analyzed in a similar manner. In conclusion, the assay is a rapid, sensitive method using paraffin sections of biopsy specimens without a tumor cell-enrichment procedure and is quite useful to select a treatment of choice in clinical practice.
During the last decade, small molecules that inhibit receptor protein kinase activity have been developed.1 Gefitinib is one such drug that targets epidermal growth factor receptor (EGFR) kinase. The EGFR, also known as HER1 or ErbB, is a 170-kd receptor tyrosine kinase (TK) that dimerizes and phosphorylates several tyrosine residues on the binding of several specific ligands.2,3 These phosphorylated tyrosines serve as binding sites for several signal transducers that initiate multiple signaling pathways, resulting in cell proliferation, migration and metastasis, evasion of apoptosis, or angiogenesis, through Ras-Raf-MEK-ERK, phosphatidylinositol-3 kinase-AKT, and PAK-JNKK-JNK pathways. EGFR is expressed in more than 80% of non-small-cell lung cancers (NSCLCs), in addition to a wide range of epithelial cancers. However, clinical trials have shown significant variability in response to gefitinib: 10 to 20% of patients respond to gefitinib treatment, and in some patients, the response is dramatic, whereas the remaining patients show no response. Although further analysis has revealed some prevalence in responders, no definite determinant of the response has been established.
Recently, it has been reported that EGFR somatic mutation can be identified in a subset of pulmonary adenocarcinomas and that tumors with EGFR mutations are highly sensitive to gefitinib.4,5 This correlation has subsequently been confirmed by our group and others,6,7,8,9 and thus the development of a rapid and sensitive assay to predict gefitinib response by means of the presence or absence of the mutation is demanded clinically. Paraffin sections are a convenient source for such an assay in practice, but most studies using immunohistochemistry failed to predict the response.10,11,12
In this study, we introduce a practical approach using a rapid screening assay of EGFR mutation to predict gefitinib response. This method uses only a single paraffin section of a small biopsy specimen and does not require a tumor cell-enrichment procedure. The result is usually obtained within 4 hours and can be applied to a large number of samples.
Materials and Methods
Patients and Tissues
A series of 195 NSCLCs, in which the mutational status of the EGFR-TK domain with both reverse transcriptase-polymerase chain reaction (RT-PCR)-coupled direct sequencing and the new assay presented here was accessible, was used for this study. Some of the mutational results by RT-PCR-coupled direct sequencing have been reported previously.13 DNA for the new assay was prepared from a section of tissue microarray blotted with 0.6-mm tissue cores of the 195 cases. To examine a correlation with the clinical response evaluated according to the guidelines of Response Evaluation Criteria in Solid Tumors (RECIST), a paraffin section of each biopsy specimen was examined for EGFR mutation in 29 patients treated with gefitinib because of the failure of first or second line therapy. To analyze the codon 790 mutation, which has been reported in association with acquired resistance to gefitinib treatment, four tissues were examined. One, reported as a rare case, was shown to have T790M, independent of gefitinib treatment.13,14 The other three presented with a recurrent tumor after gefitinib treatment, and the recurrent tumor and corresponding initial tumor tissue were examined. Appropriate approval was obtained from the institutional review committee in addition to written informed consent from the patients.
Mutation Assay by RT-PCR-Coupled Direct Sequencing
Frozen tissue from the tumor specimens was grossly dissected to pass as many tumor cells as possible into the extraction solution (at least 25% of tumor cell content), followed by the extraction of total RNA with an RNeasy kit (Qiagen, Valencia, CA). For RT-PCR-coupled direct sequencing, the EGFR tyrosine kinase domain (exon 18 to 24) was amplified, and then the products were directly sequenced with an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Foster City, CA). The primer set used was described previously.13
DNA Extraction from Paraffin-Embedded Tissues
Tumor cell-rich area in a hematoxylin and eosin-stained section was marked under a microscope, and tissues were scratched from the area of another deparaffinized unstained section. Pieces of the scratched tissue were incubated with 1× PCR buffer containing 100 μg/ml proteinase K for 1 hour at 54°C. After heat inactivation with 95°C for 3 minutes, the solution was directly used for template DNA for the assay.
EGFR Mutation Detection
To detect the point mutations at codons 858 and 790 of the EGFR gene, we used the cycleave PCR technique. This technique is based on a chimeric DNA-RNA-DNA probe labeled with a fluorescent dye and quencher at each end. The RNA sequence of the probes corresponds to that of the wild type and point mutation labeled with FMA and ROX, respectively. When mutant molecules are present in the sample and PCR-amplified DNA generates a complete hybrid with the RNA portion of the mutant probe, RNase-H digests the probe at the RNA-DNA heteroduplex into two pieces, leading to a significant increase in fluorescence intensity by separation of the fluorescent dye from the quencher. The intensity of the wild-type probe served as an internal control for the assay. This assay was performed using a cycleave PCR core kit (TAKARA, Co., Ltd., Ohtsu, Japan), and sequences of the primer set and the probes were as follows: PCR forward primer for L858R, 5′-AGGAACGTACTGGTGAAAAC-3′; PCR reverse primer for L858R, 5′-TCCCTGGTGTCAGGAAAATG-3′; wild-type probe for L858R, 5′ FAM-CCA U CCCAAAAT-Eclipse 3′; probe for L858R mutation, 5′ FAM-CCCGCCCAAAAT-Eclipse 3′; PCR forward primer for T790M, 5′-ATCTGCCTCACCTCCAC-3′; PCR reverse primer for T790M, 5′-CAATATTGTCTTTGTGTTC-3′; wild-type probe for T790M, 5′ FAM-TGCGTGATGAG-Eclipse 3′; probe for T790M mutation, 5′ FAM-TGCATGATGAG-Eclipse 3′ (italics represent RNA). Fluorescent signals were quantified with a Smart Cycler system (SC-100; Cepheid, Sunnyvale, CA).
To detect the deletion in exon 19 of the EGFR gene, common fragment analysis was used. Sample DNA was amplified with an FAM-labeled primer set as follows: forward, 5′ FAM-TCACAATTGCCAGTTAACGTCT-3′, and reverse, 5′-CAGCAAAGCAGAAACTCACATC-3′. PCR products were electrophoresed on an ABI PRISM 310. When a deletion mutation was present, PCR amplified the shorter segment of DNA, creating a new peak in an electropherogram.
Sensitivity Assay
In the preliminary examination, we prepared a mutation-positive control DNA, which contained exactly one-half each of wild-type and mutant molecules. According to the mixture ratio, the mutation-positive control DNA was mixed up with normal DNA, the concentration of which was equal to that of the mutation-positive control DNA. Therefore, 5% of tumor cells corresponded to 2.5% of mutant molecules in background of wild-type molecule. Using these mixtures of DNA, we examined the sensitivity of the assays (deletion of exon 19 and point mutation of L858R and T790M).
Statistical Analysis
The χ2 test and Fisher’s exact test for independence compared incidences of EGFR mutation, using SYSTAT software (SYSTAT Software Inc., Richmond, CA). A P value below 0.05 was considered statistically significant.
Results
Sensitivity of the New Assay
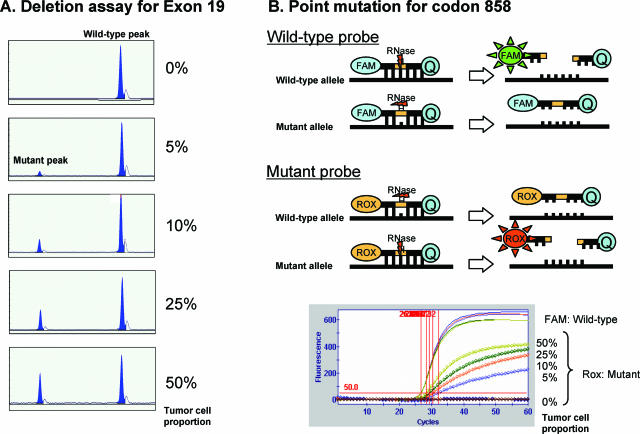
It is known that mutations in the EGFR tyrosine kinase domain are restricted to four exons, and the results of previous reports4,5,8,9,13,15 revealed that the deletion in exon 19 and the point mutation of codon 858 in exon 21 covers about 90% of cases with EGFR-TK mutation. We therefore established assays using fragment analysis for the deletion and cycleave real-time PCR for the point mutation of codon 858. The positive detection of mutated molecules makes this assay very sensitive, as shown in Figure 1. As few as ∼5% of tumor cells could be detected in this assay.
Figure 1.
Sensitivity of the new assay. A: The sensitivity of the fragment analysis in the new assay. As few as 5% of tumor cells with the deletion could be detected. In the top of B, a brief explanation of the cycleave technology is displayed. Using this technique, as few as 5% of tumor cells with point mutation at codon 858 could be detected (bottom of B).
Specificity of the New Assay and Concordance with Direct Sequencing
We evaluated the concordance of results between the new assay and conventional direct sequencing using 195 NSCLCs. The results are summarized in Table 1. Overall concordance was 186 of 195 (95%). When we excluded the seven evaluation cases, which were mutated in regions other than the targets of this assay, 99% of cases were concordant. In one case, mutation was only detected with the new assay, whereas one case was negative for mutation with the new assay but positive with direct sequencing. This disagreement resulted from the different tumor cell population in the samples examined. In the preliminary examination, at least 25% of tumor cells were required for detection of the gene mutation by direct sequencing (data not shown). Although tumor tissues in this analysis were dissected to contain more than 25% of tumor cells from most frozen sections, this case contained around 25% tumor cells, on the threshold of that detectable by the sequencing approach. In contrast, the paraffin section used for the new assay was rich in tumor cells. This difference in tumor cell content between frozen and paraffin sections may be the cause of the discrepancy. We confirmed this result by direct sequencing of the frozen section, using DNA microdissected with a laser capture microdissection system.
Table 1.
Comparison of Results between the Conventional and New Assays
| New assay | |||
|---|---|---|---|
| Direct sequencing | Wild type | Mutation at codon 858 | Deletion at exon 19 |
| Wild type | 116 | 1 | 0 |
| Point mutation at codon 858 | 0 | 32 | 0 |
| Deletion at exon 19 | 1 | 0 | 38 |
| Point mutation at codon 719 | 3 | 0 | 0 |
| Insertion at exon 20 | 3 | 0 | 0 |
| Point mutation at codon 742 | 1 | 0 | 0 |
Practical Application for the Prediction of Gefitinib Response
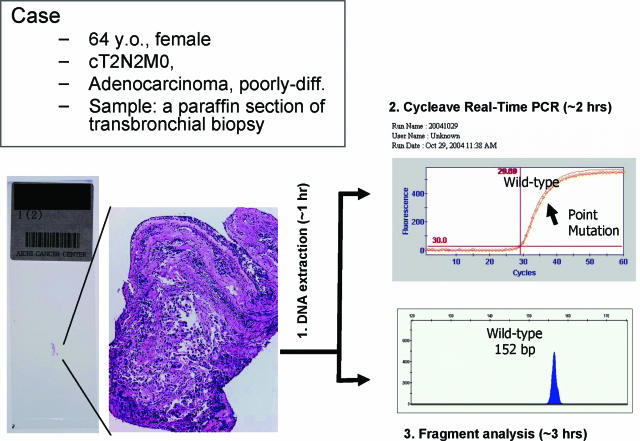
To confirm whether the new assay is useful for the prediction of gefitinib response in clinical practice, we applied the assay to 29 gefitinib-treated cases whose response had been evaluated according to RECIST. A paraffin section of the large tumor tissue, which had been surgically resected a few years before relapse, was used in seven cases, whereas DNA was extracted from a paraffin section of transbronchial biopsy or computer tomography-assisted fine needle biopsy in 20 cases (Figure 2). Partial response was achieved in 11 cases; all but one were positive for the mutation, whereas five cases with progressive disease were negative with this assay (Table 2). EGFR mutation was detected in only 2 of 13 cases evaluated as stable disease. The correlation between EGFR mutation and gefitinib response was highly significant (P = 0.0001).
Figure 2.
A representative result of the new assay. DNA was extracted from a paraffin section of the biopsy followed by simultaneous analysis using cycleave real-time PCR and fragment analysis. The entire procedure was completed within 4 hours. In this case, point mutation at codon 858 was detected, and the patient responded to gefitinib therapy.
Table 2.
Practical Application of the New Assay
| Clinical response | |||
|---|---|---|---|
| EGFR status | PD | NC | PR |
| Wild type | 5 | 11 | 1 |
| Mutated | 0 | 2 | 10 |
| Deletion in exon 19 | 0 | 2 | 5 |
| Point mutation of codon 858 | 0 | 0 | 5 |
PD, progressive disease; NC, no change; PR, partial response.
All of the 12 EGFR-mutated specimens were also examined by direct sequencing. In seven cases, identical results were obtained with both methods, whereas background noise prevented us from evaluating the results in the other five cases, all of which were small biopsy specimens. This may not indicate a lack of confirmation but rather suggests the superiority of this new assay, considering the good correlation of this result with clinical response and with the results obtained with direct sequencing using sufficient amounts of surgical tissue.
Detection of Mutation at Codon 790 Conferring Acquired Resistance to Gefitinib
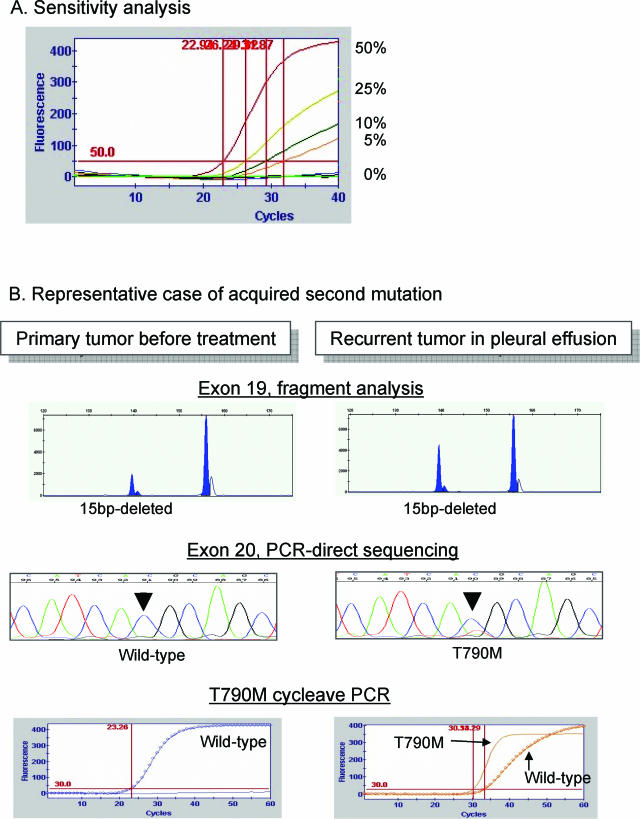
Recently, it has been reported that a second mutation, at codon 790, was associated with acquired resistance to gefitinib.16,17 On very rare occasions, the mutation was also detected independently of gefitinib treatment.13,14 An assay for this mutation, using cycleave PCR, was similarly established (Table 3). In this assay, as few as 5% of tumor cells could be detected, as shown in Figure 3. A rare case, whose tumor was known to have T790M mutation independently of gefitinib treatment,16,17 was also positive in this assay. In one of the other three recurrent tumors, this assay clearly demonstrated the mutation (Figure 3), although it was often difficult to detect the mutated signal with direct sequencing of the PCR product.
Table 3.
Detection of T790M Mutation Associated with Acquired Resistance to Gefitinib
| Patient | Gefitinib treatment | Tissue examined | T790M mutation | Comments |
|---|---|---|---|---|
| Case 1* | No | Primary tumor | Yes | A rare case, harboring T790M mutation independent of gefitinib treatment |
| Case 2 | Yes | Pleural effusion | Yes | 15-bp deletion of exon 19 in the primary and recurrent cancers (Figure 3) |
| Case 3 | Yes | Pleural effusion | No | 9-bp deletion of exon 19 in the primary and recurrent cancers |
| Case 4 | Yes | Pleural effusion | No | 15-bp deletion of exon 19 in the primary and recurrent cancers |
The mutation of codon 790 in a primary cancer, which was demonstrated with RT-PCR direct sequencing, has been reported previously.
Figure 3.
Detection of acquired mutation at codon 790. A: The sensitivity of this cycleave assay for T790M mutation. As few as 5% of tumor cells with T790M mutation could be detected. A representative result of acquired mutation at codon 790 after gefitinib treatment is displayed in B (Table 3, case 2). In contrast to the 15-bp deletion in exon 19 of the EGFR gene in both primary and recurrent tumors, T790M was detected only in the recurrent tumor, suggesting acquired mutation after gefitinib treatment. The result of the cycleave method was more obvious than that with direct sequencing.
Discussion
Paez et al5 and Lynch et al4 simultaneously published the result that somatic mutation of EGFR in lung adenocarcinoma predicts a clinical response to gefitinib. Erlotinib is another targeted small-molecule inhibitor of EGFR, and lung adenocarcinoma sensitive to erlotinib also harbored EGFR mutations. In addition, in vitro studies support the observation that EGFR mutations make tumor cells significantly sensitive to gefitinib18 and erlotinib. This increased sensitivity may be explained by the “addiction to oncogene” hypothesis proposed by Weinstein.19 Tumor cells with EGFR mutation are highly dependent on the activated EGFR pathway and are thus very susceptible to inhibition of this dependence. We have reported that patients with EGFR mutations survived longer than those without mutations after the initiation of gefitinib treatment.7 Recently, failure to show a survival benefit in the IRESSA Survival Evaluation in Lung Cancer was announced. Gefitinib may not be effective enough to kill tumor cells that are not under a state of “addiction to EGFR mutation.” Conversely, these findings suggest that selection of patients with EGFR-mutated tumors has the advantage of increasing the response rate of EGFR-targeted therapy. Furthermore, selection may also be efficient at preventing serious interstitial pneumonia occurring as a side effect.20
Although an assay using paraffin sections is very practical, immunohistochemical analysis of the tumors failed to predict the response. Currently, the microdissection of tumor cells and direct sequencing of PCR products is commonly used as a standard method. Regarding practical applications, the new assay reported here provides two benefits compared with the conventional method. First, microdissection is not necessary for the assay because a positive mutated signal makes this assay very sensitive. Second, this assay is rapid, does not require a purification step, and is usually completed within 4 hours: digestion with proteinase K for 1 hour, real-time PCR or regular PCR for 3 hours, and electrophoresis for 1 hour. In addition to paraffin sections, pleural effusion and specimens for fine needle aspiration cytology can be used. All three specimens of pleural effusion for the T790M cycleave assay were successfully analyzed, whereas direct sequencing occasionally resulted in an ambiguous result (Figure 3). The main targets for gefitinib or erlotinib therapy are recurrent and refractory tumors, and an assay using such specimens is therefore quite useful. However, the examination of limited regions of the EGFR gene appears to be a disadvantage of this study. Recent studies suggested that an insertion of exon 20 was shown to be resistant to EGFR inhibitors25, whereas the gefitinib sensitivity of cells expressing the G719S mutant was significantly less than that of cells expressing the L858R mutant form26. Therefore, these results suggest that examination of the L858R mutation and deletion in exon 19 is reasonable, because these two mutations are likely to be a major target of the EGFR inhibitors.
A few approaches for the detection of EGFR mutation have been reported recently.21,22,23 Comparing the assays, the advantage of the method presented here is its practical clinical use. Biopsy specimens frequently result in small, fragmented tissues containing only a few cancer cells. Using such biopsy specimens, the assay successfully demonstrated the EGFR mutations that correlate with gefitinib response, in contrast to failure of the direct sequencing of some biopsy specimens. Furthermore, the cycleave technique can be simultaneously applied for the detection of the K-ras mutation, which has been proposed to be an adverse prognostic marker for chemotherapy with erlotinib.24
In summary, we have introduced a new practical approach for the detection of EGFR mutations. This assay is very sensitive and useful for predicting gefitinib response. This rapid screening assay uses paraffin sections from biopsy without the need for a microdissection procedure and has significant advantages over other methods.
Acknowledgments
We thank Kaori Hayashi and Noriko Shibata for excellent technical assistance with the molecular genetic experiments on EGFR, respectively, and Hiroji Ishida for assistance in constructing the tissue array.
References
- Baselga J, Arteaga CL. Critical update and emerging trends in epidermal growth factor receptor targeting in cancer. J Clin Oncol. 2005;23:2445–2459. doi: 10.1200/JCO.2005.11.890. [DOI] [PubMed] [Google Scholar]
- Arteaga C. Targeting HER1/EGFR: a molecular approach to cancer therapy. Semin Oncol. 2003;30:3–14. [PubMed] [Google Scholar]
- Arteaga CL. Overview of epidermal growth factor receptor biology and its role as a therapeutic target in human neoplasia. Semin Oncol. 2002;29:3–9. doi: 10.1053/sonc.2002.35642. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Han SW, Kim TY, Hwang PG, Jeong S, Kim J, Choi IS, Oh DY, Kim JH, Kim DW, Chung DH, Im SA, Kim YT, Lee JS, Heo DS, Bang YJ, Kim NK. Predictive and prognostic impact of epidermal growth factor receptor mutation in non-small-cell lung cancer patients treated with gefitinib. J Clin Oncol. 2005;23:2493–2501. doi: 10.1200/JCO.2005.01.388. [DOI] [PubMed] [Google Scholar]
- Mitsudomi T, Kosaka T, Endoh H, Horio Y, Hida T, Mori S, Hatooka S, Shinoda M, Takahashi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- Marchetti A, Martella C, Felicioni L, Barassi F, Salvatore S, Chella A, Camplese PP, Iarussi T, Mucilli F, Mezzetti A, Cuccurullo F, Sacco R, Buttitta F. EGFR mutations in non-small-cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol. 2005;23:857–865. doi: 10.1200/JCO.2005.08.043. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro A, Cavina R, Latteri F, Zucali PA, Ginanni V, Campagnoli E, Ferrari B, Morenghi E, Pedicini V, Roncalli M, Alloisio M, Ravasi G, Soto Parra HJ. Activity of a specific inhibitor, gefitinib (Iressa, ZD1839), of epidermal growth factor receptor in refractory non-small-cell lung cancer. Ann Oncol. 2004;15:33–37. doi: 10.1093/annonc/mdh010. [DOI] [PubMed] [Google Scholar]
- Parra HS, Cavina R, Latteri F, Zucali PA, Campagnoli E, Morenghi E, Grimaldi GC, Roncalli M, Santoro A. Analysis of epidermal growth factor receptor expression as a predictive factor for response to gefitinib (‘Iressa’, ZD1839) in non-small-cell lung cancer. Br J Cancer. 2004;91:208–212. doi: 10.1038/sj.bjc.6601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey LR, Kris M, Wolf MK, Kay AC, Averbuch S, Askaa J, Janas M, Schmidt K, Fukuoka M: Tumor EGFR membrane staining is not clinically relevant for predicting response in patients receiving gefitinib (“Iressa”, ZD1839) monotherapy for pretreated advanced non-small-cell lung cancer: IDEAL 1 and 2. Presented at the American Association for Cancer Research annual meeting, July 11–14, 2003, Washington, DC, 2003 [Google Scholar]
- Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- Toyooka S, Kiura K, Mitsudomi T: EGFR mutation and response of lung cancer to gefitinib [letter]. N Engl J Med 2005, 352:2136; author reply, 2136 [DOI] [PubMed] [Google Scholar]
- Huang SF, Liu HP, Li LH, Ku YC, Fu YN, Tsai HY, Chen YT, Lin YF, Chang WC, Kuo HP, Wu YC, Chen YR, Tsai SF. High frequency of epidermal growth factor receptor mutations with complex patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin Cancer Res. 2004;10:8195–8203. doi: 10.1158/1078-0432.CCR-04-1245. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy S, Mukohara T, Hansen M, Meyerson M, Johnson BE, Janne PA. Gefitinib induces apoptosis in the EGFRL858R non-small-cell lung cancer cell line H3255. Cancer Res. 2004;64:7241–7244. doi: 10.1158/0008-5472.CAN-04-1905. [DOI] [PubMed] [Google Scholar]
- Weinstein IB. Cancer. Addiction to oncogenes: the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- Inoue A, Saijo Y, Maemondo M, Gomi K, Tokue Y, Kimura Y, Ebina M, Kikuchi T, Moriya T, Nukiwa T. Severe acute interstitial pneumonia and gefitinib. Lancet. 2003;361:137–139. doi: 10.1016/S0140-6736(03)12190-3. [DOI] [PubMed] [Google Scholar]
- Pan Q, Pao W, Ladanyi M. Rapid polymerase chain reaction-based detection of epidermal growth factor receptor gene mutations in lung adenocarcinomas. J Mol Diagn. 2005;7:396–403. doi: 10.1016/S1525-1578(10)60569-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Endo K, Konishi A, Takada M, Kawahara M, Iuchi K, Matsumura A, Okumura M, Tanaka H, Kawaguchi T, Shimizu T, Takeuchi H, Yano M, Fukai I, Fujii Y. EGFR mutation status in Japanese lung cancer patients: genotyping analysis using LightCycler. Clin Cancer Res. 2005;11:2924–2929. doi: 10.1158/1078-0432.CCR-04-1904. [DOI] [PubMed] [Google Scholar]
- Nagai Y, Miyazawa H, Huqun, Tanaka T, Udagawa K, Kato M, Fukuyama S, Yokote A, Kobayashi K, Kanazawa M, Hagiwara K. Genetic heterogeneity of the epidermal growth factor receptor in non-small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid-locked nucleic acid PCR clamp. Cancer Res. 2005;65:7276–7282. doi: 10.1158/0008-5472.CAN-05-0331. [DOI] [PubMed] [Google Scholar]
- Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, Ince WL, Janne PA, Januario T, Johnson DH, Klein P, Miller VA, Ostland MA, Ramies DA, Sebisanovic D, Stinson JA, Zhang YR, Seshagiri S, Hillan KJ. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- Greulich H, Chen TH, Feng W, Janne PA, Alverez JV, Zappaterra M, Bulmer SE, Frank DA, Hahn WC, Sellers WR, Meyerson M. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Greulich H, Janne PA, Sellers WR, Meyerson M, Griffin JD. Epidermal growth factor-independent transformation of Ba/F3 cells with cancer-derived epidemal growth factor receptor mutants induces gefitinib-sensitive cell cycle progression. Cancer Res. 2005;65:8968–8974. doi: 10.1158/0008-5472.CAN-05-1829. [DOI] [PubMed] [Google Scholar]